Abstract
NusG is an essential transcription factor in E. coli that is capable of increasing the overall rate of transcription. Transcript elongation by RNA polymerase (RNAP) is frequently interrupted by pauses of varying durations, and NusG is known to decrease the occupancy of at least some paused states. However, it has not been established whether NusG enhances transcription chiefly by (1) increasing the rate of elongation between pauses, (2) reducing the lifetimes of pauses, or (3) reducing the rate of entry into paused states. Here, we studied transcription by single molecules of RNAP under various conditions of NTP concentration, applied load, and temperature, using an optical trapping assay capable of distinguishing pauses as brief as 1 s. We found that NusG increases the rate of elongation, i.e., the pause-free velocity along the template. Because pauses are off-pathway states that compete with elongation, we observed a concomitant decrease in the rate of entry into short-lifetime, paused states. The effects on short pauses and elongation were comparatively modest, however. More dramatic was the effect of NusG on suppressing entry into long-lifetime (“stabilized”) pauses. Because a significant fraction of the time required for the transcription of a typical gene may be occupied by long pauses, NusG is capable of exerting a significant modulatory effect on the rates of RNA synthesis. The observed properties of NusG were consistent with a unified model where the function of this accessory factor is to promote transcriptionally downstream motion of the enzyme along the DNA template, which has the effect of forward-biasing RNAP from the pre-translocated state towards the post-translocated state.
Keywords: single-molecule biophysics, optical tweezers, NusG, transcription, RNA polymerase
Introduction
Transcript elongation by RNA polymerase is a complex, multi-step process and is a target of regulation by many cellular factors. In Escherichia coli, the accessory factor NusG is a transcription factor essential to growth and known to affect a variety of cellular and viral processes.1; 2 NusG associates with nearly all ternary elongation complexes (ECs)2, can stimulate rho-dependent termination both in vivo3 and in vitro,4; 5 and has been shown to assist rRNA6; 7 and λN8 transcription antitermination. The lifetimes of certain transcriptional pauses are reduced by NusG9; 10; 11, and it has been found to increase the overall rate of RNAP elongation.10; 11 Interestingly, evolutionarily conserved NusG in the gram-positive bacterium Bacillus subtilis enhances, rather than suppresses, pausing.12 In this study, we investigated how E. coli NusG exerts its modulatory effects on elongation by examining the transcriptional behavior of single molecules of RNAP.
Transcription by both prokaryotic and eukaryotic forms of RNA polymerase is frequently interrupted by reversible entry into elongation-incompetent states, or pauses. Such pauses represent off-pathway states, and they have been classified into various types that differ in their mechanisms, characteristic lifetimes, event triggers, and probabilities of occurrence. One frequently occurring type of short pause that has been detected by single-molecule transcription assays with E. coli RNAP interrupts elongation in a sequence-specific mechanism that is unresponsive to external loads, but even under saturating NTP conditions (~1 per 100 bp; ~3 s duration on average).13; 14; 15 These ubiquitous pauses are thought to reflect an elemental class of pause mechanism that does not itself involve backtracking, but that can be further stabilized by one or more subsequent reararrangements in the paused EC. Different modes of pause stabilization generate different classes of pause, but all eventually permit the escape of RNAP from the inactive state and allow return to the normal elongation pathway.
One mechanism for pause stabilization, only reported for prokaryotic systems, involves the formation of a hairpin structure in the nascent RNA.16 The hairpin must be energetically stable and properly spaced from the enzyme active site in order to interact with beta-flap tip helix of RNAP (a structure absent in eukaryotic polymerases).17 A well-characterized example of a such a hairpin sequence is responsible for the his pause, found near the beginning of the histidine biosynthetic operon.9 A second stabilization mechanism, common to both prokaryotic and eukaryotic polymerases, involves backtracking of the enzyme along the nucleic acid scaffold, in such a fashion that the RNA:DNA hybrid remains in register as the free RNA 3′ end is extruded into the putative NTP entry channel.9; 18; 19; 20 Backtracking of RNAP can be promoted by DNA template sequences that tend to form energetically less stable regions of RNA:DNA hybrid within the elongation complex. Backtracking displaces the RNA 3′ end from the enzyme active site, precluding further RNA synthesis until the event is resolved, either by thermally driven forward-tracking, or by cleavage of the extruded RNA, which in turn is promoted by transcript cleavage factors (GreA/B in prokaryotes or TFIIS in eukaryotes). The ops sequence from E. coli is a well-studied example of a pause site at which backtracking can occur; pausing at this site appears to involve both a shorter lifetime state that may be an elemental pause and a longer lifetime state that appears to be backtracked. The ops pause is critical for recruiting RfaH, a NusG paralog that also increases the overall rate of elongation.21
Downstream DNA sequences situated within the RNAP enzyme cleft have been shown to influence pausing strongly, although the mechanisms by which these act remain to be established.16; 22 Finally, certain transcription factors are known to act in conjunction with RNAP to further stabilize pauses. For example, RfaH21 and σ23 can independently interact with the nontemplate DNA strand at specific pause sequences to stabilize RNAP in a backtracked state. The transcription factor NusA, on the other hand, interacts with a nascent RNA hairpin to enhance pause lifetimes.9
Single-molecule transcription assays using E. coli RNAP have also identified a class of relatively infrequent (~1 per kb) but long-lived backtracked pauses, ranging in duration from 20 s to more than 30 min even under saturating NTP conditions.24 These backtracked pauses occur with a frequency that closely approximates the rate of nucleotide misincorporation by RNAP.25 Moreover, the frequency of long, backtracked pauses can be enhanced by the addition of ITP, a nucleotide that can be misincorporated in place of GTP, or decreased by the addition of GreA or GreB factors,24 which are known to increase transcription fidelity.26; 27; 28 These characteristics suggest that the majority of these pauses result from enzyme backtracking that occurs consequent to misincorporation events, and that such errors may be subsequently removed from the transcript via cleavage induced by Gre factors.24
Transcription factors that increase the elongation rate could achieve this effect, in principle, by either accelerating the main pathway for RNA synthesis or by modifying the rates into or out of off-pathway states. Ensemble, gel-based transcription assays carried out in vitro have shown that NusG decreases the enzyme occupancy at transcriptional pause sites along the DNA template, suggesting that NusG may enhance overall elongation by reducing the half-life for pausing.10; 11 However, determinations of the pause-free (on-pathway) elongation rates in such assays have been problematic because, as both ensemble and single-molecule assays have illustrated, it is a practical impossibility to identify templates that are wholly devoid of pauses. In a typical E. coli transcription unit, as represented by the rpoB gene fragment chosen for detailed single-molecule investigations,29; 30; 31 RNAP stops at a series of high-efficiency (i.e., high likelihood of entry) pause sites that function even under saturating nucleotide conditions.14 Furthermore, because pauses compete with active elongation, speeding up the nucleotide addition cycle indirectly affects not only the pause efficiency, but also the pause lifetime.14
E. coli NusG decreases the lifetime of backtracked but not hairpin-stabilized pauses, and thus is proposed to inhibit backtracking.9; 32 This model has been extended to a proposal that NusG also favors the post-translocated state over the pre-translocated state, thereby increasing the elongation rate by facilitating NTP binding.32; 33; 34 Stabilization of the post-translocated state is expected to increase the pause-free elongation rate under conditions where translocation along DNA occurs at a rate that is at least comparable to the rates of other biochemical steps in the nucleotide addition cycle. This stabilization could also affect the propensity to pause, and there is evidence to support the notion that the pause state branches off the elongation pathway from the pre-translocated state of RNAP.9; 17 A mechanism for enhancing the elongation rate by decreasing the propensity to pause from the pre-, not post-, translocated state has been proposed for RfaH.34
Here, we present results from a study of the effect of NusG on elongation measured with a “dumbbell” single-molecule transcription assay. The resolution achieved by this technique allows pauses as short as 1 s and separated by as little as ~2 bp to be reliably detected, permitting accurate determinations of pause-free elongation rates. We found that NusG increases the average pause-free velocity while decreasing the density of short-lifetime pauses. By collecting single-molecule data on transcription templates containing tandem repeat sequences, which allows individual records to be mutually aligned and thereby correlated with the underlying DNA sequence, we were able to identify and characterize specific pause sites that are differentially affected by the action of NusG. Although the NusG-induced modulation of the pause-free velocity and the short-pause density was modest, the pronounced NusG-suppression of entry into long, backtracked pauses can dramatically increase the efficiency of mRNA synthesis.
