Abstract
Background
Lateral gene transfer (LGT) appears to promote genotypic and phenotypic variation in microbial communities in a range of environments, including the mammalian intestine. However, the extent and mechanisms of LGT in intestinal microbial communities of non-mammalian hosts remains poorly understood.
Methodology/Principal Findings
We sequenced two fosmid inserts obtained from a genomic DNA library derived from an agar-degrading enrichment culture of marine iguana fecal material. The inserts harbored 16S rRNA genes that place the organism from which they originated within Clostridium cluster IV, a well documented group that habitats the mammalian intestinal tract. However, sequence analysis indicates that 52% of the protein-coding genes on the fosmids have top BLASTX hits to bacterial species that are not members of Clostridium cluster IV, and phylogenetic analysis suggests that at least 10 of 44 coding genes on the fosmids may have been transferred from Clostridium cluster XIVa to cluster IV. The fosmids encoded four transposase-encoding genes and an integrase-encoding gene, suggesting their involvement in LGT. In addition, several coding genes likely involved in sugar transport were probably acquired through LGT.
Conclusion
Our phylogenetic evidence suggests that LGT may be common among phylogenetically distinct members of the phylum Firmicutes inhabiting the intestinal tract of marine iguanas.
Introduction
There is no other quarter of the world, where this order (reptiles), replaces herbivorous mammalia in so extraordinary a manner. –Darwin [1]
During his visit to the Galápagos archipelago in 1835 Charles Darwin encountered several species of large herbivorous reptiles, and he made the intriguing observation that, with the exception of the Galápagos islands, there are few places on earth today where reptiles are the most abundant herbivores. The success of herbivorous reptiles on the Galápagos is, in part, related to their varied morphological, physiological, and behavioral adaptations [2], [3]. In addition, their effective utilization of plant material is aided by symbiotic relationships with intestinal microorganisms that hydrolyze and ferment the otherwise indigestible plant polymers [4], which is consistent with the known role of bacteria and protozoa in aiding digestion in herbivorous mammals [5] and insects [6].
In order to compete for resources and ultimately, to allow their host to survive and reproduce, intestinal microorganisms must also adapt. An emerging theme in genomic biology suggests that lateral gene transfer (LGT) is key for promoting genotypic and phenotypic variation in microorganisms [7], including those from intestinal environments [8], [9], [10], [11]. For example, Ricard et al. (2006) showed that ∼4% of genes in the genomes of ciliates common in the rumen were likely obtained from bacteria and archaea [11]. The majority of these genes were involved with carbohydrate catabolism, suggesting that their acquisition helped ciliates to successfully colonize and adapt to the rumen environment. Although the extent and control of LGT among microorganisms in the intestine of non-mammalian hosts, such as reptiles, remains unexplored, 16S rDNA clone libraries suggest that their gut bacterial communities differ in composition from those of herbivorous mammals. Firmicutes and specifically several phylogenetically defined Clostridium clusters (I, III, IV, and XIVa) are the predominant phyla in the intestine of marine iguanas (Amblyrynchus cristatus; Fig. 1) [4], land iguanas (Conolophus spp.), and giant tortoises (Testudo elephantopus) (Mackie, unpublished data). In contrast, herbivorous mammals also contain an abundance of diverse representatives of the phylum Bacteroidetes [12]. Thus if LGT is an important process in the intestine of herbivorous reptiles, it likely occurs among non-Bacteroidetes species.
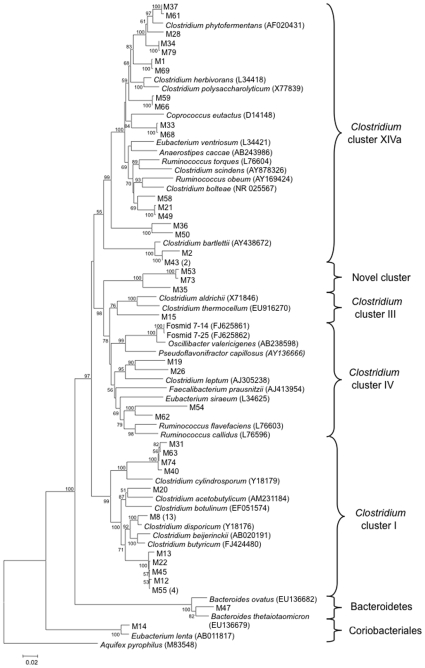
Figure 1. Phylogenetic relationships of 16S rRNA gene sequences among the fosmids, clone library, and Clostridium clusters.
Clone library sequences start with “M.” The numbers in parentheses following some of the marine iguana sequences indicate the number of times that a particular sequence was obtained. Only representatives of the major Clostridium clusters, and limited representatives of the Bacteroidetes and Coriobacteriales, are shown. The tree was inferred using the neighbor joining approach. The numbers at the nodes represent bootstrap values. The bar represents 0.02 substitutions per nucleotide position. The outgroup is Aquifex pyrophilus.
