Abstract
We measured the effects of non-nucleoside reverse transcriptase (RT) inhibitor-resistant mutations K101E+G190S, on replication fitness and EFV-resistance of HIVNL4-3. K101E+G190S reduced fitness in the absence of EFV and increased EFV-resistance, compared to either single mutant. Unexpectedly, K101E+G190S also replicated more efficiently in the presence of EFV than in its absence. Addition of the nucleoside-resistance mutations L74V or M41L+T215Y to K101E+G190S improved fitness and abolished EFV-dependent stimulation of replication. D10, a clinical RT backbone containing M41L+T215Y and K101E+G190S, also demonstrated EFV dependent stimulation that was dependent on the presence of K101E. These studies demonstrate that non-nucleoside reverse transcriptase inhibitors can stimulate replication of NNRTI-resistant HIV-1 and that nucleoside-resistant mutants can abolish this stimulation. The ability of EFV to stimulate NNRTI-resistant mutants may contribute to the selection of HIV-1 mutants in vivo. These studies have important implications regarding the treatment of HIV-1 with combination nucleoside and non-nucleoside therapies.
Keywords: HIV-1, drug resistance, drug-dependent stimulation of replication, replication fitness, non-nucleoside reverse transcriptase inhibitors, nucleoside reverse transcriptase inhibitors
Introduction
Human Immunodeficiency Virus type 1 (HIV-1) reverse transcriptase (RT) is an important target of antiretroviral therapy. Non-nucleoside reverse transcriptase inhibitors (NNRTIs) selectively bind to HIV-1 RT in a hydrophobic binding pocket adjacent to the polymerase active site, which is located in the palm subdomain of the p66 subunit (Kohlstaedt et al., 1992). NNRTI binding causes an allosteric change in RT that leads to non-productive binding of the incoming nucleotide during DNA polymerization (Spence et al., 1995). HIV-1 resistance to NNRTIs is caused by mutations in the NNRTI binding pocket, which interfere with drug binding (reviewed in (Domaoal and Demeter, 2004)).
Efavirenz (EFV) is the most commonly used NNRTI clinically, because of its demonstrated potent antiviral activity and clinical efficacy when combined as first-line therapy with two nucleoside analogs (Gulick et al., 2006; Gulick et al., 2004; Riddler et al., 2008; Robbins et al., 2003; Staszewski et al., 1999). K103N is the most frequently observed NNRTI-resistance mutation in patients failing EFV-containing regimens (Bacheler et al., 2000; Riddler et al., 2008). Other NNRTI resistance mutations, such as G190S, confer similar or greater degrees of EFV resistance compared to K103N, but develop uncommonly in patient isolates. Uncommonly occurring NNRTI resistance mutations introduced into a laboratory strain cause substantially greater reductions in replication fitness than K103N, as measured in cell culture in the absence and presence of drug, suggesting that replication fitness influences the likelihood of a mutant emerging during treatment failure of an NNRTI-containing regimen (Archer et al., 2000; Gerondelis et al., 1999; Koval et al., 2006; Wang et al., 2006).
An interesting question is why mutations that confer reductions in HIV-1 replication fitness in cell culture ever appear in clinical samples, if fitness significantly impacts mutant selection in patients. One hypothesis is that second-site mutations, either within or outside of RT, could compensate for the replication deficits conferred by these drug-resistant mutations. Another possibility is that second-site mutations could augment HIV-1 drug resistance sufficiently to favor selection of the less-fit mutant in the presence of drug. One example of this type of mutation is L74V, which confers resistance to the nucleoside analogs abacavir and didanosine and improves the replication fitness of the NNRTI-resistant mutants G190E and K103N+L100I (Kleim et al., 1996; Koval et al., 2006).
K101E has been observed in patients failing NNRTIs, including EFV, and most often occurs in combination with other known NNRTI-resistance mutations (Bacheler et al., 2001; Bacheler et al., 2000). K101E in combination with either K103N or G190S increases EFV resistance more than 10-fold (Bacheler et al., 2001; Petropoulos et al., 2000). K101E and G190S are also of interest because they are associated with HIV-1 resistance to the next-generation NNRTI, etravirine (Llibre et al., 2008) (Picchio, G., Vingerhoets, J., Staes, M., Tambuyzer, L., Bacheler, L., Pattery, T., de Bethune, M. P. [2008]. 15th Conference on Retroviral and Opportunistic Infections, Boston, MA., Abstract #866). We further explored the effects of K101E on HIV-1 replication fitness and EFV resistance alone and in combination with G190S, and evaluated the ability of other RT mutations to modulate these effects.
Materials and Methods
Reagents and cells
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Disease: the infectious molecular clone pNL4-3 was obtained from Malcolm Martin, and the PM-1 neoplastic CD4+ T cell line expressing both the CCR5 and CXCR4 co-receptors was obtained from Marvin Reitz (Adachi et al., 1986; Lusso et al., 1995). EFV was obtained from Dupont Pharmaceuticals Company; it was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 5 mg/ml (15.7 mM) and stored at -20° C. The human primary embryonal kidney cell line 293 (American Type Culture Collection; Manassas, VA) was grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum, L-glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 U/ml). PM1 cells were grown in RPMI supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 U/ml). Primary human peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll gradient centrifugation from blood that was obtained from HIV-uninfected volunteer donors, after obtaining written informed consent. PBMCs were cryopreserved and stored in liquid nitrogen until needed for studies. PBMCs were stimulated with 5 μg/ml phytohemagglutinin (PHA) and 20 units/ml interleukin-2 (IL-2) for two days before being infected, and were grown in RPMI supplemented with 20% fetal bovine serum. All cells were propagated in 5% CO2 at 37°C.
Patient specimens
Stored plasma samples were obtained from HIV-infected subjects who received EFV in combination with indinavir, after obtaining informed consent in the DMP 266-003 study; a subset of patients that initially received indinavir monotherapy subsequently received EFV in combination with the nucleoside analogs stavudine (d4T) and lamivudine (3TC) (Bacheler et al., 2000). One stored sample identified as having K101E+G190S on bulk sequence was further evaluated. These studies are compliant with federal guidelines relating to human subjects research, and were approved by the University of Rochester Research Subjects Review Board (RSRB).
Subcloning patient RT sequences
Viral RNA was extracted from patient plasma using QIAamp MinElute Virus Spin Kit (Qiagen, Inc., Valencia, CA). The HIV-1 RT sequence spanning amino acids 15-560 of RT was amplified using primers containing silent XmaI and XbaI restriction enzyme cleavage sites, as described previously (Dykes et al., 2001). PCR products were cloned into the pCR2.1 TOPO vector (TOPO TA cloning kit, Invitrogen, Inc.; Valencia, CA). After verifying the RT sequence using forward and reverse primers, plasmids were digested with XmaI and XbaI and the RT insert was cloned into pNL4-3XX as described previously (Dykes et al., 2001).
Mutagenesis
The L74V, K101E, and M41L+T215Y mutations were introduced into pNL4-3XX containing wild-type or G190S RT sequences, as previously described (Wang et al., 2006). Mutagenic primers were as follows: L74V, 5′-CAG TAC TAA ATG GAG AAA AGT AGT AGA TTT CAG AGA AC3′ (forward) and 5′ GTT CTC TGA AAT CTA CTA CTT TTC TCC ATT TAG TAC TG 3′ (reverse); K101E, 5′-GCA GGG TTA GAA AAG AAA AAA TCA G-3′ (forward) and 5′-CTG ATT TTT TCT TTT CTA ACC CTG C-3′ (reverse); M41L, 5′-GCA TTA GTA GAA ATT TGT ACA GAA TTG GAA AAG GAA GG-3′ (forward) and 5′ CC TTC CTT TTC CAA TTC TGT ACA AAT TTC TAC TAA TGC-3′ (reverse); and T215Y, 5′-GTG GGG ATT TTA CAC ACC AGA CAA AAA AC-3′ (forward) and 5′-GT TTT TTG TCT GGT GTG TAA AAT CCC CAC-3′ (reverse). M230L was introduced into wild-type pNL4-3 using the primers: 5′-CCT TTG GCT GGG TTA TGA ACT CCA TC-3′ (forward) and 5′-GAT GGA GTT CAT AAC CCA GCC AAA GG-3′ (reverse).