Results
NusG reduces the density of long-lifetime pauses
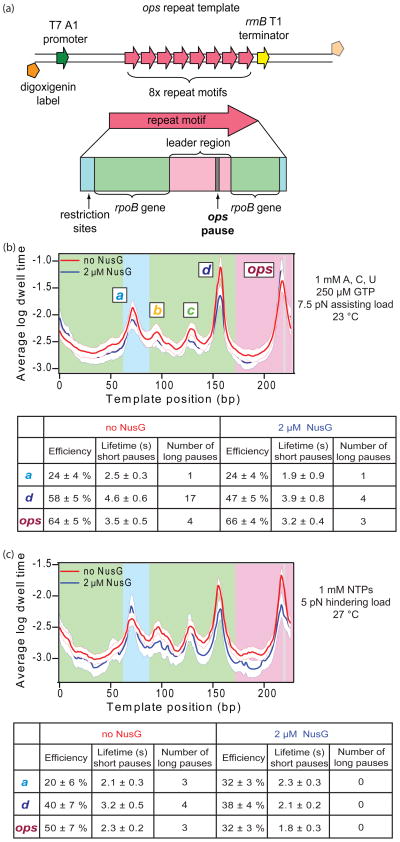
Stalled ECs were formed on digoxigenin-labeled DNA templates engineered to carry eight tandem repeats of a 227 bp sequence motif that includes an embedded ops pause sequence, as previously described.14 ECs were tethered between two polystyrene beads to create bead:RNAP:DNA:bead “dumbbells,” and transcription was reinitiated inside a microscope flow cell by the introduction of NTPs (and where indicated, 2 μM NusG). After this re-initiation, the beads were each captured in one of a pair of optical traps (Fig. 1a). By appending the digoxigenin label either to the upstream or to the downstream end of the DNA template, an assisting or hindering load could be applied to an elongating RNAP. The position of RNAP along the DNA template was determined by optically tracking the changing bead position with subnanometer accuracy as the dumbbell tether shortened (hindering load) or lengthened (assisting load). The tension on the tether was kept constant by feedback control of the position of one optical trap,24 and the temperature of the assay could be modulated by adjusting the laser intensity of the traps.35
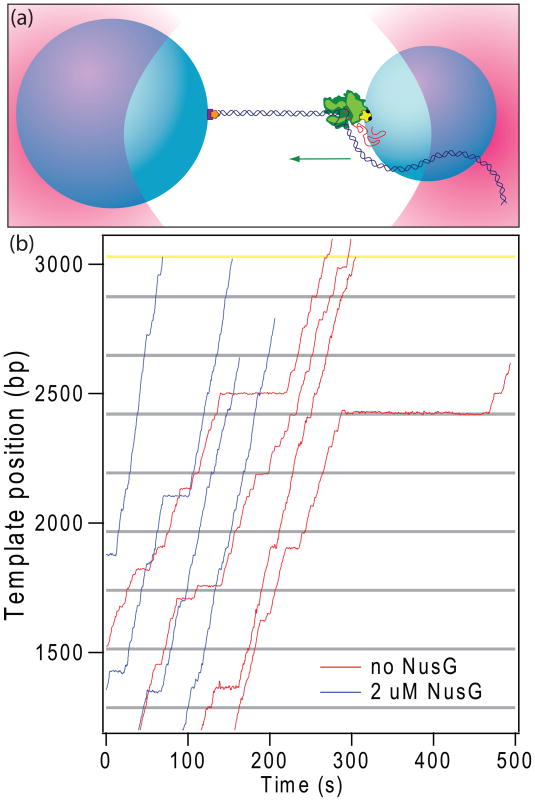
Figure 1. Single-molecule transcription assay and the effect of NusG.
(a) A cartoon showing the experimental geometry for the single-molecule assay (not to scale). Two polystyrene beads (light blue) are each held in separate optical traps (pink) above a coverglass. A biotin label (black) on RNAP (green) was used to attach RNAP to the smaller bead via an avidin linkage (yellow). The 3′-downstream end of the DNA (dark blue), labeled with digoxigenin (orange), is bound to a larger bead by an anti-digoxigenin antibody (purple). Transcription proceeds in the direction indicated (green arrow) while RNAP experiences a hindering load. By appending the digoxigenin label instead to the 3′-upstream end of the DNA, the direction of the applied load relative to transcription can be reversed.
(b) Representative records of transcription by single RNAP molecules along the ops repeat template vs. time, in the absence (red; 4 traces shown of 57) or presence of 2 μM NusG (blue; 4 traces shown of 19), shown after computer alignment. Records were obtained at ~27 °C with 1 mM NTPs under a 5 pN hindering load. The positions of ops pause sequences (horizontal grey lines) and the rrnB T1 terminator (horizontal yellow line) are indicated. Note that the blue records proceed more quickly and display fewer long pauses.