The marine iguana, which is endemic to the Galápagos Islands, is unique among herbivorous reptiles because its diet consists solely of soft macrophytic algae. A previous study of Orkney sheep consuming a diet of seaweed indicated that such a diet selected for the proliferation of Candidatus Oscillospira guilliermondii, an uncultivated, but morphologically conspicuous, member of Clostridium cluster IV [13]. More recently, a closely related member of Clostridium cluster IV, Oscillibacter valericigenes, was isolated from the alimentary canal of clams that feed on marine plankton [14]. However, the ecology and evolution of Oscillospira- and Oscillibacter-like organisms remains poorly understood. To select for gut microbes capable of degrading agar (the primary component of the algal cell walls) we established an anaerobic enrichment culture from marine iguana fecal material using agar as a sole carbon source. We then created a fosmid library from the enrichment culture to obtain genomic information from organisms actively involved in agar-degradation in the hindgut of marine iguanas. We hypothesized that members of Clostridium cluster IV would be active in the culture, given their previously demonstrated abundance in the gastrointestinal tracts of marine animals. In the course of screening the library we discovered two fosmids with 16S rRNA gene sequence similar to those of Oscillibacter valericigenes, and an initial examination of the sequences using a BLAST-based approach suggested that some of the genes on the fosmids may have been subject to LGT. Here we report these sequence data and use a phylogenetic approach to assess the extent, probable direction, and mechanisms of LGT among these members of Clostridium cluster IV.
Results and Discussion
The fosmid library that was constructed contained ∼2,000 clones. The fosmids selected for sequencing, named 7–14 and 7–25, were ∼35.3 and ∼29.5 kb in length, respectively. Each fosmid contained an RNA operon consisting of 16S rRNA, tRNA, and 23S rRNA genes (Table 1). The 16S and 23S rDNA tags on the fosmids were almost identical to each other (99.8% and 99.9% sequence similarity, respectively), and codon usage patterns on the fosmids were significantly correlated (r = 0.70, p<0.001), which indicates that the fosmids likely derive from the same species. Phylogenetic analysis, based on 16S rDNA, indicated that the fosmids were derived from members of Clostridium cluster IV with ∼97.3% 16S rDNA similarity to their nearest cultivated relative, Oscillibacter valericigenes (Fig. 1). The nearest cultivated relative with a complete genome sequence available in a public database is Bacteroides capillosus, which has now been reclassified as Pseudoflavonifractor capillosus, a member of Clostridium cluster IV, based on biochemical properties, DNA G+C content, DNA-DNA hybridization and phylogenetic position [15].
Table 1.
| Fosmid name | CDS a | Nucleotide range | Transcription direction | % GC content | Predicted function b | LGT? (probable direction of transfer) c | BLASTX analysis d | |||
| Name of top hit | Accession number | E value | % Nucleotide identity | |||||||
| 7–14 | 1 | 18–920 | + | 53.0 | Transposase for insertion sequence element ISRM5 | No | Pseudoflavonifractor capillosus | ZP_02038860 | 2.00E−136 | 92 |
| 7–14 | 2 | 1459–1929 | − | 35.6 | Teicoplanin resistance protein vanZ | Novel | Geobacillus thermodenitrificans | YP_001124564 | 7.00E−06 | 31 |
| 7–14 | 3 | 2346–3584 | − | 50.8 | Putative transposase | Yes, from Clostridium XIVa to IV | Coprococcus eutactus | ZP_02205788 | 2.00E−167 | 69 |
| 7–14 | 4 | 4214–5032 | − | 43.8 | Hypothetical protein | Novel | Anaerostipes caccae | ZP_02418355 | 1.00E−03 | 39 |
| 7–14 | 5 | 6252–6962 | − | 51.9 | Outer membrane lipoprotein-sorting protein | Novel | Caldicellulosiruptor saccharolyticus | YP_001179297 | 8.00E−06 | 31 |
| 7–14 | 6 | 6959–7480 | − | 47.3 | RNA polymerase sigma-54 factor rpoN | Unresolved | Alkaliphilus oremlandii | YP_001512203 | 1.00E−28 | 40 |
| 7–14 | 7 | 7745–7820 | 63.2 | tRNA-Pro (TGG) | Na | |||||