Generation of virus stocks
293 cells were transiently transfected with 40 μg of each plasmid DNA by lipofection (SuperFect®, Qiagen, Santa Clarita, CA); supernatants were harvested after 72 h, and stored at -80°C. HIV-1 virus capsid protein (p24) quantitation was performed on virus stocks using an ELISA (Perkin Elmer; Wellesley, MA).
Replication fitness assays
HIV-1 replication fitness was quantified using a multiple cycle growth competition assay in the PM1 cell line; a subset were also tested in the H9 T-cell line and in IL-2 and PHA-stimulated PBMC. The relative proportions of the test and reference strains were measured using direct sequence analysis, as previously described (Archer et al., 2000; Koval et al., 2006; Wang et al., 2006). Virus replication was quantified by measuring p24 antigen content in the culture supernatant. We quantified relative replication fitness using the production rate ratio (PRR), which is the ratio of the relative infection rate of the mutant and reference strains (Wu et al., 2006), using a publicly available web site calculator (http://bis.urmc.rochester.edu/vFitness/).
Drug susceptibility assays
The effects of EFV on virus replication were measured using a modification of the ACTG/DoD method (Japour et al., 1993) in both PM1 cells and PHA- and IL2-stimulated PBMC cells, as previously described (Koval et al., 2006). A virus inoculum of 150 ng p24 was used to infect 3 × 106 PM1 cells or 4 × 106 stimulated PBMCs in a total volume of 1 mL in the absence of drug. Cells were then washed and cultured either in the absence of drug or in the presence of varying concentrations of EFV, which were determined empirically for each mutant (maximum concentration was 25.6 μM, with no evidence for cytotoxicity as measured by trypan blue exclusion). Virus replication was assayed at day 6 after infection by measuring p24 antigen concentration in the culture supernatant. At a minimum, all assays were performed in triplicate.
The K101E+G190S and G190S mutants were also evaluated for EFV-dependent stimulation of replication using a modification of a previously published HIV replication fitness assay in which infected cells are detected by flow cytometry using antibodies directed against a virus-expressed Thy 1 reporter gene (Dykes et al., 2006). The K101E+G190S and G190S mutants were subcloned into the pAT2 vector, using silent XmaI and XbaI sites flanking reverse transcriptase, as previously described (Dykes et al., 2006). The pAT2 vector was derived from pNL4-3, and carries the Thy 1.2 gene in place of nef. Virus stocks derived by transient transfection of 293 cells were used to infect PM1 cells using inocula designed to give similar levels of replication in the no-drug control (5 ng p24 per million cells for G190S and 50 ng p24 per million cells for K101E+G190S). Thy 1.2 expressed on the surface of infected cells was detected using R-phycoerythrin (PE)-labeled antibody, as previously described, 6 days after infection (Dykes et al., 2006). The limit of detection of this assay is approximately 0.05% of cells.
Calculation of IC50
Traditionally, the following nonlinear model has been used to estimate the IC50: F(x) = 1 - 1/(1+[drug concentration/IC50]d), where F(x) is the proportional reduction in virus replication at a given drug concentration, relative to a no drug control; and d is a shape parameter (Chou, 1976). An important feature of this model is that the value F(x) increases from 0% to 100% as drug concentration increases. This model does not account for the possibility of virus replication in the presence of drug being greater than 100% of the no drug control (i.e., drug-dependent stimulation of virus replication).
We therefore applied a more flexible, non-parametric model to fit the observations when drug-dependent stimulation of virus replication occurs. This model can be used to describe curves determined by observations, and has been widely used in biological and biomedical research (Fan and Gijbels, 1996; King and Roth, 2003). Our approach to estimate IC50 for a mutant whose growth is also stimulated at some drug concentrations is a novel application of this approach. We assumed that the percentage Y and concentration x are related in the form Y = m(x), where m(•) is a function in the mathematical sense, but did not put any restrictions on the form of m(•), i.e. whether m(•) is linear, or nonlinear in x, etc. Hence, it is up to empirical analysis to use the observed data to find out more about m(•). We used the observations (x1, y1), … (xn, yn) to estimate m(•) by adapting local linear regression techniques (Fan and Gijbels, 1996), referred to as mn(x). The basic idea of local linear regression is that we estimate m(x) at x0, then we fit locally a straight line using the observations in a window around x0. After fitting the line, the estimation mn(x0) is provided by the value of this line at x0. By repeating this procedure for each x0, one can get the estimation function mn(x). The window width is constant. Formally, the local linear regression is computed by solving a weighted least square problem. The IC50 may be identified as the point x* which satisfies mn(x*) = 0.5. Data analysis was conducted using the statistical software R. For each experiment, we applied our nonparametric approach to obtain an IC50 value. Then we used Wilcox test for a comparison of IC50 values for each pair of mutants to obtain a p value.
Results
K101E reduces the replication fitness of G190S, except in the presence of high EFV concentrations
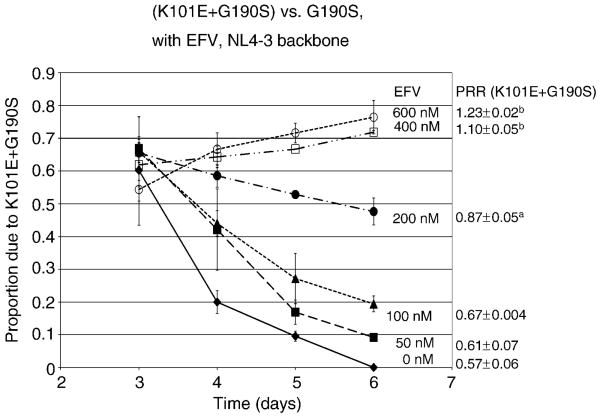
We have previously shown that the relative fitness of G190S is 40% reduced relative to wild type (Wang et al., 2006). Since K101E has been observed in combination with G190S in patients failed EFV, we wanted to determine the effect of the addition of K101E on the replication of G190S. We compared the relative fitness of recombinant viruses carrying K101E or G190S alone and in combination, in an NL4-3 backbone, using multiple-cycle growth competition assays in PM1 cells. The K101E mutation alone-reduced HIV-1 replication fitness by 20% compared to wild type (PRR=0.81±0.05) and had slightly increased fitness relative to G190S (PRR=1.07±0.01). The combination of K101E and G190S reduced replication fitness in the absence of EFV to a much greater extent than either mutation alone (PRR=0.57±0.06 versus G190S), but the fitness deficit of the double mutant relative to G190S was reversed at concentrations of EFV exceeding 200 nM (Fig. 1).
Figure 1. Relative replication fitness of K101E+G190S, in an NL4-3 backbone, in the absence and presence of EFV.
The graph represents the relative proportion of the double mutant K101E+G190S relative to the reference strain G190S NL4-3, as determined by bulk sequence analysis in growth competition experiments, as outlined in Materials and Methods. Growth competition experiments were performed in the absence of EFV (closed diamonds), or in the presence of 50 nM (closed squares), 100 nM (closed triangles), 200 nM (closed circles), 400 nM (open squares), or 600 nM (open circles) of EFV. Data represent the mean and standard deviation of results from a minimum of three independent infections. The average and standard deviation of the production rate ratio (PRR) at each EFV concentration is shown to the left of the graph. ap=0.007 and bp<0.001 compared to PRR in absence of EFV. EFV, EFV.
