Summary
Prions in Saccharomyces cerevisiae are inherited ordered aggregates reliant upon the disaggregase Hsp104 for stable maintenance. The function of other factors in the natural prion cycle is unclear. We constructed yeast-bacterial chimeric chaperones to resolve the roles of Hsp104 domains, and by extension chaperones that interact with these domains, in prion propagation. Our results show that, as with amorphous aggregate dissolution, the Hsp70/40 system recruits prion substrates to Hsp104 via its top ring. By adapting our chimera to couple to an inactive protease “trap”, we monitored the reaction products of prion propagation in vivo. We find that prion maintenance is accompanied by translocation of prion proteins through Hsp104 hexamers and that both processes critically rely upon the Hsp40 Sis1. Our data suggests that yeast prion replication is a natural extension of chaperone activity in dissolving amorphous aggregates, distinguished from its ancestral reaction by the ordered, self-propagating structure of the substrate.
Introduction
A wide array of unrelated proteins form unusually stable β-sheet-rich aggregates, or amyloid fibers, that accumulate within or between cells during the progression of neurodegenerative disorders such as Alzheimer’s and Huntington’s diseases (Luheshi et al., 2008). Molecular chaperones are intimately involved in aggregate prevention and dissolution and can modulate toxicity associated with protein misfolding in neurodegenerative disease (Muchowski and Wacker, 2005). How chaperones recognize and act on these particularly recalcitrant subtrates is largely unknown. In yeast, certain proteins form amyloid aggregates that are stably inherited through a prion-like mechanism (Chernoff, 2007; Wickner, 1994) of fiber growth and chaperone-mediated division. These yeast prions can serve as a model for understanding chaperone actions on amyloid aggregates in a cellular context.
The yeast prion [PSI+], resulting from ordered self-propagating aggregates of the translation termination factor Sup35, is a particularly attractive system for studying chaperone-amyloid interactions. Aggregation of Sup35 leads to inefficient translation termination, a phenotype readily monitored by a red/white color assay. The color of [PSI+] isolates varies with the extent of Sup35 aggregation, which in turn varies based upon the conformational differences of the Sup35 aggregates (Derkatch et al., 1996; King and Diaz-Avalos, 2004; Tanaka et al., 2004). The presence of these prion “strains” or variants allows for the study of chaperone action on multiple amyloid conformations.
Inheritance of all [PSI+] variants requires the chaperone Hsp104 (Chernoff et al., 1995), which has been proposed to fragment prion aggregates to expose new surfaces for growth (Paushkin et al., 1996). In vitro, Hsp104 has been shown to fragment fibers, as well as to promote fiber nucleation and dissolution of amyloids to noninfective aggregates (Shorter and Lindquist, 2004; Shorter and Lindquist, 2006) in the absence of cochaperones. These actions are all affected by cochaperones when present (Shorter and Lindquist, 2008). In contrast to the above studies, chemical inhibition of Hsp104 does not prevent de novo [PSI+] formation in yeast (Zhou et al., 2001), and fiber fragmentation assays using yeast extracts suggests that Hsp104 requires additional unknown factors for prion multiplication (Inoue et al., 2004). These factors are most likely other chaperones, most notably the Hsp70s and Hsp40s, as numerous genetic studies suggest (Jones and Tuite, 2005; Loovers et al., 2007; Song et al., 2005). Recently, extensive physical contacts have been reported between the Hsp70 Ssa1 and Sup35 in [PSI+] yeast (Allen et al., 2005; Bagriantsev et al., 2008) and between the Hsp40 Sis1 and another yeast prion protein, Rnq1, in [RNQ+] cells (Luke et al., 1991; Sondheimer et al., 2001).
The physical connection of Hsp70s and 40s to yeast prions along with disparities between in vitro and in vivo studies of chaperone action on amyloids underscores the need for a proximal reporter of prion propagation inside the cell. In the present study, we describe such a system based upon our design of chimeric chaperones. These chimeras allow us to identify and order cochaperone action in thermotolerance and prion propagation pathways and reveal a surprising degree of modularity in the Hsp100 family.
Results
Hsp104 recognizes prion and thermotolerance substrates through its upper ring
We began our analysis of the cellular factors required for prion propagation by determining the domains of Hsp104 required for [PSI+] maintenance. Hsp104 is necessary for induced thermotolerance (Sanchez and Lindquist, 1990) as well as [PSI+] propagation in yeast. The Hsp104 homolog, ClpB, has similar activity in bacteria but is unable to propagate [PSI+] or convey induced thermotolerance in yeast (see below). We took a systematic Hsp104-ClpB chimera approach based upon the available structure of a bacterial ClpB (Lee et al., 2003) to test which parts of Hsp104 were necessary for yeast functioning.