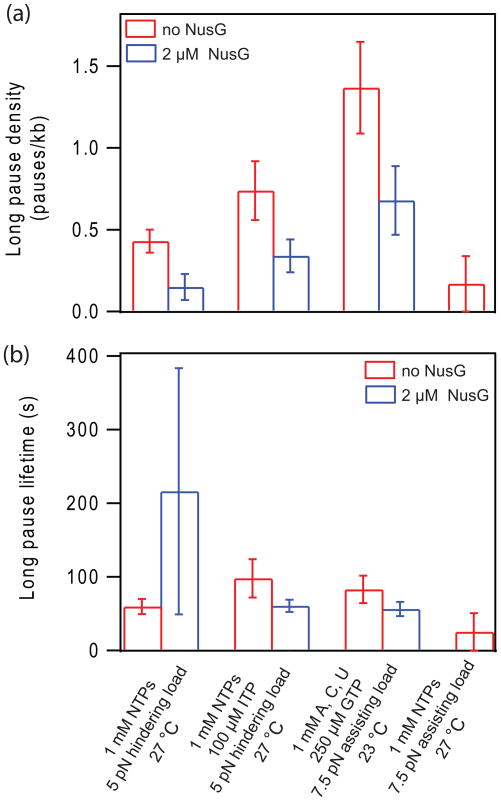
Representative records of the displacement of single RNAP molecules along the DNA template versus time illustrate the progress of transcription, which is frequently interrupted by pauses of varying duration (Fig. 1b). Most of these experiments were performed at ~27 °C with 1 mM NTPs under a 5 pN hindering load, either in the presence or absence of 2 μM NusG. Pauses were identified in individual transcription records, and only events where elongation had ceased but later recovered were scored. Pause lifetimes varied between 1 s (our detection threshold) and up to 260 s in the absence of NusG. Without NusG, long pauses constituted roughly 5% of all pauses scored, consistent with results from previous studies,24; 36 and these occurred at a density of 0.43 ± 0.07 pauses/kb (Fig. 2a and Table 1). The addition of NusG reduced the long pause fraction to just 2% of all pauses, and reduced the long pause density nearly 3-fold, to 0.15 ± 0.08 pauses/kb. (In fact, the addition of NusG reduced the number of long pauses to such an extent that any effect on the lifetimes of these pauses could not be ascertained with statistical significance; Fig. 2b.) The application of an assisting load, which acts mechanically to relieve backtracking, similarly reduced the density of long pauses, to 0.17 ± 0.17 pauses/kb.
Figure 2. Effect of NusG on long pauses.
(a) Long pause density (pauses/kb) (mean ± bootstrapped std. dev.) in the presence (blue) or absence (red) of NusG under the conditions indicated below (b).
(b) Long pause lifetime (mean ± bootstrapped std. dev.) in the presence (blue) or absence (red) of NusG under the conditions indicated.
Table 1.
Long-lifetime pause statistics.
| Conditions | [NusG] | N records | N long pauses | long-pause density (kb−1) | long-pause duration (s) |
|---|---|---|---|---|---|
| 1 mM NTPs, 27 °C hindering load | none | 57 | 30 | 0.43 ± 0.07 | 59.6 ± 10.5 |
| 2 μM | 19 | 3 | 0.15 ± 0.08 | 216.3 ± 167.3 | |
| 1 mM NTPs, 27 °C assisting load | none | 6 | 2 | 0.17 ± 0.17 | 25.5 ± 25.5 |
| 1 mM NTPs, 100 μM ITP, 27 °C, hindering load | none | 30 | 29 | 0.74 ± 0.18 | 98.1 ± 25.9 |
| 2 μM | 24 | 10 | 0.34 ± 0.10 | 60.7 ± 8.2 | |
| 1 mM ATP, CTP and UTP250 μM GTP, 23 °C, assisting load | none | 20 | 42 | 1.37 ± 0.28 | 82.9 ± 18.8 |
| 2 μM | 19 | 21 | 0.68 ± 0.21 | 56.4 ± 9.8 |
Two additional sets of assay conditions, which have previously been found to increase the long pause density, also yielded a reduction in long pauses upon addition of NusG (Fig. 2a and Table 1). The introduction of ITP into the assay (performed at ~27 °C, with 1 mM NTPs, 100 μM ITP, under 5 pN hindering load) increased the long pause density by ~60%, to 0.74 ± 0.18 pauses/kb. It also raised the mean long pause lifetime from 59.6 ± 10.5 s (without ITP) to 98.1 ± 25.9 s (with ITP) (Fig. 2b and Table 1), consistent with previous measurements.24 In the presence of NusG, however, the density of ITP-induced long pauses dropped nearly 2-fold, to 0.34 ± 0.10 pauses/kb, and the mean lifetime of such pauses was reduced to pre-ITP levels, 60.7 ± 8.2 s. A second condition known to increase the density of long pauses is limiting GTP concentration (250 μM), with saturating levels of the remaining three nucleotides (ATP, CTP, and UTP at 1 mM; performed at ~23 °C and 7.5 pN assisting load, studied previously14). Although assisting loads normally act to reduce long pauses (found here and in ref. 24), the combination of reduced temperature35 and low GTP availability37 slows the polymerase considerably (Table 2), and long pauses become more favored. Under these assay conditions and in the absence of NusG, long pauses made up ~10% of all pauses detected, with an average density of 1.37 ± 0.28 pauses/kb. The addition of 2 μM NusG reduced this density roughly 2-fold, to 0.68 ± 0.21 pauses/kb (Fig. 2a and Table 1).
Table 2.
Short-lifetime pause statistics.
| Conditions | [NusG] | N records | pause free rate (bp/s) | short-pause density (kb−1) | short-pause duration (s) |
|---|---|---|---|---|---|
| 1 mM NTPs, 27 °C, hindering load | none | 57 | 15.8 ± 0.1 | 9.6 ± 0.8 | 3.1 ± 0.1 |
| 2 μM | 19 | 17.6 ± 0.2 | 7.5 ± 0.9 | 2.7 ± 0.3 | |
| 1 mM NTPs, 27 °C assisting load | none | 6 | 17.6 ± 0.3 | 8.6 ± 3.3 | 3.1 ± 0.4 |
| 1 mM NTPs, 100 μM ITP, 27 °C, hindering load | none | 30 | 14.7 ± 0.1 | 10.7 ± 0.7 | 3.5 ± 0.2 |
| 2 μM | 24 | 17.6 ± 0.2 | 6.6 ± 0.6 | 2.9 ± 0.3 | |
| 1 mM ATP, CTP and UTP250 μM GTP, 23 °C assisting load | none | 20 | 10.9 ± 0.1 | 14.2 ± 2.3 | 3.8 ± 0.3 |
| 2 μM | 19 | 12.7 ± 0.1 | 9.7 ± 0.8 | 4.0 ± 0.3 |
NusG can increase the pause-free elongation rate
To test the effect of NusG on the nucleotide addition cycle, we determined the pause-free elongation velocities from sets of single-molecule records. The histograms of instantaneous velocities derived from records of individual RNAP molecules moving under a given set of conditions were summed in such a fashion as to give equal statistical weight to each nucleotide position traversed along the template, rather than to each time interval collected (Fig. 3). Elongation records binned over equal time intervals produce prominent peaks in histograms of pause positions, whereas those binned instead over equal position intervals eliminate these peaks, because, by definition, RNAP does not traverse the template when immobilized at a pause site. The resulting ensemble velocity distributions are well fit by single Gaussians whose central values correspond to the pause-free elongation rates, which varied with assay conditions.