The K101E+G190S mutant in an NL4-3 backbone demonstrates EFV-dependent stimulation of virus replication
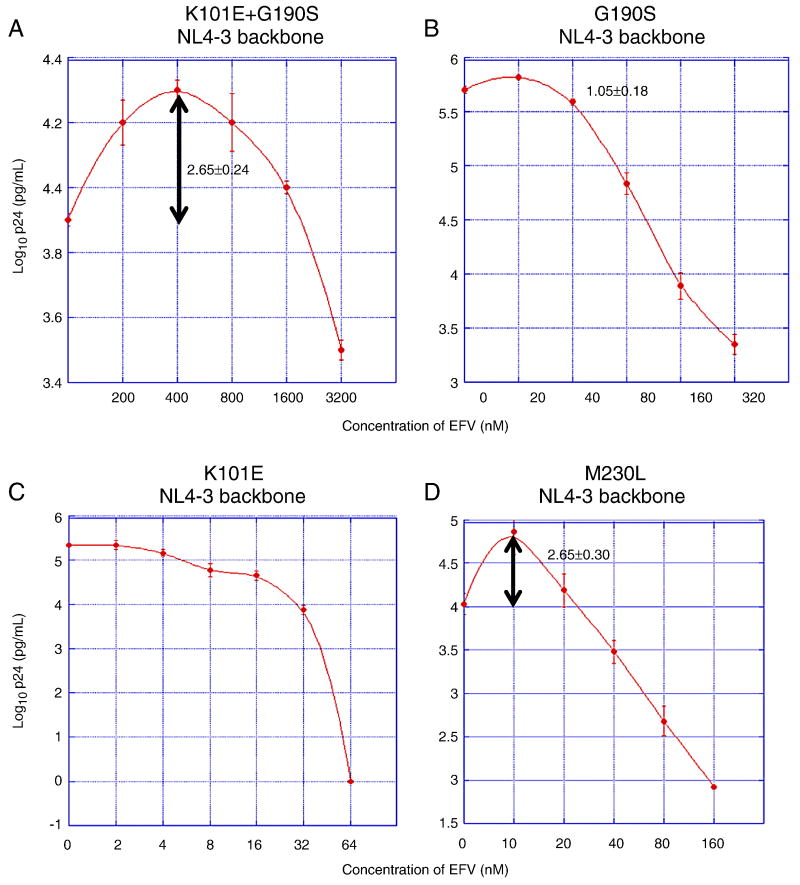
In order to better understand how K101E augments the relative fitness of G190S in the presence of EFV, we performed drug resistance assays, in which PM1 cells were infected separately by each mutant, and supernatant p24 antigen concentration was measured after six days of growth in different concentrations of EFV. The IC50 of K101E+G190S was substantially higher than either single mutant, as expected from the results of growth competition assays in the presence of EFV (Table 1). Surprisingly, when reviewing the drug resistance data, we found that the K101E+G190S double mutant's replication was 2.0 – 2.5 fold higher in 200 - 800 nM of EFV than in the absence of drug (Fig. 2A, p<0.001, comparing p24 concentration in 400 nM EFV vs. no drug). This property of EFV-dependent growth stimulation was not observed with either single mutant (Fig. 2B-C). We also measured replication of the G190S and K101E+G190S mutants using a previously published replication assay in which a reporter gene product expressed by HIV-infected cells is detected by flow cytometry (Dykes et al., 2006). Using this assay, concentrations of EFV that stimulated replication of K101E+G190S, as defined by p24 antigen production, also increased the number of infected cells compared to the no drug control (data not shown). We also observed the phenomenon of EFV-dependent stimulation in both primary PBMCs and lymphoid cell lines and at different virus inocula (data not shown), suggesting that this property is not dependent on the experimental conditions used.
Table 1. EFV susceptibilities of drug resistant mutants.
| RT backbonea | Drug Resistance Mutations | EFV IC50 (nM) b | SD | P-value |
|---|---|---|---|---|
| NL4-3 | Wild type (IC90) | 3c | 1.3 | -- |
| G190S | 66 | 1.5 | -- | |
| K101E | 13 | 1.1 | -- | |
| K101E+G190S | 3,113 | 37.7 | < 0.0001d | |
| L74V + (K101E+G190S) | 2,808 | 92.4 | NS d | |
| (M41L+T215Y) + (K101E+G190S) | 903 | 420 | < 0.0001e | |
| D10 | (M41L+T215Y) + (K101E+G190S) | 5,695 | 116.8 | -- |
| K101E+G190Sf | 17,110 | 3,315 | < 0.01g |
RT, reverse transcriptase; EFV, efavirenz; IC50, 50% inhibitory concentration; IC90, 90% inhibitory concentration; SD, standard deviation.
all viruses tested have NL4-3 sequence other than codons 15 - 560 of RT.
values represent the mean of at least 4 replicates done on at least 2 separate days.
IC90 previously reported by Bacheler Et. al. (Bacheler et al., 2001).
compared to G190S or K101E single mutants in the NL4-3 RT backbone.
compared to K101E+G190S in the NL4-3 RT backbone.
M41L and T215Y were reverted to wild-type; other D10 polymorphisms were unchanged.
compared to (M41L+T215Y) + (K101E+G190S) in the D10 RT backbone.
Figure 2. EFV susceptibilities of NNRTI-resistant mutants of NL4-3.
Graphs represent the results of EFV susceptibility assays of the NNRTI-resistant mutants K101E+G190S (panel A), G190S (panel B), K101E (panel C), and M230L (panel D). Each mutant was introduced into an NL4-3 RT backbone. X-axis represents time after infection; y-axis represents virus replication, expressed as log10 p24 antigen concentration in culture supernatant. The peak fold increases in p24 concentration compared to the p24 concentration without drug is noted on each graph at the appropriate EFV concentration. Note that the scales of the x- and y-axes differ for each mutant. Data points represent the mean and standard deviation of at least three independent infections. For K101E no increase in mean p24 antigen concentration relative to the no-drug control was observed at any of the EFV concentrations tested. For G190S, p24 concentrations of some replicate cultures of the G190S mutant in 20 nM EFV appeared to be higher than the no-drub control, however, this apparent different was not statistically significant (95% confidence intervals cross 1.00).
There are no published reports of NNRTIs stimulating HIV-1 replication, although the M230L mutant was reported to display this property in presented but unpublished work (Huang W., Parkin N.T., Lie, Y.S., et al. 4th International Workshop on HIV Drug Resistance and Treatment Strategies, June 2000, Abstract #30; in Antiviral Therapy volume 5, supplement 3, pp. 24-25). Of interest is that at least one clinical isolate in that study also contained K101E and G190S. We confirmed that the M230L mutant in an NL4-3 backbone does replicate better in the presence of low concentrations of EFV than in the absence of drug; the magnitude of EFV-dependent stimulation is similar to that observed with K101E+G190S, although the peak of growth stimulation occurred at a much lower EFV concentration than K101E+G190S (10 nM vs. 400 nM, Fig. 2D). The peak p24 concentration for the K101E+G190S double mutant in 400 nM EFV was almost ten-fold greater than the p24 concentration of G190S in a similar concentration of EFV (Fig. 2A and B), consistent with the hypothesis that the property of EFV-dependent growth stimulation contributes to the improved fitness of K101E+G190S relative to G190S in 400 and 600 nM EFV (Fig. 1). Studies using PHA- and IL-2-stimulated primary human PBMCs confirmed that the properties of the K101E+G190S mutant are also observed in primary cells (data not shown).
Identification of a clinical RT sequence containing K101E+G190S that has improved fitness compared to K101E+G190S in an NL4-3 backbone
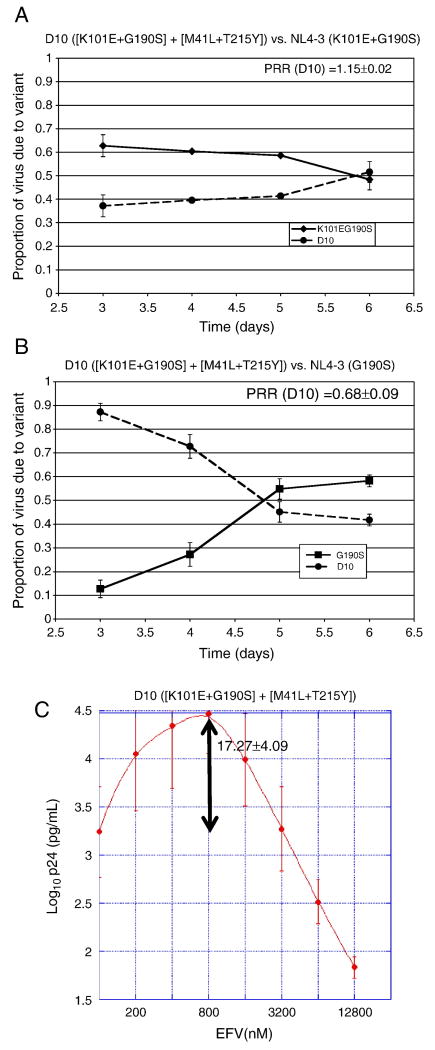
In order to determine the impact of RT backbone sequences on the properties of the K101E+G190S double mutant, we constructed a pNL4-3 clone containing an RT sequence derived from patient plasma (clone D10), which contained K101E+G190S. This clinical RT sequence also contained the nucleoside resistance mutations M41L+T215Y, in addition to 28 coding changes in RT compared to NL4-3 (Table 2). In the absence of EFV, NL4-3 virus containing the D10 RT sequence was somewhat more fit than K101E+G190S in an NL4-3 RT backbone (Fig. 3A), but still remained substantially less fit than G190S in an NL4-3 backbone (Fig. 3B).