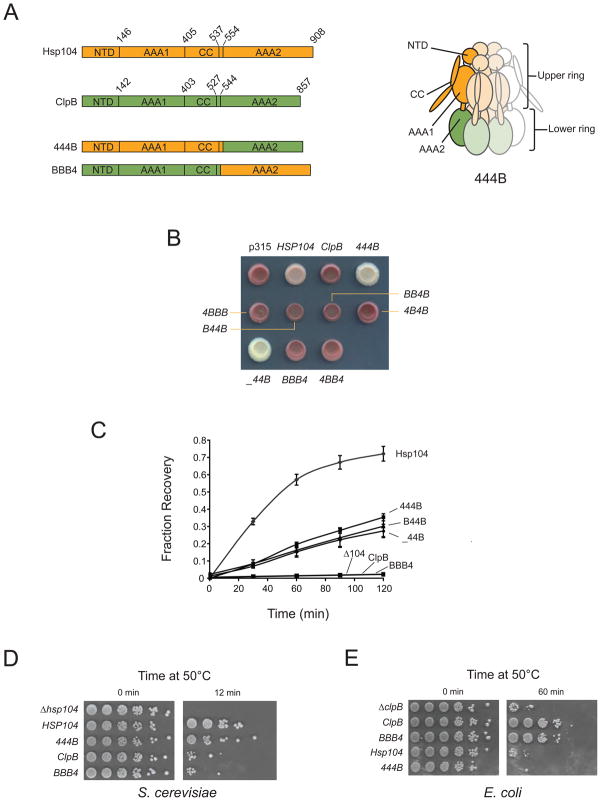
Hsp104 and ClpB belong to the Clp/Hsp100 family of AAA+ ATPase macromolecular remodeling factors (Mogk et al., 2008), which typically assemble into multimeric rings with a central pore through which substrates are threaded in an ATP-dependent fashion. Hsp104/ClpB comprises a two-tiered hexamer (Figure 1A) consisting of an N-terminal domain (NTD), a top AAA domain (AAA1), and a long coiled-coil domain (CC) with a small helix connecting this tier to AAA2. We designed a series of chimeras with different permutations of Hsp104 and ClpB domains. In naming our chimeras, we note domain origins with “4” for Hsp104, “B” for ClpB, and “_” for a domain deletion. Chimera 444B is depicted in the cartoon (Figure 1A, right) as a reference.
Figure 1. The endogenous top rings of Hsp104 and ClpB are required for thermotolerance and prion propagation.
(A) Left: Domain structures of Hsp104, ClpB and of the chimeras 444B and BBB4. Chimeras are named by domain origin (NTD, AAA1, CC and AAA2) using “4” to designate Hsp104, “B” for ClpB and “_” for a truncation. Numbers refer to amino acid sequence. Right: Cartoon of the 444B hexamer showing domain topology.
(B) [PSI+] HSP104 was replaced with the indicated Hsp100 under the control of the HSP104 promoter. Isolates are shown on low adenine media. [PSI+] appears white to pink and [psi−] appears red. [PSI+] strains were confirmed by curing with the Hsp104 inhibitor 3mM guanidine hydrochloride (not shown).
(C) Luciferase reactivation by Hsp100s in S. cerevisiae. Yeast strains constitutively expressing luciferase were grown to log phase, shifted to 37°C to induce Hsp100 expression, heat shocked at 42°C and allowed to recover at 30°C. Luciferase activity was measured at the indicated times during recovery and plotted as a fraction of pre-heat shock activity levels.
(D) Induced thermotolerance function of Hsp100 constructs in S. cerevisiae. Yeast strains were shifted from 30°C to 37°C at mid-log phase for one hour and subsequently exposed to 50°C. Shown are 1:5 stepwise dilutions.
(E) Induced thermotolerance function of Hsp100 constructs in E. coli. Strains expressing the indicated Hsp100 under the control of the ClpB promoter were grown at 30°C to mid-log, shifted to 42°C for 15 minutes and exposed to 50°C for the indicated times. Shown are 1:10 stepwise dilutions.
To assess which of our chimeras could propagate prions, we replaced native HSP104 in [PSI+] yeast with chimeras driven by the HSP104 promoter (Figure 1B). Chimeras 444B and _44B propagated [PSI+] as well or better (i.e., strengthened the original variant) than Hsp104, indicating Hsp104 AAA1 and CC are sufficient for prion recognition and processing. ClpB and the remaining chimeras were completely unable to propagate [PSI+] with the exception of very rare B44B transformants, which propagated [PSI+], albeit very inefficiently (Supplemental Figure 1).
We next tested the ability of our chimeras to refold heat-denatured luciferase in yeast, a robust activity of Hsp104 that can be recapitulated in vitro with purified Hsp104, the Hsp70 Ssa1 and the Hsp40s Ydj1 or Sis1 (Glover and Lindquist, 1998). ClpB performs the same reaction with the assistance of bacterial Hsp70 family member DnaK, Hsp40 member DnaJ and the nucleotide exchange factor GrpE (Motohashi et al., 1999; Zolkiewski, 1999). However, ClpB cannot function in vitro with yeast Hsp70 and 40s, nor can Hsp104 function with DnaKJ/GrpE (Glover and Lindquist, 1998; Krzewska et al., 2001). We introduced into Δhsp104 yeast a series of plasmids expressing HSP104, ClpB, 444B, _44B, B44B or BBB4 from the HSP104 promoter. Yeast expressing HSP104, 444B, _44B and B44B were able to rescue heat-denatured luciferase, while yeast expressing ClpB and BBB4 showed no activity over background (Figure 1C). Absolute luciferase reactivation by 444B was about 50% that of Hsp104, with cellular levels of 444B consistently around 67% that of Hsp104 (Supplemental Figure 2A). Deletion of the 444B NTD had a negligible effect on luciferase refolding, consistent with a previous study of native Hsp104 (Hung and Masison, 2006), and substitution of the ClpB NTD for the native domain had an equally negligible effect. As shown above, [PSI+] inheritance was seriously compromised by B44B but not _44B, suggesting an adaptation of Hsp104 NTD absent in ClpB to accommodate ordered aggregates.
