Abstract
Multiple Myeloma (MM) is characterized by multiple chromosomal aberrations. To assess the contribution of DNA repair to this phenotype, ionizing radiation was used to induce DNA double strand breaks in three MM cell lines. Clonogenic survival assays showed U266 (SF4= 15.3+6.4%) and RPMI 8226 (SF4=12.6.0+1.7%) were radiation sensitive while OPM2 was resistant (SF4=78.9+4.1%). Addition of the DNA-PK inhibitor NU7026 showed the expected suppression in radiation survival in OPM2 but increased survival in both radiation sensitive cell lines. To examine NHEJ repair in these lines, the ability of protein extracts to support in vitro DNA repair was measured. Among the three MM cell lines analyzed, RPMI 8226 demonstrated impaired blunt ended DNA ligation using a ligation mediated PCR technique. In a bacterial based functional assay to rejoin a DNA break within the β-galactosidase gene, RPMI 8226 demonstrated a four fold reduction in rejoining fidelity compared to U266, with OPM2 showing an intermediate capacity. Ionizing radiation induced a robust γ-H2AX response in OPM2 but only a modest increase in each radiation sensitive cell line perhaps related to the high level of γ-H2AX in freshly plated cells. Examination of γ-H2AX foci in RPMI 8226 cells confirmed data from Western blots where a significant number of foci were present in freshly plated untreated cells which diminished over 24 h of culture. Based on the clonogenic survival and functional repair assays, all three cell lines exhibited corrupt NHEJ repair. We conclude that suppression of aberrant NHEJ function using the DNA-PK inhibitor NU7026 may facilitate access of DNA ends to an intact homologous recombination repair pathway, paradoxically increasing survival after irradiation. These data provide insight into the deregulation of DNA repair at the site of DNA breaks in MM that may underpin the characteristic genomic instability of this disease.
Keywords: Multiple Myeloma, Non-homologous end joining, DNA double strand break, LM-PCR, γ-H2AX foci
1. Introduction
Malignant plasma cells in multiple myeloma (MM) often display aneuploidy and complex structural abnormalities associated with poor prognosis [1–4]. The immunoglobulin isotype switch region on chromosome 14q32 is frequently involved in translocations with several different partner chromosomes in MM [5,6]. The mechanism for this karyotypic instability is largely unknown. Recently, numeric and structural centrosomal abnormalities have been implicated in the development of karyotypic abnormalities in MM [7–10]. However abnormal DNA repair function provides an alternative explanation for aneuploidy and chromosomal rearrangements.
Under usual circumstances a DNA double strand break (DNA-DSB) sets into motion a cascade of events, collectively termed DNA damage response, DDR, which is initiated by the phosphorylation of the histone variant protein H2AX at serine 139, generating γ-H2AX which forms foci at the site of the DNA-DSB. H2AX is phosphorylated by the phosphatidylinositol-3-OH-kinase-like family of protein kinases, which includes the ataxia telangiectasia mutated (ATM) kinase as well as the DNA-protein kinase catalytic subunit. The γ-H2AX foci recruit the mediator of DNA damage checkpoint protein 1 (MDC1) which in turn interacts with several other down stream DDR mediators such as BRCA1, MRE11, RAD50 and Ku-70[11]. Once detected, DNA-DSB can be repaired by two distinct mechanisms, non-homologous end joining (NHEJ) which is predominant in the G1/S phases of the cell cycle and homologous recombination (HR) which is most active in the G2/M phases [12–14]. DNA-DSB with blunt ends as well as with 3' and 5' overhangs can be repaired by the NHEJ mechanism without requiring extensive inter-strand homology. NHEJ is also involved in the religation step of breaks introduced by the RAG endonucleases during VDJ rearrangement of the immunoglobulin heavy and light chain loci and during immunoglobulin isotype switching in B cells [15]. NHEJ is mediated by the recruitment of Ku70 and Ku80 to the DNA-DSB, which facilitates DNA-protein kinase catalytic subunit (DNA-PKcs) binding that in the presence of XRCC4/DNA-Ligase IV and Artemis, executes repair in an ATP dependent reaction [16,17]. HR, on the other hand, requires extensive inter-strand homology between the broken DNA double strand and the one serving as a repair template and is mediated by RAD51 and its paralogs. HR resolves DNA-DSBs sustained during meiosis and during DNA replication [18]. The two DNA-DSB repair pathways have been shown to compensate and cooperate in various systems in order to maintain genome integrity [19–22].
Marked genomic instability and aneuploidy is seen in NHEJ and HR core protein gene knockout mouse models [23–28]. These Ku80−/−p53−/−, XRCC4−/−p53−/−, Lig4−/−p53−/−, Ku70, RAD51, XRCC-2 deficient mice exhibit chromosomal translocations and enhanced radiation sensitivity. The NHEJ deficient mice also have abnormal lymphoid tissue development and an increased propensity to develop B and T cell lymphomas. We hypothesized that chromosomal rearrangement in MM is impacted by either inefficient or otherwise impaired DNA-DSB repair and this altered DNA-DSB repair capacity contributes to the overall aberrant karyotype observed in this disease. To explore this hypothesis we used ionizing radiation to introduce DNA-DSBs in a series of MM cell lines and analyzed their clonogenic response using a potent inhibitor of the NHEJ repair pathway, NU7026. Subsequently, the differences observed in cell survival were contrasted with a range of functional DNA repair assays measuring both intrinsic DNA rejoining and fidelity. From these results it is concluded that the MM cell lines studied are variably impaired in both DNA rejoining efficiency and fidelity. Further the defect in repair may relate to an inability to correctly assemble NHEJ repair proteins at sites of damaged DNA.
2. Materials and Methods
2.1. Cell lines
MM cell lines studied included RPMI 8226, U266 (American Type Culture Collection), and OPM2 (kind gift of Dr. Lionel Coignet, Loyola University Medical Center, Maywood, IL). The cell lines possess variable amounts of karyotypic instability that is characteristic of multiple myeloma (Table 1). These cell lines were maintained in RPMI-1640 with 10% fetal calf serum, L-glutamine, and antibiotics under standard cell culture conditions at 37°C. The DNA-PK mutant human glioblastoma cell line MO59J (NHEJ impaired) and DNA-PK wild-type cell line MO59K (NHEJ competent) were used as controls. These were cultured in DMEM:F12 (1:1) medium with 2.5 mM L-glutamine adjusted to contain 15mM HEPES, 0.5 mM sodium pyruvate, and 1.2 g/L sodium bicarbonate supplemented with 0.05 mM non-essential amino acids and 10% fetal bovine serum.
Table 1. Karyotypic analysis of cell lines used in this study.