Table 2. Codon changes in the D10 RT compared to NL4-3.
| Class of mutation | Codon changes relative to NL4-3 |
|---|---|
| nRTI resistance | M41L T215Y |
| NNRTI resistance | K101E G190S |
| RT polymorphisms | T27S K43N |
| K102Q I142V C162S Q174K | |
| Q207N R211K Q258L R277K T286A A288S V293I E298K | |
| K358R A371T A376T T386I E399D | |
| A400T T403S I435V D460N R461K V467I P468S Q480E L491S | |
nRTI, nucleoside reverse transcriptase inhibitor
NNRTI, non-nucleoside reverse transcriptase inhibitor
RT, reverse transcriptase
Figure 3. Effects of the D10 RT sequence on HIV-1 replication in the absence and presence of EFV.
Panel A, Growth competition experiment with NL4-3 virus containing the D10 RT sequence (with the resistance mutations [K101E+G190S] + [M41L+T215Y]), versus the reference strain, (K101E+G190S) in an NL4-3 RT backbone. The average and standard deviation of the production rate ratio (PRR) is shown on the graph. Panel B, Growth competition experiment with NL4-3 virus containing the D10 RT sequence versus the reference strain, G190S in an NL4-3 RT backbone. The average and standard deviation of the production rate ratio (PRR) is shown on the graph. Panel C, drug susceptibility assay using virus with the D10 RT grown in the presence of varying concentrations of EFV. The peak fold increase in p24 concentration compared to the p24 concentration without drug is noted on the graph at 800 nM EFV.
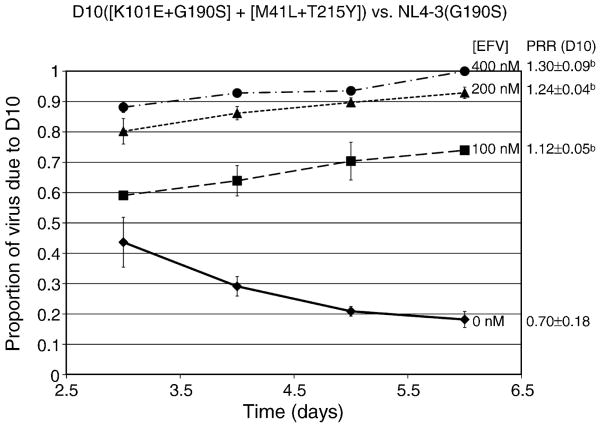
Of note is that NL4-3 virus with the D10 RT sequence demonstrated marked EFV-dependent stimulation of growth, with peak p24 concentrations 17 times higher than the no drug control in 800 nM EFV (Fig. 3C). With this level of stimulation, the p24 values of the D10 clone exceeded those of the G190S mutant beginning at an EFV concentration of 200 nM (compare Fig. 3C and 2B). Because p24 concentrations of independent cultures may not be directly comparable, we used growth competition assays to confirm that NL4-3 virus with the D10 RT sequence was more fit than G190S in the presence of EFV concentrations associated with drug-dependent stimulation of replication (Fig. 4).
Figure 4. Relative replication fitness of D10 RT versus G190S in NL4-3 in the absence and presence of EFV.
Growth competition assays of virus with the D10 RT versus G190S in NL4-3, in the presence of no drug (closed diamonds), or 100 nM (closed squares), 200 nM (closed triangles), or 400 nM (closed circles) of EFV. Bars represent standard deviations from a total of at least three independent infections. The average and standard deviation of the production rate ratio (PRR) at each EFV concentration is shown to the left of the graph. ap=0.007 and bp<0.001 compared to PRR in absence of EFV. bp<0.001 compared to PRR in absence of EFV.
Effects of the nucleoside resistance mutations M41L+T215Y on the replication fitness, EFV resistance, and EFV-dependent stimulation of K101E+G190S
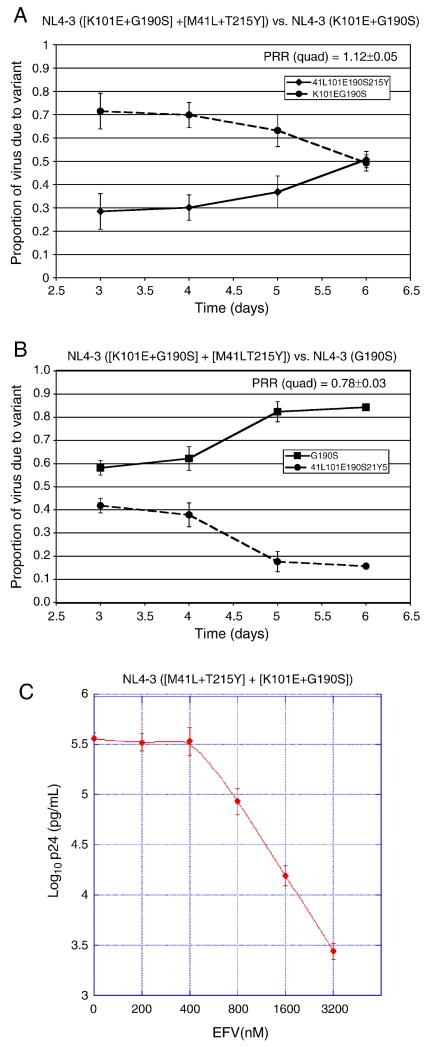
In order to understand the effects of the nucleoside resistance mutations M41L+T215Y present in the D10 RT, we introduced these two mutations into NL4-3 containing K101E+G190S. We observed that M41L+T215Y improved the replication fitness of the NNRTI-resistant K101E+G190S NL4-3 mutant, although the quadruple mutant still had substantially reduced fitness compared to G190S alone (Fig. 5A-B). However, the ([M41L+T215Y] + [K101E+G190S]) quadruple mutant in an NL4-3 RT backbone became less fit compared to K101E+G190S at higher concentrations of EFV (PRR=0.85±0.03 at 400 nM EFV), suggesting that (M41L+T215Y) reduces the EFV-dependent stimulation and/or EFV resistance of (K101E+G190S). This hypothesis was confirmed by drug susceptibility assays showing that (M41L+T215Y) reduced the EFV IC50 of (K101E+G190S) in an NL4-3 RT backbone (Table 1, p<0.0001) and abolished the EFV-dependent stimulation of viral growth phenotype of (K101E+G190S) (Fig. 5C vs. Fig. 2A). Thus, the presence of (M41L+T215Y) at least partially explains the improved fitness of NL4-3 containing the D10 RT relative to NL4-3 (K101E+G190S) in the absence of drug, but is not consistent with the observed stimulation of replication of the D10-NL4-3 virus by EFV.
Figure 5. Impact of the thymidine analog mutations (M41L+T215Y) on replication of HIV-1 containing the NNRTI resistance mutations (K101E+G190S) in an NL4-3 backbone.
Panels A and B, growth competition experiments of ([K101E+G190S] + [M41L+T215Y]) in an NL4-3 RT backbone, in the absence of drug. Reference strains are NL4-3 (K101E+G190S) in panel A, and NL4-3 (G190S) in panel B. The average and standard deviation of the production rate ratio (PRR) is shown on each the graph for the quadruple mutant. Panel C, EFV susceptibility assay.