We chose chimera 444B and its inverse, BBB4, to assay broader induced thermotolerance function in both yeast and bacteria. In agreement with our luciferase reactivation results, 444B was able to rescue yeast transiently exposed to high temperatures at about 75% the level of Hsp104, while ClpB and BBB4 showed little effect over background (Figure 1D). Conversely, BBB4 enabled bacteria to survive exposure to high temperatures at approximately the same level as ClpB, while neither Hsp104 nor 444B showed activity above the vector background (Figure 1E). We found the ratio of 444B to BBB4 to be quite similar in yeast and bacteria (1.6 versus 1.5) (Supplemental Figure 2), indicating the inactivity of 444B in bacteria and of BBB4 in yeast was not the result of expression level differences. The reciprocal functionality of 444B and BBB4 in yeast and bacteria, in the context of previous reports of species incompatibility between Hsp100s and Hsp70/40s, strongly suggests that necessary physical coupling of Hsp70 and 40 family cochaperones to Hsp104 and ClpB occurs solely upstream of the translocation process in both thermotolerance and prion propagation.
444B propagates multiple prion conformations
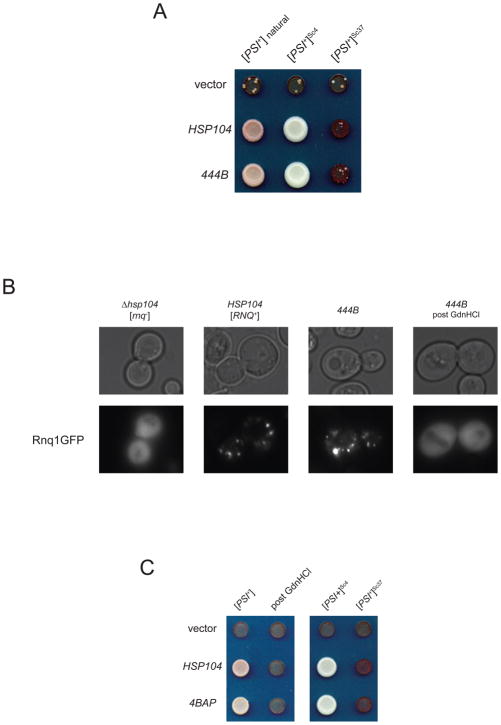
Yeast prions occur in phenotypic variants resulting from distinct physical conformations of the underlying protein aggregate (Derkatch et al., 1996; King and Diaz-Avalos, 2004; Tanaka et al., 2004). In order to test whether 444B could maintain a range of [PSI+] variants, we replaced Hsp104 in [PSI+]Sc4 and [PSI+]Sc37 strains, created by infection with different conformations of amyloid fibers (Tanaka et al., 2004). 444B was able to support naturally-occurring [PSI+] and both defined variants (Figure 2A). To establish the generality of prion maintenance function, we looked at the ability of 444B to sustain another yeast prion, [RNQ+]. [RNQ+] is readily detected by fluorescent foci in cells expressing GFP fusions to the Rnq1 protein (Aron et al., 2007). We replaced the endogenous HSP104 locus with 444B in a [PSI+] [RNQ+] yeast strain. The resulting 444B yeast formed Rnq1-GFP foci indicative of [RNQ+], and these foci disappeared after treatment with the Hsp104 inhibitor guanidine hydrochloride (Figure 2B).
Figure 2. Chimeras 444B and 4BAP propagate multiple prion conformations.
(A) Native HSP104 on a URA3-marked centromeric plasmid was replaced by an empty vector, by HSP104 or by 444B in yeast strains with the indicated [PSI+] variants. [PSI+] refers to a naturally-occurring variant, and [PSI+]Sc4 and [PSI+]Sc37 refer to variants created by infection of [psi−] yeast with recombinant Sup35 fibers polymerized at the indicated temperatures. Isolates are shown on trace adenine containing selective media. [PSI+] is pink, [PSI+]Sc4 is white, [PSI+]Sc37 appears red and displays intermediate growth, and [psi-] appears red and displays very slow growth.
(B) HSP104 was replaced in a [PSI+] [RNQ+] strain by 444B. [RNQ+/rnq−] status of the resulting 444B strain and its prion-cured derivative was assessed by a fluorescent assay in which [RNQ+] yeast display foci and [rnq−] yeast display diffuse fluorescence.
(C) Propagation by 4BAP of naturally-occurring [PSI+], its curing by 3mM GdnHCl and propagation by 4BAP of defined [PSI+]Sc4 and [PSI+]Sc37 performed as in Figure 2A.