Additions and deletions involved either whole chromosomes or parts of chromosomes. Source: HyperCLDB (http://www.biotech.ist.unige.it/cldb/indexes.html).
| Addition (whole chromosome) | Addition (partial chromosome) | Deletion (whole chromosome) | Deletion (partial chromosome) | Translocation | Derivative | Total | |
|---|---|---|---|---|---|---|---|
| OPM2 | 3 | 0 | 2 | 4 | 4 | 0 | 13 |
| RPMI 8226 | 7 | 6 | 9 | 9 | 4 | 2 | 37 |
| U266 | 0 | 8 | 4 | 0 | 3 | 0 | 15 |
2.2. Limiting Dilution Assay
Survival of all cells (± NU7026) after radiation exposure was determined by limiting dilution assay. At ~75% confluency, cells were counted, serially diluted, and plated using two cell densities per dose point into 96-well plates. Concentrations ranged from 256 to 0.5 cell/well, depending on the dose of irradiation used. Cells were treated with NU7026 [2-(morpholin-4-yl)-benzo[h]chomen-4-one] (Sigma-Aldrich, St. Louis, MI), a DNA-PK inhibitor, diluted in DMSO such that the final concentrations were 2.5 µM and 10 μM. The control group was treated with DMSO only. After two hours of drug treatment the plates were irradiated with 0, 2, 4, 6 and 8 Gy. Following irradiation, plates were placed in 37°C incubator for 14 days, and 50 µl of fresh media was added every 4 days. Growth was assessed in an “all-or-nothing” (positive or negative) manner using a phase contrast microscope. Wells were scored positive for colony formation if they contained one or more colonies of 50 or more cells. The surviving fraction was determined by the formula given below whereby plating efficiencies of control and treated cells were compared [29].
2.3. In vitro functional NHEJ assays using cellular protein extracts
Isolation of total cellular protein extracts from 1x107 cells utilized a modification of a previously published protocol [12]. Briefly, the cell pellet was suspended with hypotonic lysis buffer and centrifuged at 700 × g; the supernatant was removed. The pellet was resuspended with 2 volumes of hypotonic lysis buffer and incubated at −20°C for 20 min. Cells were lysed with a Dounce homogenizer, protease inhibitor cocktail was added with high salt buffer and the reaction mix was incubated on ice for 20 min. The extract was centrifuged at 42,000 rpm for 3 h at 4°C in a Beckman Ultracentrifuge (Beckman, Palo Alto, CA). The supernatant was dialyzed against buffer E [20 mM Tris.HCl, pH 8.0/0.1 M KOAc/20% (vol/vol) glycerol/0.5 mM EDTA/1 mM DTT] for 3 h at 4°C. Extracts were then snap-frozen in liquid nitrogen and stored at −80°C. Protein concentration was determined using the BioRad Protein Quantitation Assay (BioRad, Hercules, CA). Equal protein concentrations were used in all the experiments.
2.4. DNA Repair Assay: Ligation-mediated PCR (LM-PCR)
Five-microgram pCR-Script Amp vector was digested and linearized with 2 Units SrfI (Stratagene, La Jolla, CA). After heat inactivation, this linearized vector was purified sing a Qiaquick PCR Purification kit (Qiagen, Valencia, CA). A double-stranded linker was constructed by annealing 500 pmoles of linker-25 oligo (5′-GCG GTG ACC CGG GAG ATC TGA ATT C- 3′) with 500 pmoles linker-11 oligo (5′-GAA TTC AGA TC- 3′) in the presence of 1X T4 DNA Ligase Ligation buffer (Promega, Madison, WI) and H2O to a total volume of 5 µl. The reaction was incubated at 95°C for 5 min, then cooled to 70°C for 1 sec and then the temperature was lowered from 70°C to 25°C over 1 h. After 1 h incubation at 25°C the temperature was lowered to 4°C over 1 h. The reaction mix was stored at −20°C. The repair reaction was carried out for 2 h at 37°C using 100 ng linearized pCR-Script, 100 pmoles (1 µl) double-stranded linker, reaction buffer [Tris-HCl (pH 7.5), magnesium acetate, potassium acetate, ATP, DTT, BSA] and 10 µg protein extract with H2O to 10 µl. For samples treated with wortmannin, a PI3 kinase inhibitor, the reaction was pretreated with wortmannin to a final concentration of 25 µM and incubated at 37° C for 30 min. Linearized pCR-Script was added subsequent to pre-treatment and the reaction conducted as stated.
The capacity of cellular extracts to support ligation of the linker to the linearized vector was determined by PCR using linker and vector specific primers (5′-GGA GCC CCC GAT TTA GAG CTT GAC G-3′). PCR amplification followed a previous protocol [30]. Briefly, PCR amplification was carried out using 1X PCR buffer, 5.0 µl of the repair reaction, dNTPs, 0.5 µM linker 25 oligo, 0.5 µM rep1.1 oligo (5’-GGA GCC CCC GAT TTA GAG CTT GAC G-3’) and H2O to 25 µl. The reaction was incubated at 72°C for 3 min then 2.5U Taq Polymerase (MBI Fermentas, Hanover, MD) was added with a further incubation at 72°C for 5 min. The reaction mix was then denatured at 95°C for 4 min initially and then 30 cycles of denaturing at 95°C for 45 sec, annealing at 60°C for 60 sec, elongation at 72°C for 45 sec were performed, with a final 10 min elongation cycle at 72°C. The products were size-fractionated on a 2.0% agarose gel and visualized by ethidium bromide staining.
2.5. Repair Fidelity Assay
Cellular protein extracts were incubated with a pUC-18 plasmid Containing an Eco RI mediated single DNA-DSB within the β-galactosidase gene. Extracts that support NHEJ will result in the re-circularization of the pUC-18 plasmid with restoration of enzymatic activity [31]. For this assay cell extracts were prepared as detailed above. These protein extracts were then used to rejoin the single DNA-DSB placed within the pUC-18 plasmid. The repair reaction was similar to the one detailed above with the exception that linker oligo and PCR-Script vector were substituted with the linearized pUC-18 plasmid. T4 Ligase was used as a control. Successful repair would result in re-circularization of the plasmid with restoration of its enzymatic activity. DH5α bacteria (1x104 for each experiment) (Invitrogen-Life Technologies, Carlsbad, CA) were then transformed according to manufacturer’s protocol using the plasmids resulting from the repair reactions from each cell line protein extract, and were then cultured on X-Gal/Amp® plates. After an overnight incubation, the ratio of white (corrupt β-galactosidase gene) vs. blue (intact β-galactosidase gene) colonies was determined. Statistical analysis was performed by pooling data from all three samples for a given cell line, and the percentage of white colonies was compared between selected cell lines, two at a time, using a Chi-square test. To account for the large number of comparisons that were made we considered a P-value less than 0.01 (rather than 0.05) to be statistically significant.