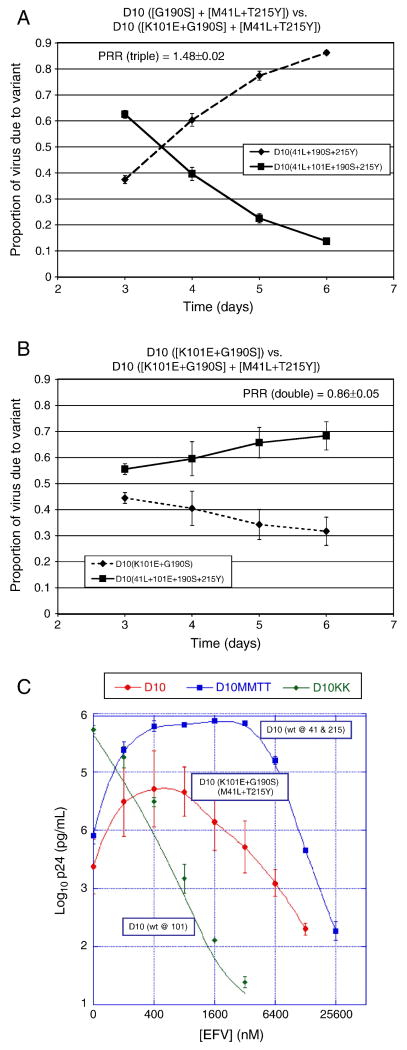
Effects of other RT polymorphisms on the replication fitness, drug resistance, and EFV-dependent growth stimulation conferred by the D10 clinical RT sequence
The above studies suggested that RT polymorphisms present in the D10 clinical RT sequence allowed the persistence of the EFV-dependent stimulation of virus replication conferred by (K101E+G190S) despite the presence of (M41L+T215Y). In order to further study the role of other RT coding sequences on the phenotypes of the D10 clinical RT sequence, we separately back-mutated the (M41L+T215Y) and K101E mutations in the D10 RT to the corresponding wild-type sequences. All resultant clones were sequenced on both strands through all of RT to ensure no spurious mutations were introduced. These studies demonstrated that in the D10 clinical RT backbone, K101E reduced replication fitness (Fig. 6A) and (M41L+T215Y) improved replication fitness in the absence of EFV (Fig. 6B), similar to the effects of these mutations in an NL4-3 RT backbone. The phenotype of EFV-dependent stimulation of virus replication was abolished by mutating K101E to wild-type (Fig. 6C). Of interest is that removal of (M41L+T215Y) enhanced the degree of EFV-dependent stimulation relative to the original D10 RT sequence, indicating that these nucleoside resistance mutations did have some suppressive effect on EFV-dependent stimulation when combined with (K101E+G190S) in the D10 backbone (Fig. 6C). However, the presence of (M41L+T215Y) in the D10 background did not fully suppress EFV-dependent growth stimulation of (K101E+G190S) (Fig. 6C), as it did in the NL4-3 backbone (Fig. 5C). Thus, the polymorphisms in D10 appear to augment the EFV-dependent growth stimulation conferred by K101E, but do not directly contribute to EFV-dependent growth stimulation in the absence of this mutation. In addition, polymorphism(s) in D10 limit the suppressive effect of (M41L+T215Y) on the EFV-dependent growth stimulation conferred by (K101E+G190S).
Figure 6. Impact of drug resistance mutations in the D10 RT backbone on HIV-1 replication in the absence and presence of EFV.
Panels A and B, growth competition assays of virus with the D10 RT in which K101E (panel A) or [M41L+T215Y] (panel B) were reverted to wild-type, competed against NL4-3 with the intact D10 RT as a reference strain. The average and standard deviation of the production rate ratio (PRR) is shown on each graph. Panel C, EFV susceptibility assays with variants of D10 in which different resistance mutations were reverted to wild-type. The peak EFV-dependent stimulation concentration for D10 with M41L+T215Y back mutated to wild-type was 3200 nM EFV and the fold stimulation was 100.73±28.8. EFV, EFV; wt, wild-type.
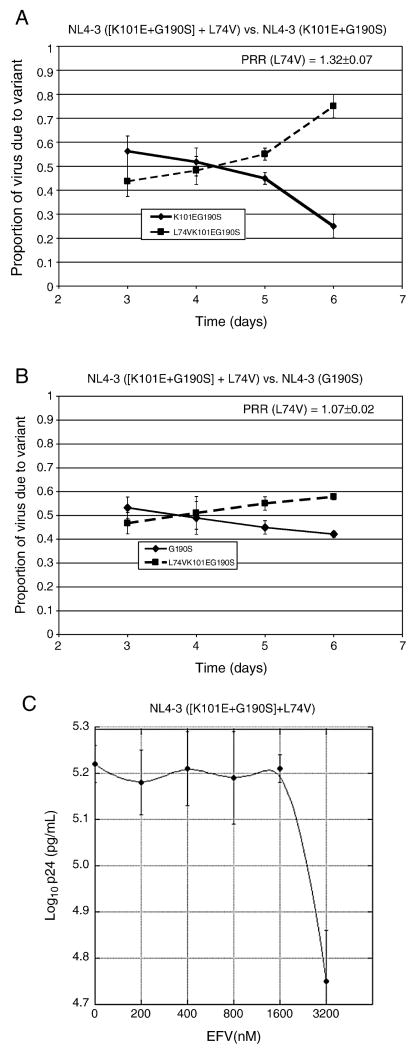
Effects of the nucleoside resistance mutation L74V on the replication fitness, EFV resistance, and EFV-dependent stimulation of (K101E+G190S)
We also evaluated whether another nucleoside resistance mutation, L74V, might affect the replication of (K101E+G190S), since L74V has been shown to improve the replication fitness of at least two other NNRTI-resistant mutants, G190E and (K103N+L100I) (Boyer, Gao, and Hughes, 1998; Kleim et al., 1996; Koval et al., 2006). In the absence of EFV, L74V substantially improved the replication fitness of (K101E+G190S) in an NL4-3 RT backbone in the absence of drug (Fig. 7A). The magnitude of improvement in relative fitness was greater than conferred by the addition of (M41L+T215Y), since the L74V + (K101E+G190S) triple mutant's fitness was as good as, if not better than, G190S alone in the absence of EFV (Fig. 7B). These data indicate that L74V fully compensates for the replication fitness impairment conferred by K101E in combination with G190S. No reduction in fitness of the L74V + (K101E+G190S) triple mutant relative to (K101E+G190S) was observed in the presence of increasing EFV concentrations, as was seen with the ([M41L+T215Y] + [K101E+G190S]) mutant (PRR of triple = 1.40±0.16 at 1200 nM EFV). Drug susceptibility assays indicated that the L74V mutation abolished the EFV-dependent growth stimulation of (K101E+G190S), without reducing its degree of EFV resistance (Fig. 7C, Table-1). Figure 8 summarizes the effects of drug resistance mutations and RT backbone on HIV-1 replication fitness, EFV-resistance, and EFV-dependent stimulation of virus replication.
Figure 7. Impact of the nucleoside resistance mutation L74V on replication by (K101E+G190S) NL4-3 in the absence and presence of EFV.
Panels A and B, growth competition experiments. The average and standard deviation of the production rate ratio (PRR) is shown on each graph. Panel C, EFV susceptibility assay. X-axis, EFV concentration; y-axis, log10 p24 concentration in culture supernatant 6 days after infection.
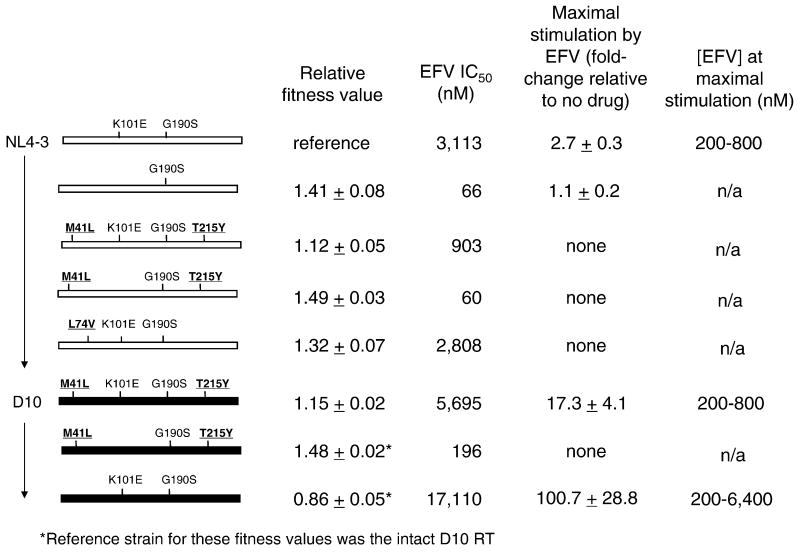
Figure 8. Summary of interactions among resistance mutations and RT polymorphisms that affect HIV-1 resistance to EFV, EFV-dependent stimulation of virus replication, and replication fitness in the absence of drug.