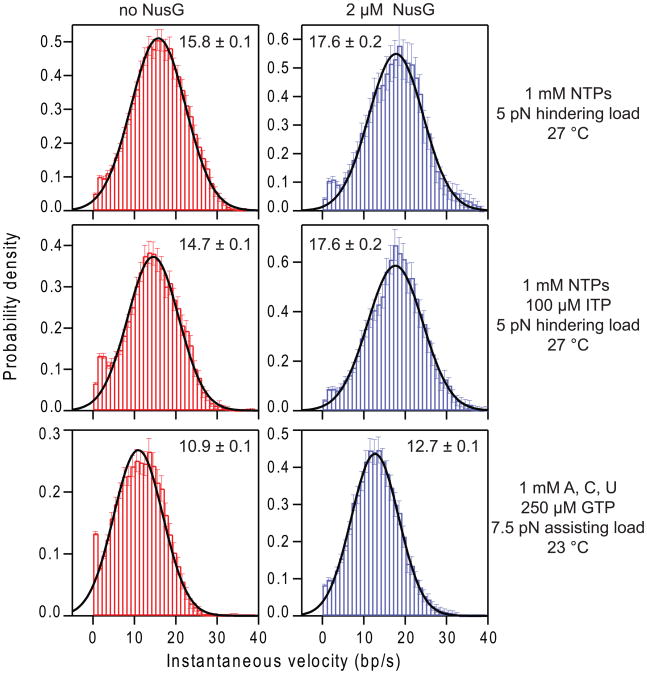
Figure 3. Ensemble distributions of instantaneous velocity.
Histogram distributions of the instantaneous velocity computed from records of individual transcribing RNAP molecules in the absence (red) or presence (blue) of 2 μM NusG were combined, a procedure that gives equal statistical weight to all positions on the template. Experimental assay conditions are shown on the right. The distributions were well fit by Gaussians (solid black lines), with the central value providing an estimate of the mean pause-free elongation velocity (mean values and std. errs. of fits are shown inset).
When transcription took place with 1 mM NTPs, at ~27 °C under a 5 pN hindering load, RNAP elongated at a pause-free rate of 15.8 ± 0.1 bp/s (Fig. 3). This rate could be increased to 17.6 ± 0.3 bp/s by reversing the direction of the applied load (Table 2). The sensitivity to external load suggests that the relative occupancy of the pre-translocated state versus the post-translocated state contributes to the overall rate of nucleotide addition under these conditions, presumably by affecting the availability of the post-translocated state for NTP binding. Introducing 2 μM NusG into the hindering load assay increased the elongation rate similarly, by ~11%, to 17.6 ± 0.2 bp/s (Fig. 3), consistent with the mode of action of NusG being to bias the equilibrium between pre- and post-translocated states in a fashion analogous to the application of mechanical load.9; 32; 33
Upon addition of 100 μM ITP to the hindering-load assay (1 mM NTPs; ~27 °C), the pause free velocity dropped to 14.7 ± 0.1 bp/s (Fig. 3). We interpret this small decrease in the velocity to indicate that ITP interferes with correct nucleotide binding (particularly for GTP), causing the NTP-binding step to become partially rate-limiting. Under these conditions, 2 μM NusG once again enhanced the pause-free elongation rate, to 17.6 ± 0.2 (Fig. 3). Finally, under the same conditions previously used for sequence-resolved measurements of pauses (1 mM ATP,CTP,UTP; 250 μM GTP; ~23 °C under 7.5 pN assisting load), the pause-free elongation velocity fell to just 10.9 ± 0.1 bp/s (Fig. 3), as the combination of reduced GTP concentration37 and temperature35 slowed the polymerase. The introduction of 2 μM NusG increased the pause-free velocity by ~17%, to 12.7 ± 0.1 bp/s (Fig. 3).
NusG reduces the density but not the lifetime of short pauses
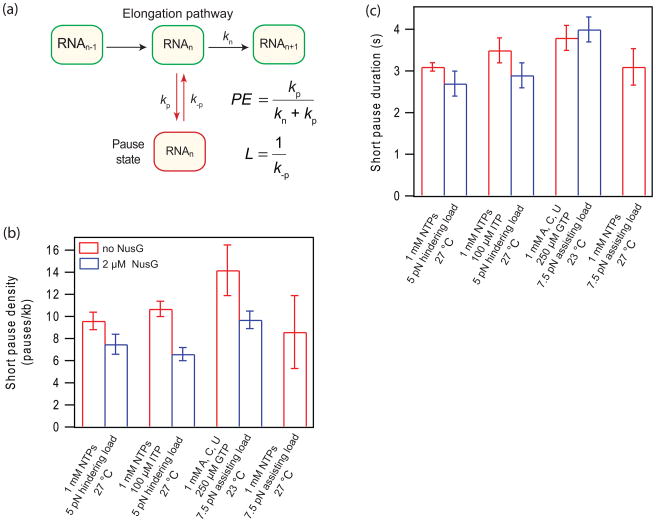
Because pausing directly competes with transcript elongation (Fig. 4a), the pause efficiency (i.e., the probability to pause) at a given template site, PE, is supplied by the branching ratio kp/(kp + kn), where kp is the rate of entry into the pause state and kn is the competing elongation rate (Fig. 4a). The average efficiency across the entire template is proportional to the pause density measured by these experiments. Any factor whose primary mode of action is to increase the on-pathway rate (e.g., forward translocation) will therefore tend to skew the branching probability in favor of the on-pathway cycle, thereby decreasing the pause density. To study this effect, short-lifetime pauses (operationally defined as pauses lasting less than 20 s) were identified in individual records using an automated detection algorithm, as previously described,35; 36 scoring only those events whose entry and exit phases were observed. Short pauses occurred at a density of 9.6 ± 0.8 pauses/kb (Fig. 4b) (assay conditions of 1 mM NTPs; ~27 °C under a 5 pN hindering load), consistent with previous measurements.36 The short pause density decreased by ~22% upon the addition of NusG, to 7.5 ± 0.9 pause/kb. This reduction compares with an 11% increase in the elongation rate under these assay conditions (Fig. 4b). The mean lifetime of pausing, supplied by the reciprocal of the rate to exit the pause state, k−p, displayed no significant difference under these conditions (2.7 ± 0.3 s with NusG and 3.1 ± 0.4 s without NusG) (Fig. 4c).
Figure 4. A simplified transcription pathway and the effect of NusG on short pauses.
(a) Kinetics of a simplified model for elongation involving one off-pathway pause state. Forward elongation is depicted by a single transition occurring at a rate kn. Pause efficiency, PE, is given by the ratio of the rate for pausing, kp, to the rate of either pausing or elongating, (kp+ kn). The pause lifetime, L, is given by the reciprocal of the rate for escaping the pause, k−p
(b) The density of short pauses was calculated for each single-molecule record by tallying the number of pauses below 20 s and dividing by the distance transcribed, measured in kilobase pairs. The mean density ± std. error is shown for the assay conditions indicated.
(c) The mean duration of short-lifetime pauses. The mean time ± std. error is shown for the assay conditions indicated.
In the presence of ITP, the short pause density was 10.7 ± 0.7 pauses/kb and decreased by ~38% in the presence of NusG, to just 6.6 ± 0.6 pauses/kb. This decrease in short pause density compares with the NusG-induced increase in elongation rate that occurred in the presence of ITP (~20%). There was no significant decrease observed in the short pause lifetime (Fig. 4c). Under the final set of assay conditions (1 mM ATP, CTP, UTP, 250 μM GTP; ~23 °C under 7.5 pN assisting load), the short pause density increased to 14.2 ± 2.3 pauses/kb. The addition of NusG reduced the short pause density by ~32%, to 9.7 ± 0.8 pauses/kb. This reduction compares with the 17% increase observed in the elongation rate under the assay conditions. The short pause lifetime was again unchanged under these conditions.