| 7–14 | 8 | 8265–10187 | − | 57.9 | Large exoproteins involved in heme utilization or adhesion | Unresolved | Herpetosiphon aurantiacus | YP_001545021 | 2.00E−58 | 35 |
| 7–14 | 9 | 10188–10862 | − | 55.4 | Hypothetical protein | Unresolved | Anaerotruncus colihominis | ZP_02440985 | 3.00E−38 | 56 |
| 7–14 | 10 | 10855–11580 | − | 54.4 | Hypothetical protein | Yes, direction unresolved | Desulfitobacterium hafniense | YP_517449 | 7.00E−67 | 56 |
| 7–14 | 11 | 11732–12193 | − | 52.8 | Iron-sulfur cluster regulator IscR | Unresolved (No) | Anaerotruncus colihominis | ZP_02443971 | 2.00E−44 | 65 |
| 7–14 | 12 | 12428–13360 | + | 59.1 | Cysteine synthase | Unresolved | Faecalibacterium prausnitzii | ZP_02090920 | 4.00E−123 | 83 |
| 7–14 | 13 | 13706–13840 | − | 46.7 | Sodium/glutamate symporter | Unresolved | Eubacterium siraeum | ZP_02423775 | 5.00E−12 | 80 |
| 7–14 | 14 | 13795–14877 | − | 56.7 | Sodium/glutamate symporter | Unresolved | Eubacterium siraeum | ZP_02423775 | 7.00E−134 | 73 |
| 7–14 | 15 | 14895–15938 | − | 61.3 | Immunogenic protein | Unresolved | Coprococcus eutactus | ZP_02205701 | 6.00E−61 | 44 |
| 7–14 | 16 | 16214–17023 | − | 57.9 | 8-oxoguanine-DNA-glycosylase | Yes, from Clostridium XIVa to IV | Ruminococcus torques | ZP_01966859 | 9.00E−55 | 43 |
| 7–14 | 17 | 17025–18026 | − | 57.6 | L-asparaginase | Unresolved | Clostridium bolteae | ZP_02087774 | 7.00E−103 | 54 |
| 7–14 | 18 | 18036–19133 | − | 62.6 | Exonuclease SbcD | No | Pseudoflavonifractor capillosus | ZP_02034908 | 3.00E−98 | 55 |
| 7–14 | 19 | 19130–19684 | − | 56.8 | EBSC protein | Unresolved | Clostridium leptum | ZP_02078782 | 2.00E−54 | 68 |
| 7–14 | 20 | 19651–20424 | − | 55.9 | Phosphoesterase family protein | No | Pseudoflavonifractor capillosus | ZP_02034909 | 4.00E−87 | 61 |
| 7–14 | 21 | 21133–21414 | − | 50.7 | Hypothetical protein | Unresolved | Eubacterium siraeum | ZP_02423392 | 2.00E−08 | 35 |
| 7–14 | 22 | 22289–23032 | + | 54.6 | Transposase for insertion sequence element ISRM5 | No | Pseudoflavonifractor capillosus | ZP_02038860 | 1.00E−110 | 87 |
| 7–14 | 23 | 23434–23508 | 58.7 | tRNA-Glu (CTC) | Na | |||||
| 7–14 | 24 | 23684–23759 | 52.6 | tRNA-Lys (CTT) | Na | |||||
| 7–14 | 25 | 23856–26709 | 52.5 | 23S rRNA gene | Na | |||||
| 7–14 | 26 | 27033–27109 | 63.6 | tRNA-Ile (GAT) | Na | |||||
| 7–14 | 27 | 27121–27196 | 53.9 | tRNA-Ala (TGC) | Na | |||||
| 7–14 | 28 | 27324–28847 | 53.5 | 16S rRNA gene | Na | |||||
| 7–14 | 29 | 29448–30365 | − | 57.4 | Germination and sporulation | Yes, from XIVa to IV (No) | Pseudoflavonifractor capillosus | ZP_02038021 | 1.00E−25 | 38 |
| 7–14 | 30 | 30362–31768 | − | 60.5 | Osmosensitive K+ channel histidine kinase kdpD | No | Pseudoflavonifractor capillosus | ZP_02038022 | 1.00E−120 | 52 |
| 7–14 | 31 | 31788–32465 | − | 55.8 | Two-component response regulator SA14-24 | No | Pseudoflavonifractor capillosus | ZP_02036840 | 7.00E−86 | 76 |
| 7–14 | 32 | 32486–32854 | − | 55.3 | Late competence protein comEA, DNA receptor | Unresolved | Cand. Desulforudis audaxviator | YP_001718193 | 3.00E−15 | 58 |
| 7–14 | 33 | 33191–34558 | + | 57.1 | D-alanyl-D-alanine carboxypeptidase | No | Pseudoflavonifractor capillosus | ZP_02036838 | 3.00E−7 | 47 |
| 7–14 | 34 | 34607–35155 | − | 59.7 | Nitroreductase family protein | Unresolved (Yes, from XIVa to IV) | Clostridium kluyveri | YP_001393744 | 9.00E−36 | 50 |
| 7–25 | 1 | 1–794 | + | 51.5 | Integrase | Yes, from Clostridium XIVa to IV | Clostridium bolteae | ZP_02083674 | 6.00E−90 | 62 |
| 7–25 | 2 | 910–2469 | − | 57.8 | GMP synthase [glutamine-hydrolyzing] | No | Pseudoflavonifractor capillosus | ZP_02035344 | 0.00E+00 | 82 |
| 7–25 | 3 | 2447–2962 | − | 56.2 | Xanthine phosphoribosyltransferase | Unresolved | Coprococcus eutactus | ZP_02207218 | 3.00E−43 | 54 |
| 7–25 | 4 | 3418–4239 | − | 60.5 | Nucleotide-binding protein | No | Eubacterium siraeum | ZP_02421312 | 4.00E−91 | 64 |
| 7–25 | 5 | 4308–6572 | − | 61.3 | Chromosome partition protein smc | No | Pseudoflavonifractor capillosus | ZP_02034907 | 6.00E−103 | 35 |