Rectangles represent the RT sequence from codons 15-560. RT backbone polymorphisms are represented by the color of the rectangle: white, NL4-3, and black, D10. NNRTI resistance mutations are in regular font, nucleoside resistance mutations are in bold. Relative fitness value is the mean PRR ± standard deviation. The value “maximal stimulation by EFV” is defined as the highest p24 antigen concentration observed in the drug susceptibility assay, divided by the p24 antigen concentration of the no-drug control. Values represent the mean ± standard deviation of at least three replicates. Viral variants for which the maximal stimulation is “none” had no p24 values in the presence of EFV that were significantly higher than the no-drug control. The concentrations of EFV at which maximal stimulation occurred are listed in the last column. n/a, not applicable.
Discussion
These studies provide evidence that the replication of some drug-resistant mutants of HIV-1 can be stimulated in the presence of an antiretroviral drug. We have observed this phenomenon with the (K101E+G190S) mutant, and have confirmed the initial report that M230L has this property (Huang W., Parkin N.T., Lie, Y.S., et al. 4th International Workshop on HIV Drug Resistance and Treatment Strategies, June 2000, Abstract #30; in Antiviral Therapy volume 5, supplement 3 pp. 24-25). We have also demonstrated that both nucleoside resistance mutations abolished EFV-dependent stimulation, despite having different effects on EFV IC50. Because an association between EFV-dependent stimulation and EFV-resistance was not consistently observed, we believe it is reasonable to make a distinction between these two phenotypes, even though they both represent changes in virus replication rates in the presence of drug. Although there appeared to be a correlation between reduced replication fitness and the presence of EFV-dependent stimulation, this also was not a consistent finding, since the D10 mutant RT sequence conferred improved fitness and augmented EFV-dependent stimulation compared to virus with an NL4-3 RT backbone containing the same resistance mutations. Moreover, we observed this phenomenon in two different RT backbones. Studies in which resistance mutations in the D10 backbone were reverted to wild type indicate that some RT polymorphisms can augment the impact of K101E+G190S on EFV-dependent stimulation of replication and reduce the ability of M41L+T215Y to reverse the drug-dependent stimulation conferred by K101E+G190S. The modulating effects of RT polymorphisms in the D10 RT backbone are dependent on the presence of K101E (and also presumably G190S, although this was not directly tested in our studies).
Another important finding of these studies is that the NNRTI-resistance mutations K101E and G190S interact to affect a variety of phenotypes, leading to increased EFV resistance, reduced replication fitness in the absence of drug, and EFV-dependent stimulation of virus replication. It is important to note that these phenotypes could not be predicted from studies of each single mutant. A limited number of previously published studies have evaluated interactions among NNRTI-resistant variants, and have also found unexpected effects on replication fitness (Collins et al., 2004; Koval et al., 2006). Understanding interactions among NNRTI-resistance mutations is important, since clinical virus isolates are evolving more complex patterns of NNRTI resistance mutations, now that next-generation NNRTIs such as etravirine are being used to treat patients who have failed first-line NNRTIs.
These studies have also demonstrated important interactions between nucleoside and NNRTI resistance mutations that affect all of the phenotypes that we evaluated: EFV resistance, replication fitness in the absence of drug, and EFV-dependent stimulation of virus replication. L74V, in addition to improving the fitness of G190E and K103N+L100I (Kleim et al., 1996; Koval et al., 2006), also has the same effect on (K101E+G190S). This finding is compatible with previously published studies of L74V and NNRTI-resistant variants (Boyer, Gao, and Hughes, 1998; Koval et al., 2006; Trivedi et al., 2008). Our study, however, is the first demonstration that thymidine analog mutations (TAMs), such as M41L+T215Y, can also augment the replication fitness of an NNRTI-resistant mutant. The TAM double mutant (M41L+T215Y) also abolishes EFV-dependent stimulation of growth, similar to L74V, but unlike L74V, sensitizes the virus to EFV. The ability of these two TAMs to sensitize HIV to EFV is consistent with a number of previously published studies demonstrating the influence of TAMs on EFV hyper-susceptibility (Clark et al., 2006; Shulman et al., 2004; Whitcomb et al., 2002). Our studies are shown to be in agreement with those of Huang et al. (Huang et al., 2003) who tested the resistance and fitness of patient RT sequences with various substitutions at the G190 position. They showed that the fitness of G190 mutations correlated with their prevalence in patients and that they were primarily responsible for the NNRTI resistance pattern. They also showed that the fitness of very poorly replicating mutants was better in the patient backbone where the mutation occurred and that L74V enhanced the replication of G190S and other mutants which is consistent with our results. They believe the reduced fitness of G190 substitutions is the result of reduced RT in the virions. They did not see any stimulation of virus replication by NNRTIs.
Our studies have demonstrated that each of these three phenotypes can influence the relative prevalence of two mutants in culture. The relative importance of EFV-dependent stimulation in a clinical setting is unclear, but could very well contribute to selection for an otherwise unfit mutant. Although the culture conditions studied do not fully replicate the situation in patients, a number of studies have found that relative replication in cell culture of drug-resistant mutants generally correlates with their prevalence in patients (Archer et al., 2000; Garcia-Lerma et al., 2000; Gerondelis et al., 1999; Harrigan, Bloor, and Larder, 1998; Hu et al., 2006; Huang et al., 2003; Kosalaraksa et al., 1999; Koval et al., 2006; Perrin and Mammano, 2003; Sugiura et al., 2002; Wang et al., 2006). One concern in extrapolating results from tissue culture experiments to patients is that the protein concentration in cell culture is lower than patient serum, and since efavirenz is highly protein-bound, free drug concentrations are likely quite different in the experimental and clinical settings. One study, which directly compared efavirenz IC90S under standard tissue culture conditions to those obtained in protein concentrations similar to that in serum, derived a protein-binding factor of 16.5 to allow comparisons of IC90s with plasma concentrations in patients (Corbett et al., 1999). Using this factor adjusts mean Cmin and Cmax values of 5.6 μM and 12.9 μM observed in patients (quoted in the efavirenz package insert) to give corresponding tissue culture values of 339 nM and 781 nM, respectively, which are in the range of the efavirenz concentrations at which we observed stimulation of the K101E+G190S mutants. Although these figures are estimates, they do suggest that efavirenz-dependent stimulation of these mutants occurs at clinically relevant efavirenz concentrations. Our studies also demonstrate that EFV-dependent stimulation contributes to a selective advantage for some mutants in certain drug concentrations, and this property may contribute to the selection of some drug-resistant mutants in clinical infection.
There is no current evidence that explains the mechanism by which EFV stimulates virus replication. However, we believe it is through its interaction with RT and not some other protein. There are several possible mechanisms for the stimulation. It could enhance RT protein incorporation into virions, polymerization, dimerization, and/or RNase H activity. We believe the later two mechanisms are most likely since EFV has already been shown to enhance these activities. EFV enhances dimerization of HIV-1 reverse transcriptase (Tachedjian et al., 2001), and the wild-type 101K residue in the p66 subunit can form a salt bridge with 138E in the p51 subunit (Ren et al., 2006; Ren and Stammers, 2008). Thus, it is possible that the K101E mutation in combination with G190S leads to destabilization of the reverse transcriptase heterodimer that is partially compensated for by weak binding of EFV, resulting in EFV-dependent stimulation of HIV-1 reverse transcription. Unlike 101K, the wild-type 230M residue does not appear to directly promote dimerization, although it possibly could have indirect effects on dimer stability. It is unclear at present how both L74V and TAMs might reduce or eliminate EFV-dependent stimulation; this effect appears independent of the effects of these nucleoside resistance mutations on EFV IC50. Our preliminary evidence shows that dimerization of K101E+G190S is not stimulated by EFV. However, we plan to more thoroughly examine the mechanism of dimerization in order to definitively determine if it is responsible for stimulation.