2.6. Analysis γ-H2AX protein
All three MM cell lines (RPMI 8226, U266, and OPM2) were grown to ~70% confluency in T25 or T75 flasks and either left un-irradiated or exposed to 4Gy. For each treatment, cell samples were collected at 0, 2, and 24 h following IR. Cells were washed with ice-cold PBS before resuspension in RIPA Lysis Buffer [150mM Sodium Chloride, 1% Triton X-100, 1% deoxycholic acid-sodium salt, 0.1% SDS, 50mM Tris-HCl, pH 7.5, 2mM EDTA, 100 µl of 100 mM sodium orthovanadate, with protease inhibitors (Complete Mini, Roche)] at a ratio of 100 µl of RIPA Lysis Buffer for every 1x106 cells. Protein lysates were measured at 595nm following the Bio-Rad Protein Assay microtiter 96-well plate protocol. Proteins were run on a 14%-SDS-PAGE Gel for 60V for 25 min, and 120V for 1.5 h then transferred to a nitrocellulose membrane at constant 0.2A for 1 h. After transfer, the membrane was washed 2x for 5 min each in TBS-0.2% Tween, blocked in 5%BSA-TBS-0.2% Tween for 30 min, washed 2x for 5 min each in TBS-0.2% Tween, and then incubated overnight at 4°C with the primary antibody (anti-phospho-histone H2AX Ser 139, Millipore, Billerica, MA) at 1:8000 diluted in 5% BSA-TBS-0.2% Tween. Following incubation with the primary antibody, the membrane was washed 5x for 4 min each in TBS-0.2% Tween, and then incubated for 1 h at room temperature with the secondary antibody, Goat-Anti-Ms IgG +IgM (H+L) HRP (Thermo Sci, Waltham, MA) at 1:20,000 diluted in 5% BSA-TBS-0.2% Tween. After incubation with the secondary antibody, the membrane was washed 5x for 4 min each, and then developed with Thermo Sci SuperSignal West Dura Extended Duration substrate.
2.7. γ-H2AX Foci Determination
RPMI 8226 cells were irradiated with 0 and 4 Gy. Aliquots (200 µl) of the cell suspension were centrifuged onto glass slides at 2200 rpm for 6 min in a cytospin 0, 1, 4, and 24 h after irradiation. Cells were fixed in methanol for 20 min at −20°C, washed 3X in PBS, and then permeabilized with 0.2% Triton X-100 in PBS/1% BSA for 10 min at room temperature. Slides were washed in PBS/1% BSA for 5 min, and then blocked with PBS/3%BSA for 30 min at room temperature. Slides were washed again in PBS/1% BSA and then incubated for 1 h at 37°C with anti-phospho-histone H2AX Ser 139 antibody (1:500; Millipore, Billerica, MA) in PBS/1%BSA/0.5% Tween. Slides were washed thoroughly 3X with PBS/1% BSA, and then incubated with a fluorescent-conjugated secondary antibody (1:1000; Fluorescein (FITC)- conjugated AffiniPure Goat Anti-Mouse IgG, Jackson Immunoresearch, West Grove, PA) for 30 min at 37°C in PBS/1%BSA/0.5% Tween. After washing with PBS 4X, the slides were mounted with Ultracruz mounting medium (Santa Cruz Biotechnology, Santa Cruz, CA). Fluorescence was viewed with Leica DM R microscope (Leica Microsystems Inc., Deerfield, IL). Digital images were captured at random using Image-Pro Plus software, and cells with γ-H2AX foci formation were manually counted.
3. Results
3.1. Radiation Survival
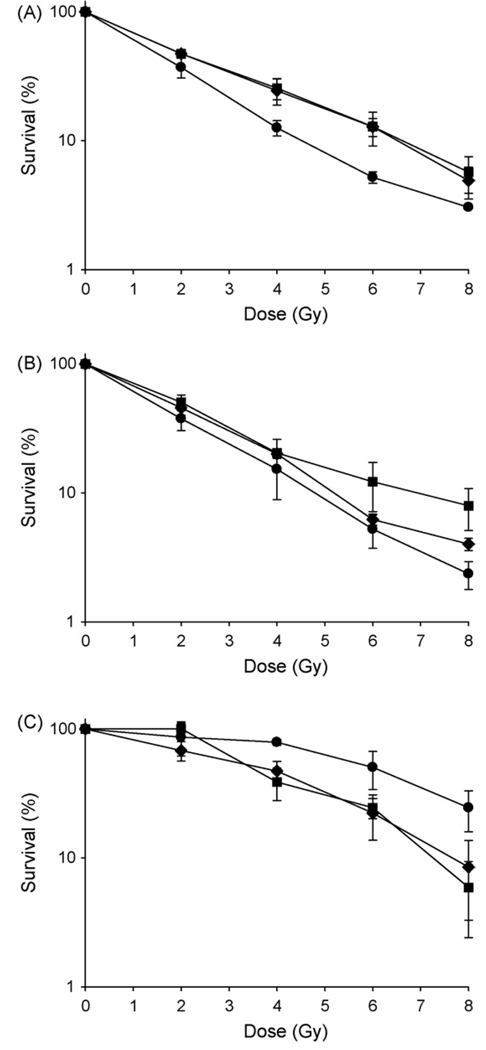
Ionizing radiation is a readily quantified method to introduce DNA-DSBs into living cells and thus gauge the efficiency of their repair systems. To determine the contribution of the NHEJ program to DNA-DSB repair within viable cells, all cell lines were exposed to graded doses of irradiation either with or without the DNA repair inhibitor, NU7026. This compound is a specific and potent inhibitor of DNA-PK and thus the NHEJ repair pathway [32]. Functional NHEJ repair that contributes to cell survival should be eradicated by incubation with the drug and thus decrease cell survival. The data obtained from all cell lines, in triplicate, shows that of the three cell lines used, OPM2 was the most radiation resistant (SF4=78.9±4.1%) while U266 (SF4= 15.3±6.4%) and RPMI 8226 (SF4= 12.6.0±1.7%) were radiation sensitive. In addition, only OPM2 cells when treated with NU7026 showed the predicted decrease in survival compared to those receiving irradiation alone (Fig. 1). In contrast, the additional two cell lines, both more radiation sensitive than the OPM2 cell line, showed the reverse effect in that the surviving fraction increased after treatment. When compared to cells treated with vehicle only, an enhancement ratio at 10% survival of between 0.65 – 0.7 was seen at the highest concentration of (10µM) of drug, indicating a protective, not enhancing effect of the NU7026. Further, treatment of cells with NU7026 alone without irradiation was toxic to the radiation sensitive cell lines but not the radiation resistant OPM2 cell line. A concentration of 10µM NU7026 killed 64% of RPMI 8226 and 55% of U266 when used as a single agent. OPM2 cells were unaffected at these concentrations of drug. These data suggest the two radiation sensitive cell lines are hypersensitive to endogenous DNA damage in the presence of NU7026.
Fig. 1. Effect of NU7026 on cell survival after irradiation.