| 7–25 | 6 | 7160–8682 | 53.6 | 16S rRNA gene | Na | |||||
| 7–25 | 7 | 8810–8885 | 55.3 | tRNA-Ala (TGC) | Na | |||||
| 7–25 | 8 | 8897–8973 | 64.9 | tRNA-Ile (GAT) | Na | |||||
| 7–25 | 9 | 9297–12150 | 52.5 | 23S rRNA gene | Na | |||||
| 7–25 | 10 | 12247–12322 | 53.9 | tRNA-Lys (CTT) | Na | |||||
| 7–25 | 11 | 12498–12572 | 58.7 | tRNA-Glu (CTC) | Na | |||||
| 7–25 | 12 | 12864–12939 | 57.9 | tRNA-Asn (GTT) | Na | |||||
| 7–25 | 13 | 12994–13070 | 61.0 | tRNA-Met (CAT) | Na | |||||
| 7–25 | 14 | 13108–13183 | 59.2 | tRNA-Trp (CCA) | Na | |||||
| 7–25 | 15 | 13246–13322 | 62.3 | tRNA-Asp (GTC) | Na | |||||
| 7–25 | 16 | 13328–13403 | 59.2 | tRNA-Thr (GGT) | Na | |||||
| 7–25 | 17 | 13613–14674 | + | 54.3 | Transposase for insertion sequence element ISRM5 | No | Pseudoflavonifractor capillosus | ZP_02038860 | 4.00E−131 | 89 |
| 7–25 | 18 | 15027–15791 | + | 40.4 | Unknown | Unresolved (No) | Bacteroides thetaiotaomicron | NP_810500 | 1.00E−117 | 78 |
| 7–25 | 19 | 15913–17193 | + | 55.2 | Hypothetical protein | Unresolved | Eubacterium siraeum | ZP_02421339 | 4.00E−86 | 44 |
| 7–25 | 20 | 17144–18088 | + | 56.9 | Hypothetical protein | Unresolved | Eubacterium ventriosum | ZP_02027484 | 5.00E−66 | 43 |
| 7–25 | 21 | 18370–19662 | + | 58.0 | Putative stomatin/prohibitin-family membrane protease subunit | Yes, direction unresolved | Clostridium acetobutylicum | NP_349972 | 2.00E−92 | 48 |
| 7–25 | 22 | 19560–20867 | + | 57.0 | Protein RtcB | No | Pseudoflavonifractor capillosus | ZP_02038919 | 2.00E−154 | 69 |
| 7–25 | 23 | 20864–21208 | + | 55.1 | Predicted nucleotidyltransferase | Unresolved | Escherichia coli | ZP_03048733 | 5.00E−16 | 59 |
| 7–25 | 24 | 22686–23951 | + | 53.7 | Hypothetical lipoprotein | Yes, from Clostridium XIVa to IV | Clostridium bolteae | ZP_02087827 | 0.00E+00 | 82 |
| 7–25 | 25 | 24010–25620 | + | 48.6 | ABC-type sugar transport system, ATP-binding protein | Yes, from Clostridium XIVa to IV | Clostridium bolteae | ZP_02087828 | 0.00E+00 | 89 |
| 7–25 | 26 | 25634–26713 | + | 54.8 | ABC transporter integral membrane protein | Yes, from Clostridium XIVa to IV | Clostridium bolteae | ZP_02087829 | 2.00E−164 | 88 |
| 7–25 | 27 | 26710–27834 | + | 54.3 | ABC transporter integral membrane protein | Yes, from Clostridium XIVa to IV | Clostridium bolteae | ZP_02087830 | 7.00E−167 | 84 |
| 7–25 | 28 | 27831–28319 | + | 54.6 | Hypothetical protein | Yes, direction unresolved | Clostridium bolteae | ZP_02087831 | 6.00E−47 | 70 |
Na, not applicable.
CDS number from 5′ to 3′ on cloned insert.
The predicted function of each gene was determined as desribed in the text. Predicted mobile elements are in bold.
The occurrence and direction of LGT was determined using phylogenetic analysis, as described in the text. Genes that likely underwent LGT are in bold.
For CDSs in which the assessment of LGT differed between the neighbor joining and maximum likelihood based approaches the neighbor joining assessment is listed first and the maximum likelihood assessment is listed in parentheses.
As compared to the protein sequence database in GenBank. Top BLASTX hits that are members of Clostridium cluster IV are in bold.
Oscillibacter 16S rRNA gene sequences were not recovered from the small clone library created from genomic DNA extracted from marine iguana fecal material. Nevertheless, we successfully amplified 16S rDNA sequences using primers unique to the fosmids from 4/5 fecal samples from 5 different marine iguanas (Fig. S1), confirming the presence of the bacteria that the fosmids represent in the original fecal material. To ensure that the primers amplified the 16S rRNA gene sequences identified in the fosmid sequences, we extracted DNA and then cloned and sequenced 16S rDNA from one sample (sample 24). As anticipated, the top BLASTN hits of two clones that were sequenced (GenBank accession numbers GQ243725 and GQ243726) were fosmids 7–14 and 7–25. These results confirm that the organisms that the fosmids represent are present in marine iguana fecal material and are commonly found in the intestinal tracts of marine iguanas (Fig. S1).