Chimera 4BAP functionally mimics 444B
The domain structure of 444B allowed us to adapt a system previously developed to study ClpB to monitor the flux of prion propagation substrates through this chimera. A synthetic version of ClpB, BAP (Weibezahn et al., 2004), was designed to couple to the bacterial peptidase ClpP or to a catalytically inactive variant, ClpPtrap (Trap) (Flynn et al., 2003). The BAP-Trap system captured known ClpB substrates that translocated through BAP in a DnaKJGrpE-dependent fashion. We used this design to create a Trap-docking variant of 444B, 4BAP, which we tested for activity in reactivating heat-denatured luciferase (Supplemental Figure 3A), conveying thermotolerance (Supplemental Figure 3B) and propagating all previously tested variants of [PSI+] and [RNQ+] (Figure 2C and data not shown). We found 4BAP behaved in all assays essentially identically to 444B.
4BAP-Trap monitors flux of prion proteins through the hexamer
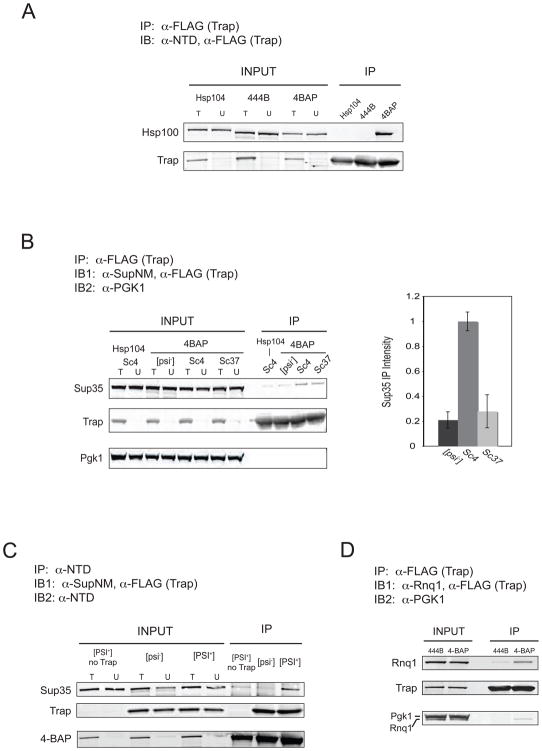
We confirmed 4BAP coupled to Trap in yeast by expressing an epitope-tagged Trap (TrapFLAG, referred to as Trap) together with 444B, HSP104 or 4BAP. We found by co-immunoprecipitation that Trap effectively coupled to 4BAP but not to 444B or Hsp104 (Figure 3A). Next, we looked at how much Sup35 was immunoprecipitated in 4BAP Trap yeast in [psi−], [PSI+]Sc4 and [PSI+]Sc37 yeast, comparing output signal with that of HSP104 Trap [PSI+]Sc4. Anti-FLAG resin co-immunoprecipited Sup35 preferentially in [PSI+] over [psi−] cells in the presence of 4BAP but not Hsp104 (Figure 3B). The capture of Sup35 was specific to 4BAP recognition, as the abundant, soluble protein Pgk1 was not detected in any eluant. This [PSI+]-specific translocation of Sup35 agrees well with a recent study using a similar ClpP-docking version of Hsp104 (termed HAP) (Tessarz et al., 2008). To address whether Sup35 bound to 4BAP independently of Trap, we used anti-Hsp104NTD coupled to Protein A/G agarose and immunoprecipitated 4BAP from 4BAP [PSI+], 4BAP Trap [psi−] and 4BAP Trap [PSI+] yeast. 4BAP was quantitatively immunoprecipitated from all yeast strains, but a clear Sup35 signal was seen only in a Trap- and [PSI+]-dependent fashion (Figure 3C).
Figure 3. 4BAP translocates prion proteins in an aggregate-dependent fashion.
(A) Co-immunoprecipitation of 4BAP but not Hsp104 or 444B with Trap. Strains expressing Trap under the control of the strong constitutive TDH3 promoter and expressing the indicated Hsp100 were grown to early stationary phase. Anti-FLAG (Trap-binding) resin was incubated with cleared extracts, washed and eluted with boiling SDS loading buffer. Total input (T), unbound extract (U) and 50X elution samples were separated by SDS-PAGE and probed with anti-Hsp104NTD and anti-FLAG (Trap-reactive). IP = immunoprecipitation, IB = immunoblot.
(B) Left: Representative blot from the co-immunoprecipitation of Sup35 with Trap in HSP104 Trap [PSI+]Sc4 and 4BAP Trap [psi−], [PSI+]Sc4 and [PSI+]Sc37 strains. Input and 50X elution samples were probed with anti-SupNM, anti-FLAG and anti-Pgk1. Right: Average elution signal intensities from three blots, one of which is shown on the left, for 4BAP Trap [psi−], [PSI+]Sc4 and [PSI+]Sc37 with HSP104 [PSI+]Sc4 background subtracted and normalized to 4BAP [PSI+]Sc4. Error: standard error of the mean.
(C) Co-immunoprecipitation of Sup35 with 4BAP in [PSI+] strains expressing Trap. Cleared yeast extracts were incubated with agarose coupled to anti-Hsp104NTD antibodies. Agarose was washed and eluted with boiling SDS loading buffer. Total input (T), unbound extract (U) and 50X elution samples were separated by SDS-PAGE and probed with anti-SupNM, anti-FLAG and anti-Hsp104NTD.