Cells were pre-treated with 0µM (●), 2.5µM (♦) or 10µM (■) of NU7026 and then irradiated with 0, 2, 4, 6, 8 Gy. A) RPMI 8226, B) U266, C) OPM2. Only OPM2 cells showed the predicted decrease in survival after treatment with the DNA-PK inhibitor. Each point on the survival curve represents the surviving fraction ± SE from three separate experiments.
One explanation for the unexpected increase in radiation resistance may be that the DNA-PK inhibitor NU7026 impacts an already compromised NHEJ pathway by affecting the correct assembly of DNA-PK and Ku70/80 at the DNA ends. This may force these cell lines to use HR repair – potentially increasing cell survival [33]. The increased toxicity of RPMI 8226 and U266 in response to NU7026 in the absence of irradiation may reflect the lack of positive induction of HR repair that irradiation provides.
3.2. Qualitative functional NHEJ assay: Ligation-mediated PCR (LM-PCR)
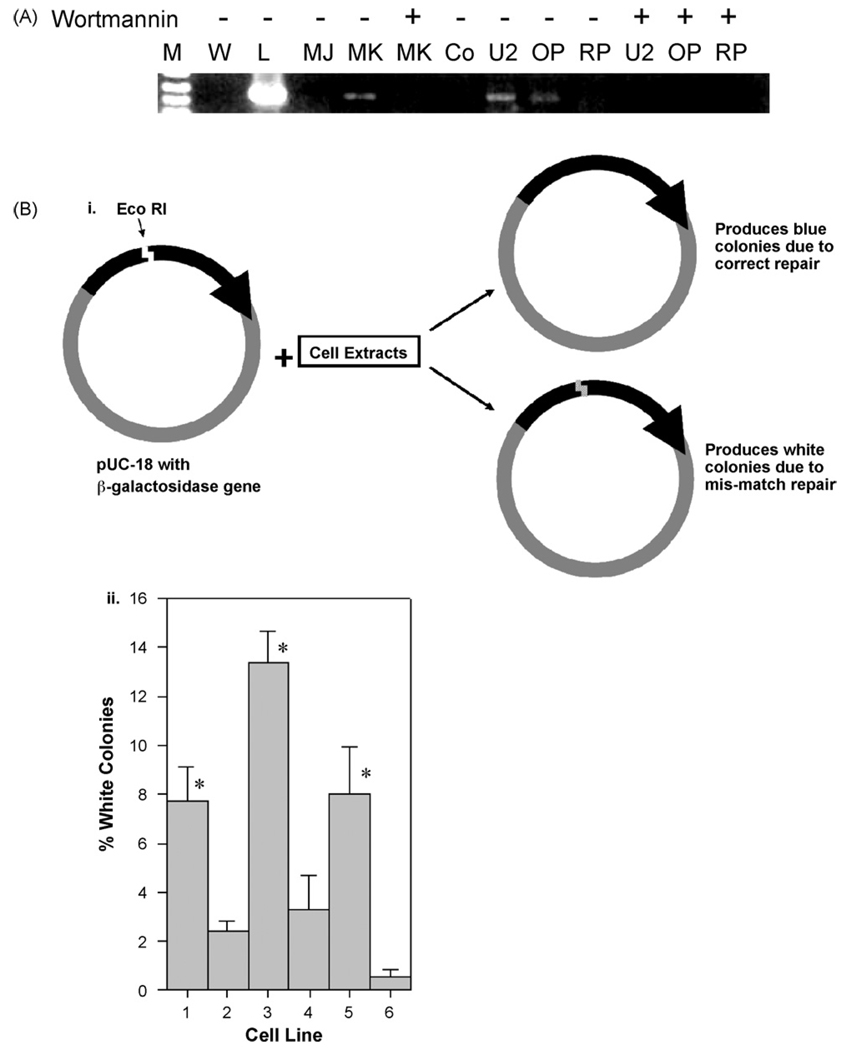
As the clonogenic survival data had implicated unexpected activity in the NHEJ repair pathway in two of the three cell lines studied, without any treatment, the capacity of these cells to engage in NHEJ repair was examined. In the first experiment a linearized plasmid was joined to a known oligonucleotide adapter in a ligation reaction using cell extracts prepared from the cell lines. Subsequently, a PCR reaction between the plasmid backbone and the ligated adapter will generate a PCR product of known size if successful ligation occurs. A unique 457 bp PCR product indicating adapter ligation was identified using extracts from the U266 and OPM2 cell line; however, no product was observed in RPMI 8226 (Fig. 2A). Additional cell lines derived from a common genetic background that were either deficient or competent in DNA repair demonstrated the predicted results: elimination of rejoining by MO59J (DNA-PK deficient) and efficient ligation by MO59K (DNA-PK proficient). Thus this pair of cell lines provides both a positive and negative control for NHEJ function. The PI-3 kinase inhibitor, wortmannin, abrogated DNA repair function in all the cell lines with an intact repair capacity supporting the notion that the DNA-DSB repair seen in this assay was mediated primarily by a DNA-PK facilitated mechanism [34]. These observations implied impairment of NHEJ function in RPMI 8226 with U266 and OPM2 showing activity comparable to that seen in the NHEJ competent MO59K cell line.
Fig. 2.
Fig. 2A. Plasmid based in-vitro rejoining assay to detect repair. Here LM-PCR is performed with cellular extracts from specific cells. W = water control, L = DNA ligase, MJ = MO59J, MK = MO59K, Co = Uncut vector, U2 = U266, O = OPM2, RP = RPMI 8226. DNA-PK deficient MO59J gives no product though DNA-PK proficient MO59K does. Only U226 and OPM2 gives product and not RPMI 8226, indicating deficit in RPMI 8226 rejoining. Wortmannin eliminates all evidence of rejoining confirming NHEJ pathway is being used. Representative reaction of 3 individual experiments is shown.
Fig. 2B. Plasmid in-vitro rejoining assay to determine repair fidelity. i) Schematic representation of the experimental design for plasmid in-vitro rejoining assay. ii) Percent of white colonies obtained from different cell lines. Here 1 = MO59J, 2 = MO59K, 3 = RPMI 8226, 4 = U266, 5 = OPM2, 6 = T4 ligase. Thus DNA-PK deficient MO59J, RPMI 8226 and OPM-2 exhibit significantly reduced repair fidelity compared to MO59K, U266 and T4 ligase groups.