NusG differentially reduces efficiencies of sequence-specific pauses
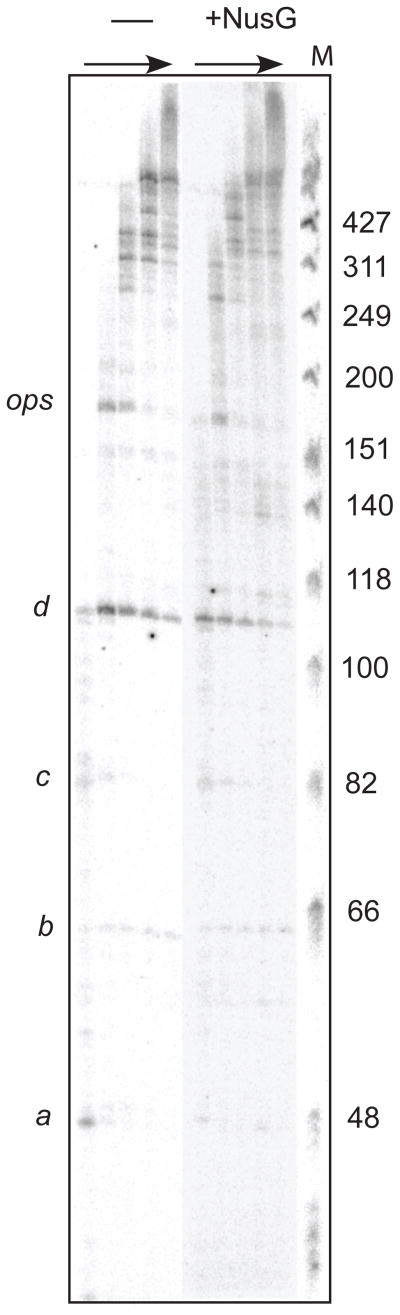
To determine whether NusG affects pausing differently at different DNA sequences, we took advantage of a technique developed previously to align single-molecule transcription records with the underlying DNA template to near-basepair accuracy.14 Records were acquired on periodic templates engineered to carry eight tandem repeats of a single short motif, which contained an ops pause sequence embedded within elements originally derived from the rpoB gene (Fig. 5a,b). These repeats were positioned just upstream of a strong termination signal. The periodic patterns of pausing behavior on such templates, taken together with the release of the template upon reaching the terminator, function as registration (fiducial) marks in the records, allowing us to employ computer-based algorithms to align ensembles of records that begin at different starting points, and thereby to identify the underlying DNA sequences. Histograms of the logarithm of the dwell time as a function of template position, compiled for all records under a given condition, were then summed for all eight identical repeats. These histograms display prominent peaks at template positions where RNAP has a high propensity to pause (Fig. 5b,c). These pause sites were assigned the same letter names (a–d plus ops) and correspond closely to the sites identified previously on this template.14
Figure 5. Sequence-resolved effects of NusG on repeating templates.
(a) Engineered transcription templates used here and in ref. 14. Templates carry a sequence of eight repeat motifs (pink) that begins ~1100 bp beyond a T7 A1 promoter (dark green), from which transcription was initiated, and ends ~80 bp before an rrnB T1 terminator (yellow). Templates were labeled with digoxigenin on the transcriptionally upstream end (for assisting loads; solid orange) or the transcriptionally downstream end (for hindering loads; transparent orange). Repeat motifs (red arrow) consist of leader sequence (pink) together with an ops pause sequence (gray), flanked by sequence elements from the rpoB gene (light green) and DNA derived from sites used in cloning (blue).
(b) Average log dwell-time histogram of aligned data (acquired at 1 mM ATP, CTP, and UTP, plus 250 μM GTP, ~23 °C under 7.5 pN assisting load) computed from all repeats of the motif in the absence (red; N = 16 molecules, 81 records) or in the presence of NusG (blue; N = 14 molecules, 81 records), plotted along with bootstrapped standard deviations (white zones). The major pause sites are labeled. The table below shows pause statistics computed for the three most prominent pause sites in the repeat (a, d, and ops) in the presence or absence of NusG.
(c) Average log dwell-time histogram for aligned data (acquired at 1 mM NTPs, ~27 °C, and 5 pN hindering load) computed from all 8 repeats of the ops repeat motif in the absence of NusG (red; N = 36 molecules, 175 records) and in the presence of NusG (blue; N = 13 molecules, 49 records), plotted with the bootstrapped standard deviations (white zones). The table below shows pause statistics computed for the three most prominent pause sites in the repeat (a, d, and ops) in the presence or absence of NusG.
From these histograms, we determined that NusG affects some pause sites more than others. Overall, the dwell plots obtained in the presence or absence of NusG displayed generally similar patterns of pausing. However, they differed noticeably at both the d and ops pause sites under hindering-load conditions (1 mM NTPs; ~27 °C with 5 pN hindering load), and at the d pause site under assisting-load conditions (1 mM ATP,CTP, UTP; 250 μM GTP, ~23 °C with 7.5 pN assisting load)(Fig. 5b). In general, single-molecule results obtained under limiting GTP concentrations resembled the findings from an ensemble transcription RNA run-off experiment, carried out under identical buffer conditions (Fig. 6), which resolves pauses as bands in a series of gel lanes obtained at successive times after triggering elongation from an initially synchronized population of ECs. In such ensemble assays, the intensity of a given band is related to the product of the pause efficiency and the pause lifetime (i.e., to the ‘pause strength’). For both the single molecule analysis under hindering load conditions and the ensemble experiment, pause strengths at the d and ops positions were reduced by the addition of NusG. To determine whether the difference in dwell time at either site was a consequence of the lifetime or the efficiency of the pause (or both), we scored pauses throughout all aligned records. The efficiency of pausing at a given site was computed from the ratio of the number of occurrences of a given pause (at a location within ±5 bp of the site) to the number of times that pause site was encountered by a transcribing molecule. Because the pause-detection algorithm cannot reliably detect pauses below 1 s, this ratio somewhat underestimates the true efficiency. Nevertheless, efficiencies computed in this fashion in the absence of NusG agreed well with efficiencies determined previously when using the same analysis.14 Pause lifetimes were scored as the median duration of short-lifetime pauses (pauses with long lifetimes were excluded from this metric because these are thought to arise by a different mechanism). None of the individual pause sites displayed a statistically significant change in the short pause lifetime upon the addition of NusG. Pause efficiencies were also unchanged at these sites. Under assisting-load conditions, NusG decreased the pause efficiency modestly at the d position (from 58% to 47%), but not at the other positions (Fig. 5b). Under hindering-load conditions, NusG decreased the pause efficiency significantly at the ops site (from 50% to 32%), increased efficiency at the a site, and had no effect at the other sites (Fig. 5c). We conclude that NusG exhibits differential, sequence-dependent effects on pause efficiency at different pause sites.
Figure 6. Effect of NusG on a transcription run-off assay for RNA.