An alternative potential explanation for EFV-dependent stimulation of virus replication is that EFV, which has been found to increase RNase H cleavage rates (Palaniappan, Fay, and Bambara, 1995; Radzio and Sluis-Cremer, 2008), could compensate for RNase H defects, which have been documented for several NNRTI-resistant mutants (Archer et al., 2000; Gerondelis et al., 1999; Wang et al., 2006). We believe this hypothesis is more likely and may depend on the particular combination of mutations. Not all NNRTI-resistant mutants that have reduced RNase H activity are stimulated by EFV, but it will be important to determine whether certain combinations do, and if nucleoside resistance mutations can affect the stimulation of RNase H activity by EFV.
Conclusions
In summary, these studies demonstrate that the replication of some NNRTI-resistant mutants is stimulated by EFV, and that nucleoside resistance mutations can reverse this phenotype. In addition, nucleoside and NNRTI-resistance mutations can interact in complex ways to affect replication fitness in the absence of drug, EFV resistance, and EFV-dependent stimulation of virus replication; and these effects can be modulated by RT polymorphisms not known to be involved in drug resistance. The mechanism(s) and clinical significance of EFV-dependent stimulation are not understood, but this phenomenon has implications for the more rational design of effective NNRTI combination regimens.
Acknowledgments
This work was supported in part by NIH R01 AI-041387 to L.M.D., NIH R01 AI-59773 to H.L, NSF grant DMS-0806097 to H.L., and the University of Rochester Developmental Center for AIDS Research (D-CFAR) P30 AI-078498. These funding sources played no role in the study design, data collection or analysis, or in the manuscript submission. We thank Kora Fox, Dongge Li, and Sue Liu for excellent technical assistance; and Robert Bambara for helpful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. Journal of Virology. 1986;59(2):284–91. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer RH, Dykes C, Gerondelis P, Lloyd A, Fay P, Reichman RC, Bambara RA, Demeter LM. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J Virol. 2000;74(18):8390–401. doi: 10.1128/jvi.74.18.8390-8401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacheler L, Jeffrey S, Hanna G, D'Aquila R, Wallace L, Logue K, Cordova B, Hertogs K, Larder B, Buckery R, Baker D, Gallagher K, Scarnati H, Tritch R, Rizzo C. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J Virol. 2001;75(11):4999–5008. doi: 10.1128/JVI.75.11.4999-5008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacheler LT, Anton ED, Kudish P, Baker D, Bunville J, Krakowski K, Bolling L, Aujay M, Wang XV, Ellis D, Becker MF, Lasut AL, George HJ, Spalding DR, Hollis G, Abremski K. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob Agents Chemother. 2000;44(9):2475–84. doi: 10.1128/aac.44.9.2475-2484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer PL, Gao HQ, Hughes SH. A mutation at position 190 of human immunodeficiency virus type 1 reverse transcriptase interacts with mutations at positions 74 and 75 via the template primer. Antimicrobial Agents & Chemotherapy. 1998;42(2):447–52. doi: 10.1128/aac.42.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC. Derivation and properties of Michaelis-Menten type and Hill type equations for reference ligands. J Theor Biol. 1976;59(2):253–76. doi: 10.1016/0022-5193(76)90169-7. [DOI] [PubMed] [Google Scholar]
- Clark SA, Shulman NS, Bosch RJ, Mellors JW. Reverse transcriptase mutations 118I, 208Y, and 215Y cause HIV-1 hypersusceptibility to non-nucleoside reverse transcriptase inhibitors. Aids. 2006;20(7):981–4. doi: 10.1097/01.aids.0000222069.14878.44. [DOI] [PubMed] [Google Scholar]
- Collins JA, Thompson MG, Paintsil E, Ricketts M, Gedzior J, Alexander L. Competitive fitness of nevirapine-resistant human immunodeficiency virus type 1 mutants. J Virol. 2004;78(2):603–11. doi: 10.1128/JVI.78.2.603-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett JW, Ko SS, Rodgers JD, Jeffrey S, Bacheler LT, Klabe RM, Diamond S, Lai CM, Rabel SR, Saye JA, Adams SP, Trainor GL, Anderson PS, Erickson-Viitanen SK. Expanded-spectrum nonnucleoside reverse transcriptase inhibitors inhibit clinically relevant mutant variants of human immunodeficiency virus type 1. Antimicrob Agents Chemother. 1999;43(12):2893–7. doi: 10.1128/aac.43.12.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domaoal RA, Demeter LM. Structural and biochemical effects of human immunodeficiency virus mutants resistant to non-nucleoside reverse transcriptase inhibitors. Int J Biochem Cell Biol. 2004;36(9):1735–51. doi: 10.1016/j.biocel.2004.02.026. [DOI] [PubMed] [Google Scholar]
- Dykes C, Fox K, Lloyd A, Chiulli M, Morse E, Demeter LM. Impact of clinical reverse transcriptase sequences on the replication capacity of HIV-1 drug-resistant mutants. Virology. 2001;285(2):193–203. doi: 10.1006/viro.2001.0920. [DOI] [PubMed] [Google Scholar]
- Dykes C, Wang J, Jin X, Planelles V, An DS, Tallo A, Huang Y, Wu H, Demeter LM. Evaluation of a multiple-cycle, recombinant virus, growth competition assay that uses flow cytometry to measure replication efficiency of human immunodeficiency virus type 1 in cell culture. J Clin Microbiol. 2006;44(6):1930–43. doi: 10.1128/JCM.02415-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Gijbels I. Monographs on statistics and applied probability 66. 1st. Chapman & Hall; London; New York: 1996. Local polynomial modelling and its applications. [Google Scholar]
- Garcia-Lerma JG, Gerrish PJ, Wright AC, Qari SH, Heneine W. Evidence of a role for the Q151L mutation and the viral background in development of multiple dideoxynucleoside-resistant human immunodeficiency virus type 1. Journal of Virology. 2000;74(20):9339–46. doi: 10.1128/jvi.74.20.9339-9346.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondelis P, Archer RH, Palaniappan C, Reichman RC, Fay PJ, Bambara RA, Demeter LM. The P236L delavirdine-resistant human immunodeficiency virus type 1 mutant is replication defective and demonstrates alterations in both RNA 5′-end- and DNA 3′-end-directed RNase H activities. J Virol. 1999;73(7):5803–13. doi: 10.1128/jvi.73.7.5803-5813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick RM, Ribaudo HJ, Shikuma CM, Lalama C, Schackman BR, Meyer WA, 3rd, Acosta EP, Schouten J, Squires KE, Pilcher CD, Murphy RL, Koletar SL, Carlson M, Reichman RC, Bastow B, Klingman KL, Kuritzkes DR. Three- vs four-drug antiretroviral regimens for the initial treatment of HIV-1 infection: a randomized controlled trial. JAMA. 2006;296(7):769–81. doi: 10.1001/jama.296.7.769. [DOI] [PubMed] [Google Scholar]
- Gulick RM, Ribaudo HJ, Shikuma CM, Lustgarten S, Squires KE, Meyer WA, 3rd, Acosta EP, Schackman BR, Pilcher CD, Murphy RL, Maher WE, Witt MD, Reichman RC, Snyder S, Klingman KL, Kuritzkes DR. Triple-nucleoside regimens versus efavirenz-containing regimens for the initial treatment of HIV-1 infection. N Engl J Med. 2004;350(18):1850–61. doi: 10.1056/NEJMoa031772. [DOI] [PubMed] [Google Scholar]