3.3. Quantitative functional NHEJ assay: Plasmid repair assay
In order to derive a quantitative evaluation of DNA-DSB rejoining, the fidelity of such repair was measured by reconstitution of the β-galactosidase gene of pUC-18, which had been cut using Eco R1 (Fig. 2B). Successful religation of the cut site restored a fully functional β-galactosidase gene and product. DH5α bacteria were transformed with the plasmids resulting from the repair reactions, the bacteria were cultured overnight and the ratio of white (corrupt gene) vs. blue (intact gene) colonies was determined. Good colony growth was seen indicating successful transformation of the DH5α bacteria. In three separate experiments a mean of 135 colonies/experiment were obtained from control cell lines MO59K and MO59J; 83 from RPMI 8226; 53 from U266; 63 from OPM2 and 121 in the T4-Ligase control. Of these, RPMI 8226 and OPM2 along with the DNA-PK mutant cell line MO59J were found to have a statistically significantly higher number of white colonies, indicative of mis-repair, when compared with MO59K, T4-Ligase and U266 cell lines (Fig. 2B). In this experiment the U266 cell line showed repair consistent with the NHEJ competent cell line, MO59K, OPM2 showed reduced rejoining fidelity while RPMI 8226 was most deficient.
3.4. Expression of γ-H2AX
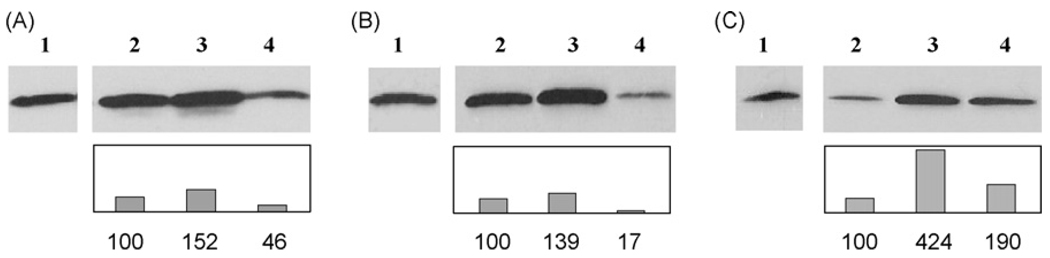
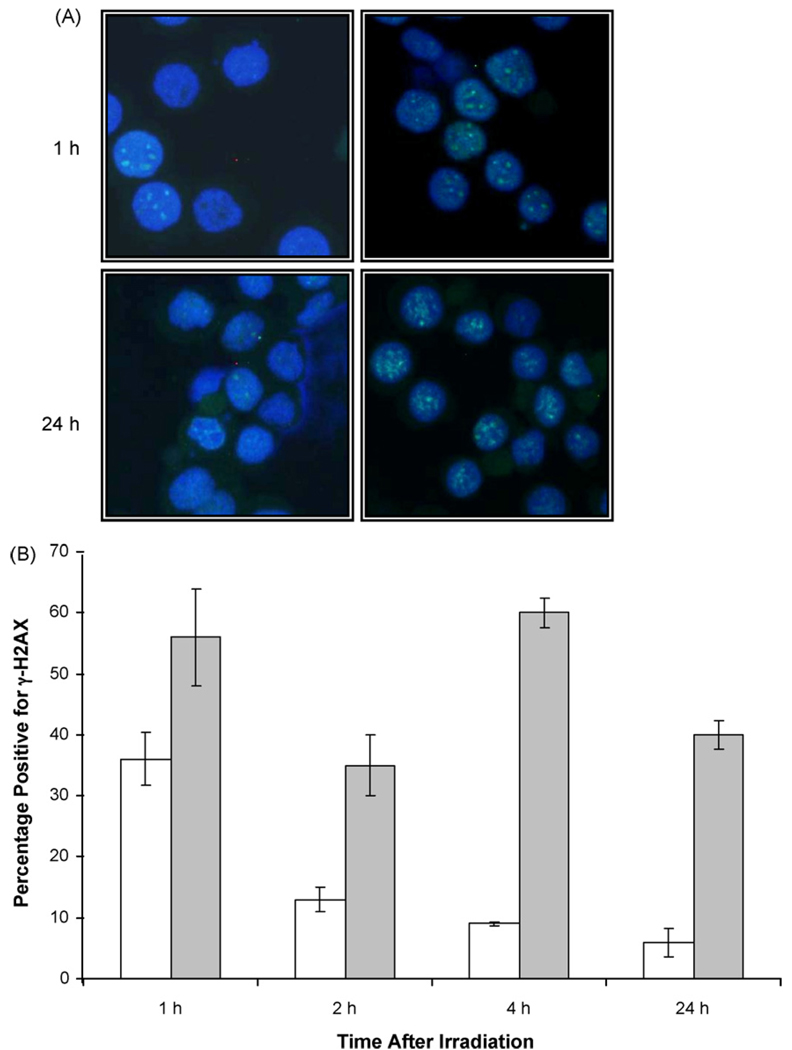
To address defects in NHEJ repair, in particular the presence of constitutive DNA damage, the presence of γ-H2AX protein was examined using a Western blot technique. H2AX becomes rapidly phosphorylated at Ser 139 in response to DNA breaks, the phosphorylation spreading in long tracts distal to the breaks and observable as discrete foci [35]. Here, all three cell lines were irradiated with 4 Gy and γ-H2AX content examined immediately after irradiation and 2 and 24 h later. As shown in Fig. 3, the two most radiation sensitive cell lines, U266 and RPMI 8226 showed a high basal level of γ-H2AX that showed a modest increase 2 h after irradiation, but decreased to below control levels by 24 h. The high basal levels of γ-H2AX are also seen in non-irradiated samples. In contrast, the most radiation resistant cell line, OPM2 exhibited the expected low endogenous level of γ-H2AX with a robust increase at 2 h followed by a decrease at 24 h. To confirm that the data obtained by Western blot reflected the presence of actual DNA-DSB, γ-H2AX foci were examined over the same time period in the RPMI 8226 cell line. The formation of γ-H2AX foci has been shown to be necessary for recruitment of other repair factors to the sites of DNA damage. As found by Western blot, RPMI 8226 showed a substantial baseline amount of γ-H2AX foci at 0 Gy, as compared to controls left in culture for an additional 24 h where the number of foci decreased. Despite a high initial baseline, persistence of the γ-H2AX foci formation following 4 Gy irradiation was observed even after 24 h following irradiation suggesting an impaired ability to resolve breaks (Fig. 4B). These data indicate that two of the three cell lines, RPMI 8226 and U266, exhibit temporary instability immediately after plating, as shown by enhanced γ-H2AX protein that is resolved over 24 h.
Fig. 3. Protein levels of γ-H2AX.
Representative western blot of γ-H2AX levels both before and after 4 Gy irradiation. A) RPMI 8226, B) U266, C) OPM2. Lane 1: No irradiation, Lane 2: 0 h, 4 Gy Lane 3: 2 h, 4 Gy, Lane 4: 24 h, 4 Gy. Histogram beneath each lane shows fold changes in γ-H2AX expression, determined by densitometry, for each data point compared to actin controls run with each experiment.
Fig. 4. Effect of radiation on γ-H2AX foci formation in RPMI 8226 cells.