Transcription of RNA was initiated by mixing 25 nM of a short template carrying a single copy of the repeat motif region, 40 nM biotin-tagged RNAP, and 10 mM ATP, GTP, and 32P-CTP and incubated for 15 min at 37°C. If present, 100 nM NusG was added to halted complexes and incubated for 3 min at 37°C. Halted complexes were equilibrated to room temperature and elongation was restarted by adjustment to 1 mM ATP, CTP, and UTP, and 250 μM GTP with 100 mg/ml rifampicin. Samples were removed at 5, 15, 30, 60, and 120 s intervals. The marker lane (M) is fX714 DNA digested with HinfI; sizes of fragments (in basepairs) are indicated on the right, pauses are identified on the left.
We also observed effects on NusG on entry into long, backtracked pauses. Long, backtrack pauses are rare events and consequently present a challenge for systematic study due to their limited statistics. In the absence of NusG, based on the density of long pauses across the template, we expect approximately one long pause per site in these data sets, assuming that long pauses are randomly distributed. In fact, under low GTP conditions, a single long pause was detected at each of the a, b, and c sites (data not shown). However, more long pause events were observed at the ops and d sites (4 and 17, respectively). NusG reduced the long pauses at the d site to just 4 for the same number of encounters (Fig. 5b, bottom). This reduction is largely responsible for the observed decrease in the peak height of the dwell plot at the d site, because neither the short-pause lifetime nor efficiency were significantly affected at this position. Under saturating NTP concentrations at 27 °C, fewer long pauses were recorded overall, but in the absence of NusG, the a, d, and ops sites each produced more than one long pause (3, 4, and 3, respectively; Fig. 5c, bottom). However, NusG eliminated all long pauses at the a, d, and ops sites under hindering load (Fig. 5c). The reduction in long pauses explains the decrease in the heights of the dwell-plot peaks at the ops and d pause sites, although the ops site displayed a reduction in efficiency that also acts to reduce the dwell time. No decrease in peak height occurred at the a site due to a compensating increase in pause efficiency at that site. These results indicate that NusG decreases the propensity of RNAP to enter into long pauses.
Discussion
NusG modestly increases pause-free elongation rates while decreasing the likelihood of pausing
In addition to its roles in stimulating Rho-dependent termination or anti-termination depending on interaction partners,3; 4; 5; 6; 7; 8 NusG on its own has been shown to accelerate transcript elongation in vivo and in vitro.10; 11 Because NusG decreases the occupancy of RNAP at a subset of transcriptional pause sites, it has generally been assumed that NusG achieves this acceleration by reducing the lifetimes of off-pathway pause states.9; 10; 11 However, using a single-molecule assay, we have found that NusG directly enhances the pause-free elongation rate of E. coli RNAP by 10% to 20% under a variety of NTP, temperature and load conditions, including under near-physiological NTP concentrations. This activity is consistent with the suggestions that NusG inhibits backtracking9; 32 and biases RNAP towards the post-translocated state,33 provided that equilibrium between the pre- and post-translocated states contributes to the overall elongation rate.
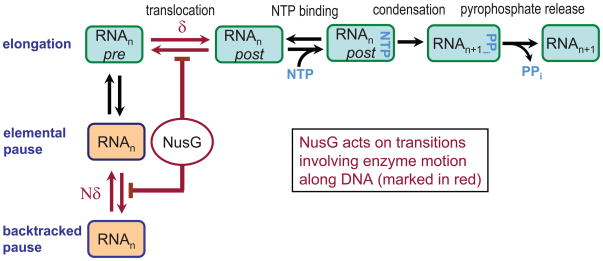
A unified model for transcript elongation and pausing incorporates a Brownian ratchet-type translocation mechanism, where rapid, stochastic fluctuations of RNAP between its pre- and post-translocated states along the DNA template are rectified by the nucleotide condensation reaction, together with an isomerization to offline, pause states (Fig. 7).33; 37; 38; 39 The elemental pause state is shown branching from the pre-translocated state, an assignment that is supported by results from cross-linking and pyrophosphorolysis experiments9; 17; 34; 40. Single-molecule studies have further shown that this paused state does not involve the translocation of RNA polymerase along the DNA template.13; 14 The elemental pause state can be further stabilized by several mechanisms, with RNAP backtracking being among the most important. Backtracking plays key roles in sensing nucleotide misincorporation28, in DNA repair,41 and in transcriptional regulation.42 We note that recent findings suggest that at least non-backtracked pauses may not revert to the elongation pathway until after translocation and NTP binding.43; 44; 45
Figure 7. A unified pathway for elongation and pausing.
The main pathway for transcript elongation is shown (light blue boxes; top row; adapted from ref. 38). In a Brownian ratchet mechanism, RNAP oscillates stochastically between pre- and post-translocated states prior to the reversible binding of NTP followed by the (nearly) irreversible condensation reaction and pyrophosphate release, which rectify this motion in the transcriptionally downstream direction. The displacement associated with translocation, δ, corresponds to the longitudinal distance subtended by a single base pair. The elemental pause state is depicted (middle row, orange box; adapted from ref 14), shown branching from the pre-translocated state: entry into this state does not involve translocation. The long-lifetime, backtracked pause state (bottom row; orange box) is entered via the elemental pause state, and involves the upstream translocation of RNAP through one or more base pairs, Nδ. Our modeling suggests that the addition of NusG promotes the downstream motion of RNAP, affecting those transitions that involve translocation (red arrows).
Because transcriptional pausing competes directly with elongation, one anticipates that, in the simplest possible branching scheme, increasing the pause-free elongation rate would decrease the corresponding rate of entry into pause states. In a simplified pathway model of Fig. 4a, the extent of this pause suppression depends on the relative values of kp and kn, but in no case will the fractional decrease in pausing exceed the fractional increase in elongation, and these two fractional changes become equal in the limit kp ≪ kn. The addition of NusG in these experiments, however, suppressed short pauses to a somewhat greater extent than that predicted simply from the acceleration of elongation rate alone in the simplified model. For example, the 20% increase in the pause-free elongation rate induced by NusG (under hindering load conditions in the presence of ITP) predicts a pause density of 9.6 pauses/kb, whereas we measured 6.6 ± 0.6 pauses/kb. While the additional pause suppression is not especially large, it is statistically significant, and a similar effect was consistently observed under all assay conditions with NusG (Table 2). The additional suppression of short pauses cannot be trivially explained by the reduction in long pause density produced by NusG, because long pauses (whose distribution is assumed to be exponential) constitute such a tiny fraction (<5%) of all pause events. However, this finding is consistent with the suggestion that isomerization to the elemental pause occurs (mainly) from the pre-translocated state.34; 45 In the more complex kinetic scheme depicted in Fig. 7, the forward bias induced by NusG, towards the comparatively more pause-resistant, post-translocated state could qualitatively explain the small additional short pause suppression we observed.
Another possible contribution to this phenomenon is the reduction by NusG of long-lifetime, high-efficiency pauses. The pause density value across the template is dominated by only a few such pauses, for example the d and ops pauses, whose efficiency was found to be significantly reduced by NusG (Fig. 5b,c). However, on the basis of our data, we cannot exclude the alternative possibility that NusG jointly affects both kp and kn, rather than kn alone, modifying or stabilizing RNAP so that it becomes less likely to pause, in addition to influencing the translocation rate. However, such an explanation is less parsimonious, invoking two separate but correlated effects on rates, and any structural basis for such a mechanism is presently unknown.