(D) Co-immunoprecipitation of Rnq1 with Trap in a 4BAP Trap [RNQ+] strain. IP was performed as in (B) and blot was probed with anti-Rnq1, anti-FLAG and anti-Pgk1.
We extended the use of 4BAP-Trap to explore action on different amyloid conformations of [PSI+] and on the prion [RNQ+]. Significantly, the amount of Sup35 captured by 4BAP Trap was typically 3-fold higher in [PSI+]Sc4 cells than in [PSI+]Sc37 cells (Figure 3B, right), a result that could explain the finding that [PSI+]Sc4 yeast contain more amyloid fibers than [PSI+]Sc37 (Tanaka et al., 2004; Tanaka et al., 2006). We also found Rnq1 protein was translocated into Trap in [RNQ+] (Figure 3C) but not in [rnq−] cells (data not shown). Our results using [PSI+] variants and [RNQ+] show that Hsp104 recognizes multiple yeast prions and that translocation efficiency differs with the conformation of the aggregate substrate.
Prion propagation and translocation is dependent on Sis1
Having shown prion-dependent translocation of Sup35 and Rnq1 through 4BAP, we turned to the question of whether the Hsp70/40 system was acting upstream of the Hsp100, as we suspected from the architecture of 444B. Previous work used a doxycycline-repressible allele of the Hsp40 gene SIS1 (Δsis1-TETrSIS1) to show Sis1 involvement in [RNQ+] aggregate size and maintenance (Aron et al., 2007). However, these experiments do not identify the point in the propagation cycle at which Sis1 acts. We sought to do this by using the repressible SIS1 in our 4BAP Trap yeast.
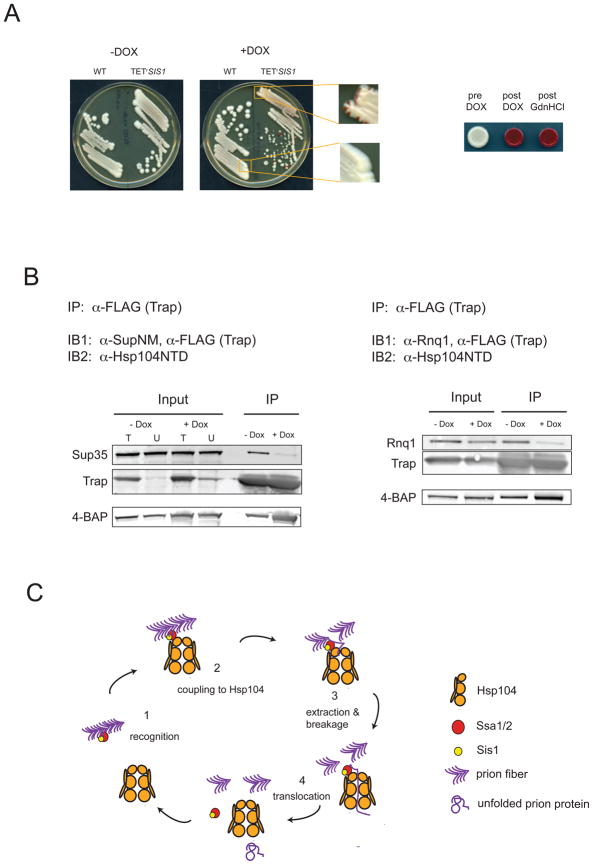
Our initial test of Sis1 involvement in [PSI+] maintenance was performed by streaking HSP104 Δsis1-TETrSIS1 [PSI+]Sc4 strains on doxycycline-containing media. We found that repression of SIS1 resulted in permanent loss of [PSI+], shown by the appearance of red sectoring yeast (Figure 4A, left and enlargements) which retained the [psi−] phenotype after SIS1 repression was alleviated (Figure 4A, right). Our finding that [PSI+] relies upon Sis1 makes it clear that [PSI+] is not simply affected by, but critically depends upon, a chaperone other than Hsp104 for its maintenance.
Figure 4. [PSI+] propagation and prion protein translocation depend upon the Hsp40 Sis1.
(A) [PSI+] was introduced into Δsis1-[TETrSIS1]. SIS1 wild-type and Δsis1-[TETrSIS1] sister spores were plated on rich media with or without doxycycline (left). Blowups of strains on doxycyline show red sectors indicative of [PSI+] loss in the Δsis1-[TETrSIS1] but not in the SIS1 strain. Right: Δsis1-[TETrSIS1] [PSI+] strains before and after passage on doxycyline or 3mM guanidine hydrochloride.
(B) Immunoprecipitation of Sup35 (left) and Rnq1 (right) from prion aggregates requires Sis1. Strains 4BAP Δsis1-[TETrSIS1] [PSI+] and 4BAP Δsis1-[TETrSIS1] [RNQ+] were transformed with a high-copy plasmid expressing Trap from the inducible GAL1 promoter. Overnight cultures were grown in dextrose-containing media, diluted into raffinose-containing media and grown to early log phase. Trap expression was induced by addition of galactose and cultures were split into flasks with or without doxycyline (Dox). Strains were harvested around OD600 = 1, and immunoprecipitations were performed as in Figure 3B and 3D.