A) Cells were irradiated with either 0 or 4 Gy and fixed after different time points for immunocytochemical analysis of γ-H2AX foci. A. Foci detected either 1 h or 24 h after either no treatment (left) or exposure to 4 Gy (right). B) Numerical analysis of foci numbers after either no treatment (white) or exposure to 4 Gy (gray). Data illustrates average of three separate experiments.
4. Discussion
Multiple Myeloma is a disease that is characterized by gross and diverse genomic instability involving multiple chromosomes (1–6). To investigate repair pathway utilization in the three MM cell lines under study, we used the DNA-PK specific inhibitor, NU7026, to completely suppress the NHEJ pathway using a clonogenic cell survival as readout [32]. Our hypothesis was that if the NHEJ pathway was functioning, even if at a low level, inhibition of the key component DNA-PK should lower cell survival after irradiation, as such survival is functionally linked to efficient resolution of DNA-DSBs. In contrast, the results of triplicate experiments were a uniform increase in cell survival after application of the inhibitor in two of the three cell lines (RPMI 8226 and U266) with an enhancement ratio at 10% survival of between 0.65 – 0.7 at the highest concentrations of NU7026 used. The most radiation resistant cell line, OPM2, showed the predicted reduction in survival after both concentrations of drug. Others have shown very similar clonogenic survival curves after irradiation for the two sensitive cell lines used, indicating no gross differences in their intrinsic radiation response in this study [36].
One interpretation of these data is the possibility that NHEJ repair is corrupt in the two radiation sensitive cell lines, an observation potentially underpinning the genomic instability of MM disease in general. In this case, suppression of DNA-PK catalytic function by drug may further suppress an already impaired NHEJ repair system, allowing better or more efficient access of HR repair components to the sites of DNA repair. Support for the hypothesis of preexisting defect(s) within the NHEJ pathway is observed by the toxicity of NU7026 alone in the two radiation sensitive cell lines but not in the resistant OPM2 system. In this setting, decreased survival is observed as untreated cells lack the stimulation for HR provided by irradiation. A key role for DNA-PK in regulating DNA repair pathway access has been observed before in other systems where chemical inhibition of DNA-PK permits unproductive occupation of DNA break sites by it and/or additional NHEJ components, restricting access to HR components thus increasing toxicity [22]. However, these studies utilized cell lines competent for the NHEJ pathway and it is possible with an already dysfunctional NHEJ repair pathway, suppression of DNA-PK offers a net increase in survival due to a more effective utilization of the HR repair pathway.
Clearly, the cell lines studied here do differ in their capacity to execute NHEJ and that difference may be related to the anomalous behavior of the DNA-PK component. In an attempt to dissect the effectiveness of the NHEJ repair pathways in these cell lines, a range of biochemical endpoints were studied. This approach may also address the karyotypic chaos observed in MM potentially linked to DNA repair abnormalities; specifically NHEJ deficits, given its central role in B cell ontogeny [37]. We first evaluated the capacity of cellular protein extracts to ligate double stranded DNA oligonucleotides in an in vitro system. Of the three MM cell lines studied only RPMI 8226 was incapable of rejoining DNA fragments in vitro. The other two cell lines showed a rejoining capacity at least as good as that of MO59K cells, a DNA-PK proficient cell line. To test rejoining fidelity, a plasmid rejoining assay was employed. Here the β-galactosidase gene is cleaved within a plasmid and the ability to exactly rejoin the break and transcribe the gene tested by its productive expression (blue color) in a bacterial system. In this system also, the RPMI 8226 and OPM2 cell lines exhibited a reduced capacity to accurately repair such breaks, equivalent to the DNA-PK deficient MO59J cell line. Thus using in vitro rejoining assays both the efficiency and fidelity of DNA-DSB repair is suppressed in RPMI 8226 though defects are also observed in the OPM2 cell line. In the assays used here the NHEJ pathway is the most likely repair pathway involved, as demonstrated in the LM-PCR reaction where all rejoining is lost when the broadspectrum PI-3 kinase inhibitor wortmannin is used. NHEJ is the dominant DNA repair pathway in mammalian cells except in the G2/M phases of the cell cycle or in meiosis [13]. These functional studies suggest, in the RPMI 8226 cell line in particular, an impairment of the capacity to repair DNA-DSB via the NHEJ pathway in response to clastogenic insults, such as radiation.
To address the status of DNA-DSB repair in these cell lines the presence of γ-H2AX was studied after irradiation by Western blot. Only the more radiation resistant cell line OPM2 showed a substantial increase in γ-H2AX protein after irradiation and subsequent return to normal levels. Both RPMI 8226 and U266 cell lines showed a higher initial level of protein than OPM2. In order to address in more detail the DNA repair defects in the most affected cell line (RPMI 8226), the ability to resolve γ-H2AX foci was studied. Such foci are a widely accepted measure of DNA-DSBs. Here, exposure to 4 Gy induced substantial levels (50–60%) of foci positive cells that were minimally resolved over the 24 h of the study. Also, without treatment other than plating, the most radiation sensitive cell line, RPMI 8226, exhibited a substantial number of foci that were gradually resolved over the 24 h period of cell culture. This suggests that these cells are sensitive to the stress of initial plating in culture that diminished over time. In this context, Amrein et al. have shown that inhibition of DNA-PK phosphorylation leads to accumulation of γ-H2AX foci for extended periods of time, indicating that disruption to the assembly of the catalytic complex leaves DNA breaks unresolved [38]. The persistence of γ-H2AX foci, 24 h following radiation, clearly indicates defects in repair, or its regulation possibly involving disruption to the NHEJ pathway.
This observed alteration of the DNA-DSB repair pathways in myeloma cell lines may have implications for the generation of chromosomal translocations in MM in response to clastogenic stimuli. In eukaryotes, NHEJ predominates over HR for repairing DNA-DSB in the G1/S phase of the cell cycle, while HR is active in the later S/G2/M phases when a sister chromatid is available to provide a template for repair [13]. Impairment of NHEJ, such as through XRCC4 mutation or by use of drugs such as NU7026 alters DNA-DSB repair pathways favoring HR in the CHO cell line [20]. Ku70−/− mouse embryonic stem cell lines have also been shown to have a 5–6 fold increase in homology directed repair [21]. Additionally, Adachi et al. demonstrated that HR can partially substitute for the loss of NHEJ in Ku70-deficient chicken B-lymphocyte cell line [19]. Reciprocally, DNA-PKcs complementation in the DNA-PK deficient CHO-V3 cell line suppresses HR [39]. Interestingly, using the ISce-I endonuclease DSB repair system, cooperation between NHEJ and HR has been demonstrated, such that repair is initiated by a gene conversion event where one strand of the disrupted double chromosome invades an area of homology (such as a DNA repeat sequence) on a heterologous chromosome and is rejoined to its partner by NHEJ after replicative repair [40]. In such a system one may postulate that lack of functional NHEJ may result in the development of chromosomal translocations.