NusG modulates overall transcription rate both directly and by decreasing long-lifetime, backtracked pauses
Under physiological buffer conditions, only a tiny minority of pauses have lifetimes in excess of 20 s. Despite the fact that this population represents a comparatively low percentage of all pauses, they subtend up a significant fraction of the time for transcribing a typical gene. Based on the measured values for elongation rates, densities and lifetimes of short-lifetime pauses in the assisting-load assay, we estimate that the mean time taken to transcribe a given length of DNA would be reduced by about 20% in the presence of NusG, due solely to its effects on the elongation rate and entry into short pauses. However, if we also take into account the additional time spent in long pause states, NusG is found to decrease the transcription time by 40%. We estimated comparable levels of rate improvement using pause and elongation statistics for our other assay conditions. Recently, a theoretical model for transcription elongation has indicated that rare, long-lifetime pauses could lead to blockages followed by episodic bursts in mRNA production.46 A major contribution of NusG to overall mRNA synthesis may result from reducing such fluctuations during mRNA synthesis.
The same effects of NusG on the longitudinal mobility of RNAP not only will alter the equilibrium between pre- and post-translocated states, but also will alter the propensity of RNAP to backtrack,9; 32 likely through contacts to both the enzyme and nucleic acid scaffold.4; 47; 48 Single-molecule experiments with near-basepair resolution have shown that at least some long-lifetime pauses (namely, those induced by nucleotide misincorporation, and possibly others) lead directly to backtracking of the enzyme.24 The present finding that long pauses are suppressed by the addition of NusG is fully consistent with a model in which NusG promotes the forward translocation state of RNAP and disfavors backtracking.
NusG exerts sequence-specific effects on pausing
Previous work indicated that NusG can decrease the pause lifetime at certain sites while not significantly affecting others.9 Here, we extend this finding and confirm that NusG exerts sequence-specific effects on pausing. In tandem repeat templates, we found a significant reduction in the numbers of long pauses at the d site by NusG: the enrichment in long pauses at this template position suggests that the probability of backtracking at this site is higher than elsewhere. We do not observe a NusG effect on pausing under assisting-load conditions at the ops site, which is classified as a backtracked pause, based on load-free, ensemble experiments. Biochemical studies have revealed that the ops pause site is complex, and actually consists of three different, closely spaced pause positions and at least two kinetically distinguishable pause states (with shorter and longer lifetimes). The longer lifetime state is thought to result from backtracking, and to be shortened upon addition of NusG.9 It seems likely that under the single-molecule assay conditions used here, which apply an assisting load that opposes backtracking, we only observe the shorter lifetime state, which does not backtrack and is independent of NusG. This interpretation would also explain results from our previous study, performed under similar conditions of assisting load, where we also did not observe any RNAP backtracking at the ops pause site.14 In support of this interpretation, when the direction of load was reversed to the hindering direction, we found that pause efficiencies at the ops site became sensitive to NusG.
NusG affects transitions that involve translocations
Here, we find that NusG can both enhance pause-free velocity and decrease the entry into long-lifetime, backtracked pause states. Both these processes involve polymerase translocation with respect to the DNA template. At the same time, NusG did not affect the rates of exiting short-lifetime (elemental) pause states, which is not rate-limited by translocation.43; 44 Altogether, these findings suggest that NusG serves to influence the translocation of RNAP along DNA. In a thermal ratchet model, biasing RNAP towards its post-translocated state tends to increase the pause-free elongation rate while reducing the tendency to enter pause states and to slide upstream into any backtracked states. Such a bias is consistent with previous experiments showing that NusG can affect the RNAP equilibrium between pre- and post-translocated positions33 and can decrease pausing at backtracking-type sites.9; 21 Recent work has shown that the NusG paralog RfaH (and likely also NusG) binds to the non-template strand in the transcription bubble, and interacts at least indirectly (and probably directly) with the β′-subunit clamp helices.47; 49 It remains to be determined whether similar interactions of NusG occur with a transcription complex. It further remains to be established whether these are sufficient to stabilize the forward-translocated state by promoting re-annealing at the the upstream end of the transcription bubble, or, if as proposed,33 NusG action is mediated by the RNAP trigger loop within the enzyme active site.
Methods
Cloning of a plasmid for assisting load experiments
The pKH2 plasmid, used to create the ops tandem repeat templates for a previous single-molecule transcription study using assisting load,14 was modified and adapted here for hindering-load assays by moving the AlwNI site from a location ~1000 bp downstream of the rrnBT1 terminator to a position ~1000 bp upstream of the T7A1 promoter using two rounds of site-directed mutagenesis (Qiagen Quickchange), creating pKH4.
Proteins
Biotin-tagged RNAP was purified as described previously.50 NusG was purified as described previously.2
Transcription templates for optical trapping assays
Linear, end-labeled templates were constructed from both pKH2 and pKH4 plasmids by digesting each plasmid at its unique AlwNI site. The resulting 3′ ends were labeled using DIG-ddUTP and terminal transferase (Roche DIG Oligonucleotide 3′-End Labeling Kit). Subsequent digestion of pKH2 at a unique SapI site removed the label at the transcriptionally downstream end, leaving a single label at the upstream end for tethering to anti-digoxigenin antibody-coated polystyrene beads for assisting load templates. Digestion of pKH4 at its unique Eco109I site removed the upstream end, leaving a single label on the downstream end, and this fragment was used to create hindering load templates.
Optical trapping assay
Biotin-labeled RNAP was initiated for transcription, then stalled 29 base pairs past the T7A1 promoter24, on labeled, assisting (or hindering) load templates, constructed as described above. Avidin-coated polystyrene beads (600 nm diameter) and anti-digoxigenin antibody-coated polystyrene beads (730 nm diameter) beads were prepared, and stalled ECs were bound to the beads, creating bead:RNAP:DNA:bead dumbbells.24 Experiments were performed as described previously14, using the NTP conditions, load directions, and temperatures specified in the main text. Where indicated, 2 μM NusG was added. For experiments at 23 °C, the resting tension in the DNA was maintained approximately constant at 7.5 ± 2.4 pN (mean ± std.dev.) by moving the trap holding the larger bead in 50-nm increments whenever the tension on the DNA fell below 5 pN.24 The experiments at 27 °C were implemented using a force clamp that continuously updated the position of the trap holding the larger bead using an acousto-optic deflector, in order to maintain the smaller bead at a fixed offset from its trap center.
Data analysis
The contour length of the DNA tether between the two beads of the dumbbell was calculated as described.24 The template position of the RNAP enzyme was computed from the measured DNA contour length during transcription by subtracting the contour length of the segment of DNA template between the transcription initiation site and the position of the 730-nm bead (2,607 bp for assisting load or 5,467 bp for hindering load, based on a pitch of 0.338 nm/bp). Pauses were identified using an algorithm similar to that of ref. 24, except to avoid scoring a single pause multiple times (a consequence of drift), any extra pauses found within a 2 bp region downstream of a given prior pause were concatenated. Transcription record alignments were performed as previously described.14 Analysis was performed in Igor Pro (Wavemetrics) and by programs written in C.
Acknowledgments
This work was supported by grants from the NIGMS to R.L. and S.M.B. We thank Jeff Gelles and members of the Gelles laboratory for useful discussions, and M. Valentine, N. Guydosh, M. Larson, P. Anthony, K. Frieda for critical reading of the manuscript.