- Harrigan PR, Bloor S, Larder BA. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J Virol. 1998;72(5):3773–8. doi: 10.1128/jvi.72.5.3773-3778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Giguel F, Hatano H, Reid P, Lu J, Kuritzkes DR. Fitness comparison of thymidine analog resistance pathways in human immunodeficiency virus type 1. Journal of Virology. 2006;80(14):7020–7027. doi: 10.1128/JVI.02747-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Gamarnik A, Limoli K, Petropoulos CJ, Whitcomb JM. Amino acid substitutions at position 190 of human immunodeficiency virus type 1 reverse transcriptase increase susceptibility to delavirdine and impair virus replication. J Virol. 2003;77(2):1512–23. doi: 10.1128/JVI.77.2.1512-1523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japour AJ, Mayers DL, Johnson VA, Kuritzkes DR, Beckett LA, Arduino JM, Lane J, Black RJ, Reichelderfer PS, D'Aquila RT, et al. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. The RV-43 Study Group, the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Antimicrob Agents Chemother. 1993;37(5):1095–101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Roth FP. A non-parametric model for transcription factor binding sites. Nucleic Acids Res. 2003;31(19):e116. doi: 10.1093/nar/gng117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleim JP, Rosner M, Winkler I, Paessens A, Kirsch R, Hsiou Y, Arnold E, Riess G. Selective pressure of a quinoxaline nonnucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on HIV-1 replication results in the emergence of nucleoside RT-inhibitor-specific (RT Leu-74-->Val or Ile and Val-75-->Leu or Ile) HIV-1 mutants. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(1):34–8. doi: 10.1073/pnas.93.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256(5065):1783–90. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Kosalaraksa P, Kavlick MF, Maroun V, Le R, Mitsuya H. Comparative fitness of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 (HIV-1) in an In vitro competitive HIV-1 replication assay. J Virol. 1999;73(7):5356–63. doi: 10.1128/jvi.73.7.5356-5363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koval CE, Dykes C, Wang J, Demeter LM. Relative replication fitness of efavirenz-resistant mutants of HIV-1: correlation with frequency during clinical therapy and evidence of compensation for the reduced fitness of K103N + L100I by the nucleoside resistance mutation L74V. Virology. 2006;353(1):184–92. doi: 10.1016/j.virol.2006.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llibre JM, Santos JR, Puig T, Molto J, Ruiz L, Paredes R, Clotet B. Prevalence of etravirine-associated mutations in clinical samples with resistance to nevirapine and efavirenz. J Antimicrob Chemother. 2008;62(5):909–13. doi: 10.1093/jac/dkn297. [DOI] [PubMed] [Google Scholar]
- Lusso P, Cocchi F, Balotta C, Markham PD, Louie A, Farci P, Pal R, Gallo RC, Reitz MS., Jr Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J Virol. 1995;69(6):3712–20. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniappan C, Fay PJ, Bambara RA. Nevirapine alters the cleavage specificity of ribonuclease H of human immunodeficiency virus 1 reverse transcriptase. J Biol Chem. 1995;270(9):4861–9. doi: 10.1074/jbc.270.9.4861. [DOI] [PubMed] [Google Scholar]
- Perrin V, Mammano F. Parameters driving the selection of nelfinavir-resistant human immunodeficiency virus type 1 variants. J Virol. 2003;77(18):10172–5. doi: 10.1128/JVI.77.18.10172-10175.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos CJ, Parkin NT, Limoli KL, Lie YS, Wrin T, Huang W, Tian H, Smith D, Winslow GA, Capon DJ, Whitcomb JM. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44(4):920–8. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzio J, Sluis-Cremer N. Efavirenz accelerates HIV-1 reverse transcriptase ribonuclease H cleavage, leading to diminished zidovudine excision. Mol Pharmacol. 2008;73(2):601–6. doi: 10.1124/mol.107.038596. [DOI] [PubMed] [Google Scholar]
- Ren J, Nichols CE, Stamp A, Chamberlain PP, Ferris R, Weaver KL, Short SA, Stammers DK. Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138. FEBS J. 2006;273(16):3850–60. doi: 10.1111/j.1742-4658.2006.05392.x. [DOI] [PubMed] [Google Scholar]
- Ren J, Stammers DK. Structural basis for drug resistance mechanisms for non-nucleoside inhibitors of HIV reverse transcriptase. Virus Res. 2008;134(1-2):157–70. doi: 10.1016/j.virusres.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Riddler SA, Haubrich R, DiRienzo AG, Peeples L, Powderly WG, Klingman KL, Garren KW, George T, Rooney JF, Brizz B, Lalloo UG, Murphy RL, Swindells S, Havlir D, Mellors JW. Class-sparing regimens for initial treatment of HIV-1 infection. N Engl J Med. 2008;358(20):2095–106. doi: 10.1056/NEJMoa074609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins GK, De Gruttola V, Shafer RW, Smeaton LM, Snyder SW, Pettinelli C, Dube MP, Fischl MA, Pollard RB, Delapenha R, Gedeon L, van der Horst C, Murphy RL, Becker MI, D'Aquila RT, Vella S, Merigan TC, Hirsch MS. Comparison of sequential three-drug regimens as initial therapy for HIV-1 infection. N Engl J Med. 2003;349(24):2293–303. doi: 10.1056/NEJMoa030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman NS, Bosch RJ, Mellors JW, Albrecht MA, Katzenstein DA. Genetic correlates of efavirenz hypersusceptibility. Aids. 2004;18(13):1781–5. doi: 10.1097/00002030-200409030-00006. [DOI] [PubMed] [Google Scholar]
- Spence RA, Kati WM, Anderson KS, Johnson KA. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science. 1995;267(5200):988–93. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staszewski S, Morales-Ramirez J, Tashima KT, Rachlis A, Skiest D, Stanford J, Stryker R, Johnson P, Labriola DF, Farina D, Manion DJ, Ruiz NM. Efavirenz plus zidovudine and lamivudine, efavirenz plus indinavir, and indinavir plus zidovudine and lamivudine in the treatment of HIV-1 infection in adults. Study 006 Team. New England Journal of Medicine. 1999;341(25):1865–73. doi: 10.1056/NEJM199912163412501. [DOI] [PubMed] [Google Scholar]
- Sugiura W, Matsuda Z, Yokomaku Y, Hertogs K, Larder B, Oishi T, Kano A, Shiino T, Tatsumi M, Matsuda M, Abumi H, Takata N, Shirahata S, Yamada K, Yoshikura H, Nagai Y. Interference between D30N and L90M in selection and development of protease inhibitor-resistant human immunodeficiency virus type 1. Antimicrobial Agents and Chemotherapy. 2002;46(3):708–715. doi: 10.1128/AAC.46.3.708-715.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachedjian G, Orlova M, Sarafianos SG, Arnold E, Goff SP. Nonnucleoside reverse transcriptase inhibitors are chemical enhancers of dimerization of the HIV type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 2001;98(13):7188–93. doi: 10.1073/pnas.121055998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi V, Von Lindern J, Montes-Walters M, Rojo DR, Shell EJ, Parkin N, O'Brien WA, Ferguson MR. Impact of human immunodeficiency virus type 1 reverse transcriptase inhibitor drug resistance mutation interactions on phenotypic susceptibility. AIDS Res Hum Retroviruses. 2008;24(10):1291–300. doi: 10.1089/aid.2007.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Dykes C, Domaoal RA, Koval CE, Bambara RA, Demeter LM. The HIV-1 reverse transcriptase mutants G190S and G190A, which confer resistance to non-nucleoside reverse transcriptase inhibitors, demonstrate reductions in RNase H activity and DNA synthesis from tRNA(Lys, 3) that correlate with reductions in replication efficiency. Virology. 2006;348(2):462–74. doi: 10.1016/j.virol.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb JM, Huang W, Limoli K, Paxinos E, Wrin T, Skowron G, Deeks SG, Bates M, Hellmann NS, Petropoulos CJ. Hypersusceptibility to non-nucleoside reverse transcriptase inhibitors in HIV-1: clinical, phenotypic and genotypic correlates. Aids. 2002;16(15):F41–7. doi: 10.1097/00002030-200210180-00002. [DOI] [PubMed] [Google Scholar]
- Wu H, Huang Y, Dykes C, Liu D, Ma J, Perelson AS, Demeter LM. Modeling and estimation of replication fitness of human immunodeficiency virus type 1 in vitro experiments by using a growth competition assay. J Virol. 2006;80(5):2380–9. doi: 10.1128/JVI.80.5.2380-2389.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]