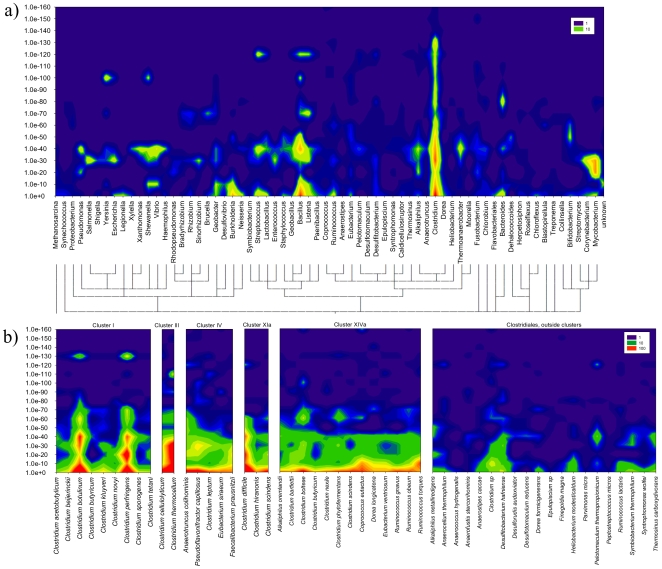
Consistent with the fact that their 16S rRNA genes indicate that the fosmids are members of Clostridium cluster IV (Fig. 1), the most dominant flare in a BLAST Heat Map of the coding genes was observed compared to the genus Clostridium. However, zones of relatively conserved sequences (e-values<1e-80) are also evident in comparison with other genera (Fig. 2a), and a concatenated BLAST Heat Map, created using custom databases of Clostridium clusters, displays flares with members outside of Clostridium cluster IV (Fig. 2b). In addition, over half (52%) of the protein-coding genes on the fosmids have top BLASTX hits to bacteria that are not members of Clostridium cluster IV (Table 1). These results contrast with phylogenetic relationships based on 16S rRNA gene sequences, and they suggest potential exchange of genetic material between phylogenetically distinct groups of bacteria.
Figure 2. BLASTP result distribution across fosmids 7–14 and 7–25.
a) The X-axis indicates genera with at least 10 BLASTP hits throughout the ORFeome of the analyzed fosmids. Using a previously published approach [33] the organism distribution on a genus level was identified for each coding gene, e-values were grouped into ranges, and threshold levels were defined for minimum overall frequency. Genera are phylogenetically sorted. The Y-axis indicates respective e-value ranges. The frequency of hits for each genus in each e-value range (log scale) is shown by color coding and corresponding values are indicated in the figure. All BLASTP hits per genus per ORF were accepted. b) Same as a), except that custom databases of species from phylogenetically defined Clostridium clusters were used.
To more rigorously assess which genes may have been subject to LGT we used phylogenetic analysis, in conjunction with parsimony analysis [9]. Assessments of LGT using neighbor-joining (NJ) and approximately maximum-likelihood (ML) trees were congruent for all but four of the coding genes. In two cases (CDS 11 on fosmid 7–14 and CDS 18 on fosmid 7–25, Figs. S2 and S3, respectively) the NJ trees could not resolve the occurrence of LGT and the ML trees suggested that LGT did not occur (Table 1). For one coding gene (CDS 29 on fosmid 7–14, Fig. S2) the NJ tree suggested LGT, whereas the ML tree indicated no LGT (Table 1). Bootstrap support for the NJ tree was low (61) and thus this gene was unlikely to have experienced recent LGT. For one coding gene (CDS 34 on fosmid 7–14, Fig. 2) LGT was unresolved with the NJ approach, whereas the ML approach suggested the occurrence of LGT. Thus we conservatively estimate that at least 10 of 44 coding genes on the fosmids (Table 1, Figs. S2 and S3) had been subject to LGT, which confirms that LGT is an important process in the evolution of intestinal microorganisms in marine iguanas. Although the precise proportion of genes subject to LGT on fosmids 7–14 and 7–25 may differ from the extent of LGT in the genome from which the fosmids derive, these results nevertheless indicate the occurrence of LGT. For all cases in which the direction of LGT could be resolved (i.e. 7 of 10 cases), the transfers likely occurred from Clostridium cluster XIVa to cluster IV. Representatives of Clostridium clusters XIVa and IV are predominated by intestinal bacteria and are common in marine iguana fecal material (Fig. 1). Thus our finding of the acquisition of genetic material by organisms in Clostridium cluster IV from those in cluster XIVa is reasonable.
The presumed split of Clostridium clusters XIVa and IV cannot explain the occurrence of genes from Clostridium cluster XIVa within the fosmids, because the phylogenetic results indicate that the gene transfers likely occurred after these organisms diverged. When phylogenetic analysis based on the NJ and ML trees indicated that a particular gene was likely not subject to LGT, the top BLASTX hit was from Clostridium cluster IV, whereas when phylogenetic analysis indicated the gene was subject to LGT the top BLAST hit was not from Clostridium cluster IV (Table 1). It is possible that some coding genes for which the occurrence of LGT could not be resolved using phylogenetic analysis were also subject to LGT as indicted by their lack of top BLASTX hits to members of Clostridium cluster IV (Table 1). These results suggest an occurrence of the exchange of genetic material between phylogenetically distinct groups of intestinal microbes in herbivorous reptiles, consistent with recent evidence for the occurrence of LGT among microorganisms in the intestine of mammalian herbivores [9], [11], [16], as well as in marine water and sediment [7], [17].
In addition, our sequence analysis indicated a total of four transposase-encoding genes and an integrase-encoding gene on the fosmids, which provides circumstantial evidence of a potential mechanism for facilitation of LGT (Table 1). Three of the transposase-encoding genes are native to Clostridium cluster IV (not subject to LGT), and two of these appear to have interrupted the RNA operons on their respective fosmids. One transposase-encoding gene and an integrase-encoding gene appear to have originated from Clostridium cluster XIVa, suggesting their potential role in transferring genes specific to Clostridium cluster XIVa into the genomes of members of cluster IV.