A large number of murine studies have demonstrated the critical role of NHEJ core proteins as well as HR mediators in maintaining genomic integrity, one or more of which may underpin the defect(s) in NHEJ repair observed in the RPMI 8226 cell line. For example the NHEJ core protein/P53 double knockout mice developed by Alt et al. develop lymphomas with IgH/myc translocations and aneuploidy, and rapidly succumb to lymphoproliferative disorders [26]. Not surprisingly, lymphoproliferative disorders in general and MM in particular are associated with translocations. In particular aberrations involving chromosome 14q32, the IgH locus, have been shown with several different partners in MM (bcl-1, FGF-R3/MMSET, c-maf) [5]. In MM, these abnormalities involve the immunoglobulin isotype switch recombination loci [6], and interest has focused on DNA repair proteins. Polymorphisms in NHEJ protein DNA Ligase 4 (LIG4) have been shown to impact the likelihood of the development of MM and alanine-3-valine & threonine-9-iso-leucine polymorphisms in LIG4 being associated with a reduction in risk of developing MM [41]. Intriguingly, the gene for LIG4 is located telomeric to the D13S319 marker on chromosome 13q14, the most frequently deleted marker in patients with –13q MM [42]. The other protein of the NHEJ pathway that has been implicated in the development of MM is a variant of Ku86 (Ku86v). It was proposed by Tai et al. that Ku86v had impaired ability to form complexes with DNA-PKcs making patients more susceptible to DNA damage from radiation and chemotherapy [43]. This theory of Ku86v involvement was later nullified by a study conducted by Kato et al. in which they did not find the variant of the Ku86 in either 16 MM cell lines or six marrow samples of the MM patients [44]. They argued that the Ku86v found in the previous study was the result of an artifact of protein preparation. More recently this issue has re-emerged with a study indicating that the presence of Ku86v is of biological relevance[45]. Despite these contradictory studies it has been shown that the presence of Ku70/Ku86 complex is important for the NHEJ pathway activity following induction of DNA-DSBs in myeloid leukemia cells [46]. There is thus some evidence of protein alterations within the NHEJ pathway influencing myeloma biology.
Of the three MM cell lines studied here, both U266 and RPMI 8226, exhibited defects in NHEJ repair after suppression of DNA-PK without any applied stimuli. This suggests the NHEJ pathway is grossly dysfunctional in these cell lines. In addition, RPMI 8226 was deficient in all NHEJ-linked assays of DNA-DSB repair. The anomalous increase in radiation survival after suppression of DNA-PK by NU7026 further implies that suppression of NHEJ activity functionally enhances alternative repair pathways such as HR. It is proposed that a competitive imbalance between the two major DNA-DSB repair pathways, at the site of DNA-DSBs, may contribute to the karyotypic instability observed in MM [47]. Specifically, the most repair deficient cell line, RPMI 8226, is the most karyotypically unstable linking the observations made here to the phenotype of an individual’s myeloma disease (Table 1). NHEJ deficits may potentially be relevant in terms of myeloma therapy since NHEJ is involved in the repair of both radiation and chemotherapy induced DNA-DSBs, and intrinsic defects in DNA break processing may contribute to a more proliferative and resistant tumor phenotype emerging at the time of disease recurrence.
Acknowledgements
We would like to acknowledge Steven Creech MS for the statistical analysis of the repair fidelity assay.
(Work supported by Sharon Newman Research Award, International Myeloma Foundation and the Veterans Administration).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None.
Contributor Information
Clara Yang, Department of Radiation Oncology, University of California, Davis, California.
Amir A. Toor, VCU Health Systems, Richmond, VA
Sheetal Singh, Department of Radiation Oncology, University of California, Davis, California
Christopher Betti, Department of Radiation Oncology, Stritch School of Medicine, Loyola University Medical Center, Maywood, Illinois.
Andrew Vaughan, Department of Radiation Oncology, University of California, Davis, California
References
- 1.Calasanz MJ, Cigudosa JC, Odero MD, Ferreira C, Ardanaz MT, Fraile A, Carrasco JL, Sole F, Cuesta B, Gullon A. Cytogenetic analysis of 280 patients with multiple myeloma and related disorders: primary breakpoints and clinical correlations. Genes Chromosomes Cancer. 1997;18:84–93. [PubMed] [Google Scholar]
- 2.Rajkumar S, Fonseca R, Lacy M, Witzig T, Lust J, Greipp P, Therneau T, Kyle R, Litzow M, Gertz M. Abnormal cytogenetics predict poor survival after high-dose therapy and autologous blood cell transplantation in multiple myeloma. Bone Marrow Transplant. 1999;24:497–503. doi: 10.1038/sj.bmt.1701943. [DOI] [PubMed] [Google Scholar]
- 3.San Miguel JF, Garcia-Sanz R, Gonzalez M, Orfao A. DNA cell content studies in multiple myeloma. Leuk. Lymphoma. 1996;23:33–41. doi: 10.3109/10428199609054799. [DOI] [PubMed] [Google Scholar]
- 4.Smadja NV, Bastard C, Brigaudeau C, Leroux D, Fruchart C. Hypodiploidy is a major prognostic factor in multiple myeloma. Blood. 2001;98:2229–2238. doi: 10.1182/blood.v98.7.2229. [DOI] [PubMed] [Google Scholar]
- 5.Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001;20:5611–5622. doi: 10.1038/sj.onc.1204641. [DOI] [PubMed] [Google Scholar]
- 6.Ho PJ, Brown RD, Pelka GJ, Basten A, Gibson J, Joshua DE. Illegitimate switch recombinations are present in approximately half of primary myeloma tumors, but do not relate to known prognostic indicators or survival. Blood. 2001;97:490–495. doi: 10.1182/blood.v97.2.490. [DOI] [PubMed] [Google Scholar]
- 7.Ahmann GJ, Henderson KJ, Salisbury JL, Greipp PR, Fonseca R. Centrosomal expression and amplification in bone marrow plasma cells from healthy individuals and patients with plasma proliferative disorders. Blood. 2003;102:2522a. [Google Scholar]
- 8.Crainie M, Belch AR, Mant MJ, Pilarski LM. Overexpression of the receptor for hyaluronan-mediated motility (RHAMM) characterizes the malignant clone in multiple myeloma: identification of three distinct RHAMM variants. Blood. 1999;93:1684–1696. [PubMed] [Google Scholar]
- 9.Maxwell CA, Reiman T, Ye M, Belch AR, Pilarski LM. RHAMM overexpression: a potential mechanism to generate extensive centrosomal abnormalities in multiple myeloma. Blood. 2003;102:102a. [Google Scholar]