(C) Model of Hsp104 cooperation with the Hsp70/40 system in propagating yeast prions. Prion aggregates (purple arrowheads) are recognized by Sis1 (yellow) and Ssa1 (red) (1) and are recruited to Hsp104 (2). A prion subunit is transferred to Hsp104 at the pore opening of the hexamer (3), and the ATP-dependent translocation activity extracts the monomer from the fiber, generating two new ends for prion growth (4).
To determine whether Sis1 was required for 4BAP translocation of prion proteins into Trap, we controlled the timing of substrate capture in 4BAP Δsis1-TETrSIS1 strains by introducing an inducible Trap on a high-copy plasmid. We found that Sup35 was specifically and efficiently captured in [PSI+] strains in the presence of Sis1 but that Sup35 signals sharply decreased (12-fold) with an 80% drop in Sis1 levels (Figure 4B, left, Supplemental Figure 4A). For the duration of SIS1 repression, [PSI+] was maintained (Supplemental Figure 4B), indicating the amyloid substrate was present but not recognized and processed by 4BAP. In a [RNQ+] 4BAP Δsis1-TETrSIS1 isolate, we similarly observed a 3-fold decrease in Rnq1 immunoprecipitated resulting from a 70% decrease in Sis1 levels (Figure 4B, right; Supplemental Figure 4C). Taken together, our results strongly suggest the natural yeast prion propagation cycle is an Hsp70/40-dependent process involving a Sis1-dependent, and likely Ssa1-dependent, delivery of prion substrates to Hsp104.
Discussion
Through the construction of the yeast-bacterial chimeras 444B and 4BAP, we developed a system for dissecting the pathway of yeast prion propagation in vivo, and we used this system to identify Sis1 as a critical pathway component upstream of Hsp104. We designed our studies of the yeast prion replication cycle based upon current knowledge of other Hsp104-mediated reactions and on successful strategies for monitoring substrates of AAA+ ATPases in the Hsp104 family. Mounting evidence indicates the initial step in the disaggregation of amorphous aggregates in bacteria is mediated by DnaK (Weibezahn et al., 2004; Zietkiewicz et al., 2004; Zietkiewicz et al., 2006), which interacts with the top ring of ClpB (Schlee et al., 2004) via the coiled-coil, which couples ClpB motor activity to DnaK activity (Haslberger et al., 2007), to hand off the substrate for translocation through the hexamer (Weibezahn et al., 2004). Whether this order of operations was also true for Hsp104 was not known (Bosl et al., 2006), nor was it known whether the Hsp70/40 system coupled to the exit of the hexamer to assist substrate refolding in either amorphous aggregate processing or in prion propagation. Our results indicate that the emerging model for the ClpB-mediated disaggregation pathway holds for Hsp104 as well, both on amorphous and ordered aggregates, and that these reactions are directed by their top rings, with AAA2 likely operating as a generic ATP motor. Furthermore, the generic contribution of AAA2 predicts no necessary downstream physical coupling to the Hsp70/40 system occurs during these activities. Successful replacement of Hsp104 AAA2 also suggests the Hsp90 system, found to couple to Hsp104 through its C-terminal TPR-binding motif (Abbas-Terki et al., 2001), plays no obligatory role in thermotolerance or prion propagation.
Recent studies showing residues critical for AAA+ translocation function are necessary for [PSI+] propagation (Hung and Masison, 2006) and Hsp104 translocation of prion aggregates as well as amorphous aggregates (Tessarz et al., 2008) emphasized the similarity of thermotolerance and prion propagation pathways. However, it remained unclear whether the prion propagation reaction was unassisted and therefore distinct from thermotolerance functions in substrate recognition and engagement. The question of whether Hsp104 is necessarily assisted by cochaperones in replicating prions in vivo is particularly relevant given the in vitro evidence of unassisted prion replication (Shorter and Lindquist, 2004; Shorter and Lindquist, 2006; Shorter and Lindquist, 2008). Our results show that Hsp104 relies upon co-chaperones for prion replication in vivo and emphasize the importance of considering the cellular milieu in understanding chaperone action on amyloids.
Together, our data suggests a model of prion propagation (Figure 4C) where prion aggregates (purple) are recognized by Ssa1/2 (red) and/or Sis1 (yellow), allowing their recruitment to the top ring of Hsp104 (gold). Although we believe it highly probable that Sis1 delivers prion substrates in conjunction with Ssa1, technical limitations stemming from the high degree of redundancy between the four Ssa homolgs made direct tests of this unfeasible. We propose the cycle likely proceeds by the transfer of one or more monomers to Hsp104, where complete or partial translocation would separate the engaged monomer from the fiber substrate, generating two new surfaces for fiber growth. The end of a completed translocation cycle would result in the release of an unfolded prion protein monomer, which might fold with or without assistance from chaperones, or might add back on to the end of a growing fiber. Our model of prion propagation is consistent with the recent finding that [PSI+] aggregates include large amounts of Ssa1 and trace amounts of Hsp104, Sis1 and the Ssa1 nucleotide exchange factor, Sse1(Bagriantsev et al., 2008).