Together, these results indicate the exchange of genetic material between phylogenetically distinct Clostridia found within the intestine of the marine iguana that are potentially involved in agar degradation and are the subject of ongoing research. Some of the transferred genes may have functions particularly valuable for enabling bacteria to colonize and survive in the marine iguana intestine. For example, a primary role of bacteria in the intestine of the marine iguana is to degrade algal polysaccharides (e.g. agar and agaropectin), found in the cell walls of macrophytic algae, into simple sugars. These sugars may then be transported into bacterial cells. The acquisition of new types of transporters through LGT may increase the types of sugars from which microorganisms may obtain energy, and Oscillospira has been shown to rapidly associate with freshly ingested forage [18]. Although the precise timing of the LGT events revealed on the fosmids is unclear, there is evidence that some transfers may have occurred recently in evolutionary history. For example, the ABC transporters on fosmid 7–25 have synteny with, and high nucleotide-level sequence similarity to, sequences found in Clostridium bolteae (Table 1 and Fig. 1), suggesting little divergence and a relatively recent transfer event. Indeed, a recent review concludes that transfers of complex protein-encoding genes, many of which are located on operons and gene clusters, could be very common [19].
Conversely, other genes subject to LGT have less nucleotide-level sequence similarity, suggesting more ancient transfers (Table 1). Thus LGT appears to be a means for microorganisms in the intestine of herbivorous reptiles to acquire new functions and adapt to changing environmental conditions. These results, combined with other recent studies, indicate that the high microbial density and diversity of the rumen and other intestinal ecosystems create an environment conducive to LGT [8].
Materials and Methods
Fresh fecal material from 5 individual marine iguanas was collected and stored at −20°C. All procedures were non-invasive and conducted in accordance with guidelines from the American Society of Icthyologists and Herpetologists, approved by the Charles Darwin Research Station and covered under University of Illinois Urbana-Champaign LACAC #03041 and Princeton University IACUC #1428. To assess overall bacterial community composition in feces a 16S rDNA clone library was created from pooled genomic DNA. DNA was extracted using the UltraClean Soil DNA kit (MO BIO Laboratories, Carlsbad, CA). The primers used for PCR amplification of DNA from the pooled fecal samples were 27f and 1525r [20]. Amplicons were directly cloned into the PCRII-TOPO cloning vector (Invitrogen, Carlsbad, CA), and recombinant plasmids were extracted using the Wizard® Plus Minipreps DNA Purification System (Promega, Madison, WI). Sequencing was performed by the W.M. Keck Center for Comparative and Functional Genomics at the University of Illinois Urbana-Champaign.
An enrichment culture from the fecal material was created using agar as the sole carbon source in anaerobic medium [21]. The culture actively degraded agar as evidenced by rapid liquefaction. However, repeated attempts to isolate pure cultures of organisms capable of agar-degradation failed. A fosmid library was created from the fecal material using previously described methods [22]. The fosmid library was screened using PCR for those harboring inserts with a phylogenetic tag, the 16S rRNA gene, from Clostridium cluster IV [23], a heterogeneous group that includes non-clostridial species and is abundant in the intestine of the marine iguana (Fig. 1). The complete sequences of two of these fosmids, named 7–14 and 7–25, were obtained using Sanger sequencing and a “primer walking” approach. The sequences of fosmids 7–14 and 7–25 (GenBank accession numbers FJ625861 and FJ625862, respectively) were analyzed in the SEED Annotation Engine in RAST (http://rast.nmpdr.org/, Version 2.0) in order to identify genes and determine their predicted function [24]. Putative tRNA genes were folded using tRNA-scan [25] to confirm their identity.
The 16S rRNA gene sequences from the fosmids, the clone library, and representatives of the major Clostridium clusters, and limited representatives of the Bacteroidetes and Coriobacteriales were aligned using CLUSTAL W [26]. Evolutionary distances were calculated using the method of Kimura [27], and phylogenetic trees were inferred using the NJ [28] and maximum parsimony [29] methods in the MEGA 3.1 software package [30]. An approximately maximum-likelihood (ML) phylogenetic tree was also inferred using FastTree 2.1.2 [31]. All trees were concordant with each other. We also aligned the sequences using a core set of 16S rRNA gene sequences (i.e. http://greengenes.lbl.gov) and the resulting phylogenetic trees were concordant with those derived from sequences that were aligned using CLUSTAL W. To verify the presence of the 16S rDNA sequences of fosmids 7–14 and 7–25 in marine iguana fecal samples we randomly selected and extracted genomic DNA from 5 other samples of fresh fecal material. Primers unique to the 16S rDNA sequence of the fosmids (99f, 5′-AATGTTTAGTGGCGGACTGG-3′, and 1503r, 5′-ACCTTCCGATACGGCTACCT-3′) were designed and used to amplify the genomic DNA.