References
- 1.Downing WL, Sullivan SL, Gottesman ME, Dennis PP. Sequence and transcriptional pattern of the essential Escherichia coli secE-nusG operon. J Bacteriol. 1990;172:1621–7. doi: 10.1128/jb.172.3.1621-1627.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mooney RA, Schweimer K, Rosch P, Gottesman M, Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol. 2009;391:341–58. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan SL, Gottesman ME. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell. 1992;68:989–94. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Mason SW, Greenblatt J. Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes Dev. 1993;7:161–72. doi: 10.1101/gad.7.1.161. [DOI] [PubMed] [Google Scholar]
- 5.Nehrke KW, Platt T. A quaternary transcription termination complex. Reciprocal stabilization by Rho factor and NusG protein. J Mol Biol. 1994;243:830–9. doi: 10.1006/jmbi.1994.1685. [DOI] [PubMed] [Google Scholar]
- 6.Torres M, Balada JM, Zellars M, Squires C, Squires CL. In vivo effect of NusB and NusG on rRNA transcription antitermination. J Bacteriol. 2004;186:1304–10. doi: 10.1128/JB.186.5.1304-1310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zellars M, Squires CL. Antiterminator-dependent modulation of transcription elongation rates by NusB and NusG. Mol Microbiol. 1999;32:1296–304. doi: 10.1046/j.1365-2958.1999.01442.x. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Horwitz R, McCracken S, Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J Biol Chem. 1992;267:6012–9. [PubMed] [Google Scholar]
- 9.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci U S A. 2000;97:7090–5. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns CM, Richardson LV, Richardson JP. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J Mol Biol. 1998;278:307–16. doi: 10.1006/jmbi.1998.1691. [DOI] [PubMed] [Google Scholar]
- 11.Burova E, Hung SC, Sagitov V, Stitt BL, Gottesman ME. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J Bacteriol. 1995;177:1388–92. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yakhnin AV, Yakhnin H, Babitzke P. Function of the Bacillus subtilis transcription elongation factor NusG in hairpin-dependent RNA polymerase pausing in the trp leader. Proc Natl Acad Sci U S A. 2008;105:16131–6. doi: 10.1073/pnas.0808842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalal RV, Larson MH, Neuman KC, Gelles J, Landick R, Block SM. Pulling on the nascent RNA during transcription does not alter kinetics of elongation or ubiquitous pausing. Mol Cell. 2006;23:231–9. doi: 10.1016/j.molcel.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–94. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neuman KC, Block SM. Optical trapping. Rev Sci Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–6. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 17.Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell. 2003;12:1125–36. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- 18.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci U S A. 1997;94:1755–60. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palangat M, Landick R. Roles of RNA:DNA hybrid stability, RNA structure, and active site conformation in pausing by human RNA polymerase II. J Mol Biol. 2001;311:265–82. doi: 10.1006/jmbi.2001.4842. [DOI] [PubMed] [Google Scholar]
- 20.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 21.Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 22.Palangat M, Hittinger CT, Landick R. Downstream DNA selectively affects a paused conformation of human RNA polymerase II. J Mol Biol. 2004;341:429–42. doi: 10.1016/j.jmb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Roberts JW, Yarnell W, Bartlett E, Guo J, Marr M, Ko DC, Sun H, Roberts CW. Antitermination by bacteriophage lambda Q protein. Cold Spring Harb Symp Quant Biol. 1998;63:319–25. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- 24.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–7. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erie DA, Yager TD, von Hippel PH. The single-nucleotide addition cycle in transcription: a biophysical and biochemical perspective. Annu Rev Biophys Biomol Struct. 1992;21:379–415. doi: 10.1146/annurev.bb.21.060192.002115. [DOI] [PubMed] [Google Scholar]
- 26.Erie DA, Hajiseyedjavadi O, Young MC, von Hippel PH. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993;262:867–73. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 27.Jeon C, Agarwal K. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc Natl Acad Sci U S A. 1996;93:13677–82. doi: 10.1073/pnas.93.24.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas MJ, Platas AA, Hawley DK. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93:627–37. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 29.Schafer DA, Gelles J, Sheetz MP, Landick R. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature. 1991;352:444–8. doi: 10.1038/352444a0. [DOI] [PubMed] [Google Scholar]
- 30.Yin H, Wang MD, Svoboda K, Landick R, Block SM, Gelles J. Transcription against an applied force. Science. 1995;270:1653–7. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- 31.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–7. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 32.Pasman Z, von Hippel PH. Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry. 2000;39:5573–85. doi: 10.1021/bi992658z. [DOI] [PubMed] [Google Scholar]
- 33.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–93. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 34.Svetlov V, Belogurov GA, Shabrova E, Vassylyev DG, Artsimovitch I. Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 2007;35:5694–705. doi: 10.1093/nar/gkm600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abbondanzieri EA, Shaevitz JW, Block SM. Picocalorimetry of transcription by RNA polymerase. Biophys J. 2005;89:L61–3. doi: 10.1529/biophysj.105.074195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–47. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 37.Bai L, Fulbright RM, Wang MD. Mechanochemical kinetics of transcription elongation. Phys Rev Lett. 2007;98:068103. doi: 10.1103/PhysRevLett.98.068103. [DOI] [PubMed] [Google Scholar]
- 38.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–5. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo Q, Sousa R. Translocation by T7 RNA polymerase: a sensitively poised Brownian ratchet. J Mol Biol. 2006;358:241–54. doi: 10.1016/j.jmb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–3. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 41.Donahue BA, Yin S, Taylor JS, Reines D, Hanawalt PC. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci U S A. 1994;91:8502–6. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stepanova E, Wang M, Severinov K, Borukhov S. Early transcriptional arrest at Escherichia coli rplN and ompX promoters. J Biol Chem. 2009;284:35702–13. doi: 10.1074/jbc.M109.053983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci U S A. 2009;106:8900–5. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landick R. Transcriptional pausing without backtracking. Proc Natl Acad Sci U S A. 2009;106:8797–8. doi: 10.1073/pnas.0904373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–19. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 46.Voliotis M, Cohen N, Molina-Paris C, Liverpool TB. Fluctuations, pauses, and backtracking in DNA transcription. Biophys J. 2008;94:334–48. doi: 10.1529/biophysj.107.105767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26:117–29. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao D, Lurz R, Dobrinski B, Dennis PP. A NusG-like protein from Thermotoga maritima binds to DNA and RNA. J Bacteriol. 1996;178:4089–98. doi: 10.1128/jb.178.14.4089-4098.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sevostyanova A, Svetlov V, Vassylyev DG, Artsimovitch I. The elongation factor RfaH and the initiation factor sigma bind to the same site on the transcription elongation complex. Proc Natl Acad Sci U S A. 2008;105:865–70. doi: 10.1073/pnas.0708432105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tolic-Norrelykke SF, Engh AM, Landick R, Gelles J. Diversity in the rates of transcript elongation by single RNA polymerase molecules. J Biol Chem. 2004;279:3292–9. doi: 10.1074/jbc.M310290200. [DOI] [PubMed] [Google Scholar]