- 10.Neben K, Liebisch P, Dohner H, Lichter P, Kramer A. High frequency of centrosome aberrations in advanced-stage multiple myeloma. Blood. 2003;102:2520a. [Google Scholar]
- 11.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumann P, West SC. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soulas-Sprauel P, Rivera-Munoz P, Malivert L, Le Guyader G, Abramowski V, Revy P, de Villartay JP. V(D)J and immunoglobulin class switch recombinations: a paradigm to study the regulation of DNA end-joining. Oncogene. 2007;26:7780–7791. doi: 10.1038/sj.onc.1210875. [DOI] [PubMed] [Google Scholar]
- 16.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber MR. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol. Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 17.Rooney S, Alt FW, Lombard D, Whitlow S, Eckersdorff M, Fleming J, Fugmann S, Ferguson DO, Schatz DG, Sekiguchi J. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 2003;197:553–565. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adachi N, Ishino T, Ishii Y, Takeda S, Koyama H. DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku70: implications for DNA double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12109–12113. doi: 10.1073/pnas.201271098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delacote F, Han M, Stamato TD, Jasin M, Lopez BS. An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002;30:3454–3463. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 23.Deans B, Griffin CS, O’Regan P, Jasin M, Thacker J. Homologous recombination deficiency leads to profound genetic instability in cells derived from Xrcc2-knockout mice. Cancer Res. 2003;63:8181–8187. [PubMed] [Google Scholar]
- 24.Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferguson DO, Sekiguchi JM, Chang S, Frank KM, Gao Y, DePinho RA, Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 27.Li GC, Ouyang H, Li X, Nagasawa H, Little JB, Chen DJ, Ling CC, Fuks Z, Cordon-Cardo C. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol. Cell. 1998;2:1–8. doi: 10.1016/s1097-2765(00)80108-2. [DOI] [PubMed] [Google Scholar]
- 28.Lim DS, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pekkola-Heino K, Kulmala J, Klemi P, Lakkala T, Aitasalo K, Joensuu H, Grenman R. Effects of radiation fractionation on four squamous cell carcinoma lines with dissimilar inherent radiation sensitivity. J. Cancer Res. Clin. Oncol. 1991;117:597–602. doi: 10.1007/BF01613295. [DOI] [PubMed] [Google Scholar]
- 30.Betti CJ, Villalobos MJ, Diaz MO, Vaughan AT. Apoptotic stimuli initiate MLL-AF9 translocations that are transcribed in cells capable of division. Cancer Res. 2003;63:1377–1381. [PubMed] [Google Scholar]
- 31.Powell SN, McMillan TJ. The repair fidelity of restriction enzyme-induced double strand breaks in plasmid DNA correlates with radioresistance in human tumor cell lines. Int. J. Radiat. Oncol. Biol. Phys. 1994;29:1035–1040. doi: 10.1016/0360-3016(94)90399-9. [DOI] [PubMed] [Google Scholar]
- 32.Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008–6015. [PubMed] [Google Scholar]
- 33.Deriano L, Guipaud O, Merle-Beral H, Binet JL, Ricoul M, PotocKi-Veronese G, Favaudon V, Maciorowski Z, Muller C, Salles B, Sabatier L, Delic J. Human chronic lymphocytic leukemia B cells can escape DNA damage-induced apoptosis through the nonhomologous end-joining DNA repair pathway. Blood. 2005;105:4776–4783. doi: 10.1182/blood-2004-07-2888. [DOI] [PubMed] [Google Scholar]
- 34.Boulton S, Kyle S, Yalcintepe L, Durkacz BW. Wortmannin is a potent inhibitor of DNA double strand break but not single strand break repair in Chinese hamster ovary cells. Carcinogenesis. 1996;17:2285–2290. doi: 10.1093/carcin/17.11.2285. [DOI] [PubMed] [Google Scholar]
- 35.Pilch DR, Sedelnikova OA, Redon C, Celeste A, Nussenzweig A, Bonner WM. Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem. Cell Biol. 2003;81:123–129. doi: 10.1139/o03-042. [DOI] [PubMed] [Google Scholar]
- 36.Supiot S, Thillays F, Rio E, Gouard S, Morgenstern A, Bruchertseifer F, Mahe MA, Chatal JF, Davodeau F, Cherel M. Gemcitabine radiosensitizes multiple myeloma cells to low let, but not high let, irradiation. Radiother. Oncol. 2007;83:97–101. doi: 10.1016/j.radonc.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst.) 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 38.Amrein L, Loignon M, Goulet AC, Dunn M, Jean-Claude B, Aloyz R, Panasci L. Chlorambucil cytotoxicity in malignant B lymphocytes is synergistically increased by 2-(morpholin-4-yl)-benzo[h]chomen-4-one (NU7026)-mediated inhibition of DNA double-strand break repair via inhibition of DNA-dependent protein kinase. J. Pharmacol. Exp. Ther. 2007;321:848–855. doi: 10.1124/jpet.106.118356. [DOI] [PubMed] [Google Scholar]
- 39.Allen C, Kurimasa A, Brenneman MA, Chen DJ, Nickoloff JA. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3758–3763. doi: 10.1073/pnas.052545899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 41.Roddam PL, Rollinson S, O’Driscoll M, Jeggo PA, Jack A, Morgan GJ. Genetic variants of NHEJ DNA ligase IV can affect the risk of developing multiple myeloma, a tumour characterised by aberrant class switch recombination. J. Med. Genet. 2002;39:900–905. doi: 10.1136/jmg.39.12.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang H, Bouman D, Boerkoel CF, Stewart AK, Squire JA. Frequent monoallelic loss of D13S319 in multiple myeloma patients shown by interphase fluorescence in situ hybridization. Leukemia. 1999;13:105–109. doi: 10.1038/sj.leu.2401208. [DOI] [PubMed] [Google Scholar]
- 43.Tai YT, Teoh G, Lin B, Davies FE, Chauhan D, Treon SP, Raje N, Hideshima T, Shima Y, Podar K, Anderson KC. Ku86 variant expression and function in multiple myeloma cells is associated with increased sensitivity to DNA damage. J. Immunol. 2000;165:6347–6355. doi: 10.4049/jimmunol.165.11.6347. [DOI] [PubMed] [Google Scholar]
- 44.Kato M, Iida S, Komatsu H, Ueda R. Lack of Ku80 alteration in multiple myeloma. Jpn. J. Cancer Res. 2002;93:359–362. doi: 10.1111/j.1349-7006.2002.tb01264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gullo CA, Ge F, Cow G, Teoh G. Ku86 exists as both a full-length and a protease-sensitive natural variant in multiple myeloma cells. Cancer Cell Int. 2008;8:4. doi: 10.1186/1475-2867-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brady N, Gaymes TJ, Cheung M, Mufti GJ, Rassool FV. Increased error-prone NHEJ activity in myeloid leukemias is associated with DNA damage at sites that recruit key nonhomologous end-joining proteins. Cancer Res. 2003;63:1798–1805. [PubMed] [Google Scholar]
- 47.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]