NJ and approximately ML phylogenetic trees of amino acid sequences from fosmids 7–14 and 7–25 were used to assess LGT, as described by Xu et al. [9]. Briefly, genes were marked as “novel” if they had e-values>10−6. If e-values were<10−6 we started at the query sequence and then stepped back in the tree until a bootstrap-supported node (>60) that contained sequences from a different species was found. If the node had decedents only from Clostridium cluster IV the gene was marked as “no LGT.” If the node had decedents from within and outside of Clostridium cluster IV the gene was marked as “unresolved.” If the node had decedents only from outside of Clostridium cluster IV the gene was marked as laterally transferred. Genes marked as laterally transferred were then subject to Fitch parsimony analysis [32] in order to determine the ancestral state of each node and the probable direction of transfer, when possible.
Supporting Information
PCR assessment of the presence of fosmid 7–14 and 7–25 16S rDNA sequences in marine iguana fecal samples from five different marine iguanas (named 24, 10, 18, 26, and 19). Arrows point to the 1.4 and 1.5 kb markers, between which is the expected PCR product size. Lanes 1 and 10 are molecular weight ladders (M). Lanes 2–6 represent the samples. A faint band of the expected size is present in sample 10, whereas no bad is visible in sample 18. Lanes 7–8 are positive controls (DNA from fosmids 7–14 and 7–25), and lane 9 is negative control (−).
(0.15 MB PDF)
Neighbor-joining phylogenetic trees of amino acid sequences from fosmid 7–14 that were used to assess LGT, as described in the text. Sequences in bold represent those from Clostridium cluster IV. For CDS 11, 29, and 34 the conclusion of LGT based upon the neighbor-joining trees differed from that based upon maximum likelihood trees (as listed in Table 1). Thus for these CDSs we also show the maximum likelihood trees.
(4.49 MB PDF)
Neighbor-joining phylogenetic trees of amino acid sequences from fosmid 7–25 that were used to assess LGT, as described in the text. Sequences in bold represent those from Clostridium cluster IV. For CDS 18 the conclusion of LGT based upon the neighbor-joining tree differed from that based upon the maximum likelihood tree (as listed in Table 1). Thus for this CDS we also show the maximum likelihood tree.
(3.02 MB PDF)
Acknowledgments
We thank M. Wikelski for assistance with sample collection, R. Aminov and S. Kocherginskaya for creating the 16S rDNA clone library, M. Mori for establishing the enrichment culture, and W. Metcalf and J.K. Zhang for assistance with creating the fosmid library. We appreciate the logistical assistance of the Charles Darwin Research Station in conducting field work.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Support for this research was provided by National Science Foundation grant #0238422 (R.I.M. and I.K.O.C.), a postdoctoral fellowship from the Institute for Genomic Biology (D.M.N.), and United States Department of Agriculture grant #2008-35107-18664 (D.M.N.). E.A. is a current employee of AgResearch Limited (www.agresearch.co.nz), a New Zealand Crown Research Institute. The funders had no role in study design, data collection and analysis, or preparation of the manuscript.
References
- 1.Browne J, Neve M, editors. The Voyage of the Beagle: Charles Darwin's Journal of Researches. 1989. 448. Penguin Classics (abridged edition)
- 2.Wikelski M, Thom C. Marine iguanas shrink to survive El Niño-Changes in bone metabolism enable these adult lizards to reversibly alter their length. Nature. 2000;403:37–38. doi: 10.1038/47396. [DOI] [PubMed] [Google Scholar]
- 3.Throckmorton GS. Oral food-processing in 2 herbivorous lizards, Iguana iguana (Iguanidae) and Uromastix aegyptius (Agamidae). Journal of Morphology. 1976;148:363–390. doi: 10.1002/jmor.1051480307. [DOI] [PubMed] [Google Scholar]
- 4.Mackie RI, Nelson DM, Wheeler E, Wikelski M, Cann IKO. Fermentative digestion in herbivorous lizards: bacterial population analysis in the intestinal tract of free-living land (Conolophus pallidus) and marine iguanas (Amblyrynchus cristatus) on the Galápagos archipelago. In: Morris S, Vosloo A, editors. 4th Comparative Physiology and Biochemistry Meeting in Africa: Mara 2008 Molecules to Migration: The Pressures of Life. Bologna, Italy: Medimond Publishing Co; 2008. pp. 193–202. [Google Scholar]
- 5.Mackie RI, White BA, Isaacson RE, editors. New York: Springer-Verlag; 1997. Gastrointestinal Microbiology: Gastrointestinal Microbes and Host Interactions.676 [Google Scholar]
- 6.Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560-U517. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 7.Nesbo CL, Boucher Y, Dlutek M, Doolittle WF. Lateral gene transfer and phylogenetic assignment of environmental fosmid clones. Environmental Microbiology. 2005;7:2011–2026. doi: 10.1111/j.1462-2920.2005.00918.x. [DOI] [PubMed] [Google Scholar]
- 8.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, et al. Evolution of symbiotic bacteria in the distal human intestine. PloS Biology. 2007;5:1574–1586. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Vallve S, Romeu A, Palau J. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Research. 2000;10:1719–1725. doi: 10.1101/gr.130000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ricard G, McEwan NR, Dutilh BE, Jouany JP, Macheboeuf D, et al. Horizontal gene transfer from Bacteria to rumen Ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genomics. 2006;7:22. doi: 10.1186/1471-2164-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Reviews Microbiology. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orpin CG, Greenwood Y, Hall FJ, Paterson IW. The rumen microbiology of seaweed digestion in Orkney sheep. Journal of Applied Bacteriology. 1985;58:585–596. doi: 10.1111/j.1365-2672.1985.tb01715.x. [DOI] [PubMed] [Google Scholar]