A condition for being a prion is the ability to be coopted into aggregates even under nonstress conditions, a property that is conferred by the presence of natively unfolded, amyloidogenic regions (such as the glutamine/asparagines-rich domains of Sup35 and Rnq1). Once that condition is met, coupling with the endogenous chaperone machinery allows the aggregate to undergo a self-sustaining growth and division cycle. These conditions have been met by multiple proteins in yeast as well as by multiple, but not all, amyloid conformations of the same protein. Although it has been convincingly shown that the infective unit of [PSI+] is Sup35 amyloid (King and Diaz-Avalos, 2004; Tanaka et al., 2004), it has also been shown that not all Sup35 amyloids are heritable (Salnikova et al., 2005). Additionally, scrambling the primary sequence on the prion domain of Sup35 yields many heritable amyloids of different strengths (Ross et al., 2004). In our studies, the increase we consistently observed in the amount of Sup35 captured in the [PSI+]Sc4 variant compared with the [PSI+]Sc37 variant agrees well with both the physical characterization of the fibers of [PSI+]Sc4 as more fragile (Tanaka et al., 2006) and the structural data indicating potential chaperone binding sites in Sc4 fibers are less protected than in Sc37 fibers (Toyama et al., 2007).
Many questions remain about how amyloid aggregates are recognized by Hsp70s and Hsp40s and delivered to and processed by Hsp104. Understanding these processes will be crucial for determining what spectrum of aggregate sequences and structures allow amyloids to be prions. With the tools we develop here, we can begin to address these questions in vivo and in vitro by creating a system that represents the full propagation cycle of yeast prions.
Experimental Procedures
Strains and Plasmids
All yeast strains are derived from W303 and are described in the Supplement except for the parent strain of the TETrSIS1 strains, which was generously provided by E. Craig (Aron et al., 2007). Bacterial strains MC4100 wild-type and ΔclpB::kan were provided by B. Bukau and the E. coli Genetic Resource Center. Plasmids are listed in the Supplement along with details of their construction.
Yeast prion propagation
[PSI+] propagation was determined by replacing pKAT136 (pRS316 HSP104) in YKT12, 24 or 26 with pRS315 HSP104, ClpB, 444B, etc. under the control of the HSP104 promoter. [PSI] status was determined by color phenotype on low adenine media and growth on trace adenine or no adenine media. [RNQ+] propagation was determined by foci formation in strains transformed with 416CUP1pRNQ-GFP (gift of E. Craig) after a 4 hour 50uM CuSO4 induction or by [PSI+] conversion by 50uM CuSO4 induction of 426CUP1pSupNM.
Thermotolerance
Induced thermotolerance assays in S. cerevisiae were performed essentially as described (Sanchez and Lindquist, 1990) and induced thermotolerance assays in E. coli were performed as described (Weibezahn et al., 2004). Chaperone levels in S. cerevisiae and E. coli were confirmed by SDS-PAGE of soluble lysates followed by coomassie staining and/or immunoblotting. See supplement for details.
Luciferase Refolding
In S. cerevisiae, YKT52 (Δhsp104) transformed with p316GPDlux (gift of B. Bukau) and p315 carrying the indicated chaperone were grown to mid-log at 30°C, shifted to 37°C for 60min, shifted to 44°C for 60min with 20μg/mL cycloheximide added at 50min, and shifted back to 30°C for 120min. Luciferase activity was measured before heat shock and at the indicated time points by adding 50μL 1mM Beetle Luciferin (Promega) to 100mu;L cells and taking 10sec integrated luminescence signal in an EG&G Berthold Microlumat Plus luminometer.
Immunoprecipitations (IPs)
For the prion capture experiments in uninduced conditions, cells were grown to OD600 ~2.0 and harvested. For Sup35 induction experiments, cells were grown overnight in synthetic media lacking uracil (SD-Ura), backdiluted to OD600 = 0.15 in SD-Ura + 10uM CuSO4 and harvested at OD ~ 1–2. For Sis1 shutoff experiments, cells were grown in SD-Ura overnight, backdiluted to OD600 = 0.15 in 2% raffinose-containing media (SR-Ura) and grown to OD600 ~ 0.4. Cultures were supplemented with 2% galactose to induce Trap, split and half were treated with 5μg/mL Doxycyline. Cells were harvested at OD600 ~ 1.
For all IPs, cells were lysed at 4°C in IP buffer (see supplement). Lysates were cleared by centrifugation at 20,000g for 10min at 4°C and the supernatants were bound at 4°C to pre-equilibrated anti-FLAG beads (Sigma #F2426). Beads were washed at 4°C with 3× IP buffer, 1× IP buffer + 1M NaCl, 2× IP buffer and were eluted with with boiling 2× SDS-PAGE loading buffer. IPs using active ClpP-FLAG were performed as above and showed no evidence of 4BAP proteolysis (data not shown).
Western Blots
Immunoblotting was performed according to standard procedures. See supplement for details.