- 14.Lino T, Mori K, Tanaka K, Suzuki KI, Harayama S. Oscillibacter valericigenes gen. nov., sp nov., a valerate-producing anaerobic bacterium isolated from the alimentary canal of a Japanese corbicula clam. International Journal of Systematic and Evolutionary Microbiology. 2007;57:1840–1845. doi: 10.1099/ijs.0.64717-0. [DOI] [PubMed] [Google Scholar]
- 15.Carlier J-P, Bedora-Faure M, K'ouas G, Alauzet C, Mory F. Proposal to unify Clostridium orbiscindens Winter et al. 1991 and Eubacterium plautii (Séguin 1928) Hofstad and Aasjord 1982 with description of Flavonifractor plautii gen. nov., comb. nov. and reassignment of Bacteroides capillosus to Pseudoflavonifractor capillosus gen. nov., comb. nov. International Journal of Systematic and Evolutionary Microbiology. 2010;60:585–590. doi: 10.1099/ijs.0.016725-0. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Vallve S, Romeu A, Palau J. Horizontal gene transfer of glycosyl hydrolases of the rumen fungi. Molecular Biology and Evolution. 2000;17:352–361. doi: 10.1093/oxfordjournals.molbev.a026315. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert JA, Muhling M, Joint I. A rare SAR11 fosmid clone confirming genetic variability in the ‘Candidatus Pelagibacter ubique’ genome. ISME Journal. 2008;2:790–793. doi: 10.1038/ismej.2008.49. [DOI] [PubMed] [Google Scholar]
- 18.Mackie RI, Aminov RI, Hu WP, Klieve AV, Ouwerkerk D, et al. Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Applied and Environmental Microbiology. 2003;69:6808–6815. doi: 10.1128/AEM.69.11.6808-6815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gophna U. Complexity apparently is not a barrier to lateral gene transfer. Microbe. 2009;4:549–553. [Google Scholar]
- 20.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. New York: Wiley; 1991. pp. 115–175. [Google Scholar]
- 21.Mackie RI, Rycyk M, Ruemmeler RL, Aminov RI, Wikelski M. Biochemical and microbiological evidence for fermentative digestion in frre-living land iguanas (Conolophus pallidus) and marine iguanas (Amblyrhynchus cristatus) on the Galapagos Archipelago. Physiological and Biochemical Zoology. 2004;77:127–138. doi: 10.1086/383498. [DOI] [PubMed] [Google Scholar]
- 22.Eliot AC, Griffin BM, Thomas PM, Johannes TW, Kelleher NL, et al. Cloning, expression, and biochemical characterization of Streptomyces rubellomurinus genes required for biosynthesis of antimalarial compound FR900098. Chemistry & Biology. 2008;15:765–770. doi: 10.1016/j.chembiol.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Dyke MI, McCarthy AJ. Molecular biological detection and characterization of Clostridium populations in municipal landfill sites. Applied and Environmental Microbiology. 2002;68:2049–2053. doi: 10.1128/AEM.68.4.2049-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Research. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 28.Saitou N, Nei M. The neighbor-joining method - a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Nei M, Kumar S. New York: Oxford University Press; 2000. Molecular evolution and phylogenetics.333 [Google Scholar]
- 30.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 31.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fitch WM. Distinguishing homologous from analogous proteins. Systematic Zoology. 1970;19:99–113. [PubMed] [Google Scholar]
- 33.Azcarate-Peril MA, Altermann E, Goh YJ, Tallon R, Sanozky-Dawes RB, et al. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Applied and Environmental Microbiology. 2008;74:4610–4625. doi: 10.1128/AEM.00054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR assessment of the presence of fosmid 7–14 and 7–25 16S rDNA sequences in marine iguana fecal samples from five different marine iguanas (named 24, 10, 18, 26, and 19). Arrows point to the 1.4 and 1.5 kb markers, between which is the expected PCR product size. Lanes 1 and 10 are molecular weight ladders (M). Lanes 2–6 represent the samples. A faint band of the expected size is present in sample 10, whereas no bad is visible in sample 18. Lanes 7–8 are positive controls (DNA from fosmids 7–14 and 7–25), and lane 9 is negative control (−).
(0.15 MB PDF)
Neighbor-joining phylogenetic trees of amino acid sequences from fosmid 7–14 that were used to assess LGT, as described in the text. Sequences in bold represent those from Clostridium cluster IV. For CDS 11, 29, and 34 the conclusion of LGT based upon the neighbor-joining trees differed from that based upon maximum likelihood trees (as listed in Table 1). Thus for these CDSs we also show the maximum likelihood trees.
(4.49 MB PDF)
Neighbor-joining phylogenetic trees of amino acid sequences from fosmid 7–25 that were used to assess LGT, as described in the text. Sequences in bold represent those from Clostridium cluster IV. For CDS 18 the conclusion of LGT based upon the neighbor-joining tree differed from that based upon the maximum likelihood tree (as listed in Table 1). Thus for this CDS we also show the maximum likelihood tree.
(3.02 MB PDF)