Supplementary Material
Acknowledgments
We would like to thank P. Tessarz, A. Mogk and D. Siegele for advice on E. coli strains; D. Agard, C. Gross and D. Mullins for helpful discussions; C. Chu for purification of the Hsp104NTD and experimental advice; J. Weibezahn, S. Collins, B. Toyama, M. Schuldiner, D. Cameron and members of the Weissman lab for critical reading of the manuscript; B. Toyama for help with graphics, S. Braun for equipment and advice and B. Bukau and E. Craig for reagents. This work was funded by the Howard Hughes Medical Institute (J.S.W.), the National Science Foundation Graduate Resarch Fellowship program (K.A.T. and K.J.V.), and the Hillblom Foundation (K.A.T.).
References
- Abbas-Terki T, Donze O, Briand PA, Picard D. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol Cell Biol. 2001;21:7569–7575. doi: 10.1128/MCB.21.22.7569-7575.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron R, Higurashi T, Sahi C, Craig EA. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. Embo J. 2007;26:3794–3803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] Infection is Transmitted by Sup35 Polymers within [PSI+] Aggregates with Heterogeneous Protein Composition. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosl B, Grimminger V, Walter S. The molecular chaperone Hsp104--a molecular machine for protein disaggregation. J Struct Biol. 2006;156:139–148. doi: 10.1016/j.jsb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Chernoff YO. Stress and prions: lessons from the yeast model. FEBS Lett. 2007;581:3695–3701. doi: 10.1016/j.febslet.2007.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Haslberger T, Weibezahn J, Zahn R, Lee S, Tsai FT, Bukau B, Mogk A. M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell. 2007;25:247–260. doi: 10.1016/j.molcel.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173:611–620. doi: 10.1534/genetics.106.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Taguchi H, Kishimoto A, Yoshida M. Hsp104 binds to yeast Sup35 prion fiber but needs other factor(s) to sever it. J Biol Chem. 2004;279:52319–52323. doi: 10.1074/jbc.M408159200. [DOI] [PubMed] [Google Scholar]
- Jones GW, Tuite MF. Chaperoning prions: the cellular machinery for propagating an infectious protein? Bioessays. 2005;27:823–832. doi: 10.1002/bies.20267. [DOI] [PubMed] [Google Scholar]
- King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- Krzewska J, Langer T, Liberek K. Mitochondrial Hsp78, a member of the Clp/Hsp100 family in Saccharomyces cerevisiae, cooperates with Hsp70 in protein refolding. FEBS Lett. 2001;489:92–96. doi: 10.1016/s0014-5793(00)02423-6. [DOI] [PubMed] [Google Scholar]
- Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, Tsai FT. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- Loovers HM, Guinan E, Jones GW. Importance of the Hsp70 ATPase domain in yeast prion propagation. Genetics. 2007;175:621–630. doi: 10.1534/genetics.106.066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luheshi LM, Crowther DC, Dobson CM. Protein misfolding and disease: from the test tube to the organism. Curr Opin Chem Biol. 2008;12:25–31. doi: 10.1016/j.cbpa.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Luke MM, Sutton A, Arndt KT. Characterization of SIS1, a Saccharomyces cerevisiae homologue of bacterial dnaJ proteins. J Cell Biol. 1991;114:623–638. doi: 10.1083/jcb.114.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Haslberger T, Tessarz P, Bukau B. Common and specific mechanisms of AAA+ proteins involved in protein quality control. Biochem Soc Trans. 2008;36:120–125. doi: 10.1042/BST0360120. [DOI] [PubMed] [Google Scholar]
- Motohashi K, Watanabe Y, Yohda M, Yoshida M. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc Natl Acad Sci U S A. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. Embo J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Baxa U, Wickner RB. Scrambled prion domains form prions and amyloid. Mol Cell Biol. 2004;24:7206–7213. doi: 10.1128/MCB.24.16.7206-7213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikova AB, Kryndushkin DS, Smirnov VN, Kushnirov VV, Ter-Avanesyan MD. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J Biol Chem. 2005;280:8808–8812. doi: 10.1074/jbc.M410150200. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- Schlee S, Beinker P, Akhrymuk A, Reinstein J. A chaperone network for the resolubilization of protein aggregates: direct interaction of ClpB and DnaK. J Mol Biol. 2004;336:275–285. doi: 10.1016/j.jmb.2003.12.013. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. Embo J. 2008 doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. Embo J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Wu YX, Jung G, Tutar Y, Eisenberg E, Greene LE, Masison DC. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot Cell. 2005;4:289–297. doi: 10.1128/EC.4.2.289-297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Zhou P, Derkatch IL, Liebman SW. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)] Mol Microbiol. 2001;39:37–46. doi: 10.1046/j.1365-2958.2001.02224.x. [DOI] [PubMed] [Google Scholar]
- Zietkiewicz S, Krzewska J, Liberek K. Successive and synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. J Biol Chem. 2004;279:44376–44383. doi: 10.1074/jbc.M402405200. [DOI] [PubMed] [Google Scholar]
- Zietkiewicz S, Lewandowska A, Stocki P, Liberek K. Hsp70 chaperone machine remodels protein aggregates at the initial step of Hsp70-Hsp100-dependent disaggregation. J Biol Chem. 2006;281:7022–7029. doi: 10.1074/jbc.M507893200. [DOI] [PubMed] [Google Scholar]
- Zolkiewski M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J Biol Chem. 1999;274:28083–28086. doi: 10.1074/jbc.274.40.28083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.