Abstract
Mammalian sirtuin 1 (SIRT1) may control fatty acid homeostasis in liver. However, this possibility and underlying mechanism remain to be established. In this study, we addressed the issues by examining the metabolic phenotypes of SIRT1 heterozygous knockout (SIRT1+/−) mice. The study was conducted in the mice on three different diets including a low-fat diet (5% fat wt/wt), mediate-fat diet (11% fat wt/wt), and high-fat diet (HFD, 36% fat wt/wt). On low-fat diet, the mice did not exhibit any abnormality. On mediate-fat diet, the mice exhibited a significant increase in hepatic steatosis with elevated liver/body ratio, liver size, liver lipid (triglyceride, glycerol, and cholesterol) content, and liver inflammation. The hepatic steatosis was deteriorated in the mice by HFD. In the liver, lipogenesis was increased, fat export was reduced, and β-oxidation was not significantly changed. Body weight and fat content were increased in response to the dietary fat. Fat was mainly increased in sc adipose tissue and liver. Inflammation was also elevated in epididymal fat. Whole body energy expenditure and substrate utilization were reduced. Food intake, locomotor activity, and fat absorption were not changed. These data suggest that a reduction in the SIRT1 activity increases the risk of fatty liver in response to dietary fat. The liver steatosis may be a result of increased lipogenesis and reduced liver fat export. The inflammation may contribute to the pathogenesis of hepatic steatosis as well. A reduction in lipid mobilization may contribute to the hepatic steatosis and low energy expenditure.
In response to SIRT1 deficiency, an increase in PPARγ and NF-κB activities may play a critical role in the molecular mechanisms of energy metabolism and hepatic steatosis in the SIRT1+/− mice.
Mammalian sirtuin 1 (SIRT1), the ortholog of yeast gene Sir2 (silent information regulator 2), is a class III histone deacetylase (HDAC) whose activation is dependent on nicotinamide adenine dinucleotide in the nucleus (1,2). Genetic studies suggest that an increase in the SIRT1 activity promotes longevity in organisms ranging from yeast to worms (3,4,5). SIRT1 may act by regulation of energy metabolism (6). In response to nutritional and hormonal signals, SIRT1 inhibits fatty acid synthesis/storage (7), stimulates fatty acid β-oxidation (8), and induces gluconeogenesis (9). Induction of SIRT1 activity by gene overexpression or by the chemical activator resveratrol leads to energy expenditure and prevention of obesity in mice (8,10,11). SIRT1 activation by resveratrol also prevented liver steatosis in mice on high-fat diet (HFD) (11) or treated with alcohol (12). These studies suggest that SIRT1 has two separate activities in the regulation of fatty acid metabolism in liver: inhibiting fat storage and stimulating fatty acid oxidation. However, it is not clear which is the primary activity of SIRT1. We hypothesize that inhibition of fatty acid storage is the primary activity of SIRT1. Fatty acid oxidation may be secondary to the inhibition of fatty acid storage that leads to increased supply of fatty acid to mitochondria.
Several lines of SIRT1 transgenic (Tg) mice have been reported. These include both gain-of-function and loss-of-function mice. Global overexpression of SIRT1 by cDNA knock-in into the β-actin locus generates some phenotypes in mice similar to those on calorie restriction (10). The knock-in mice are leaner, more metabolically active, and more sensitive to insulin. Overexpression of SIRT1 from a bacterial artificial chromosome leads to a complex phenotype with an increase in energy efficiency and protection against HFD-induced insulin resistance (13). Complete deletion of the SIRT1 gene leads to developmental defects and postnatal lethality (14,15). The SIRT1-null (SIRT1−/−) mice die shortly after birth on the 129/J inbred background; however, on an outbreed gene background, some of them can survive to adulthood. The surviving SIRT1-null (SIRT1−/−) mice look normal but are small, sterile, and have craniofacial abnormalities (14). Thus, SIRT1−/− mice are not appropriate for the study of energy metabolism. The SIRT1 heterozygous mice (SIRT1+/−) were normal in development and reproductivity (14,15). In these models of SIRT1 Tg mice, liver steatosis was not investigated. It is not clear if the reduction of SIRT1 activity by genetic modification alters fatty acid homeostasis in the liver.
In the current study, we examined the metabolic phenotype of heterozygous SIRT1 KO (SIRT1+/−) mice in the C57BL/6 gene background. We observed that their energy expenditure was reduced, and body fat content was increased while their physical activities and food intake were not altered. The mice had severe hepatic steatosis on diets with mediate- or high-fat content. Their peroxisome proliferator-activated receptor-γ (PPARγ) and sterol regulatory element-binding protein (SREBP) activities were increased in the liver in response to the dietary fat. β-Oxidation-related genes were not changed in the SIRT1+/− mice. The gene expression data support our hypothesis that the primary metabolic activity of SIRT1 is to promote fatty acid mobilization. Fatty acid oxidation is a consequence of fatty acid mobilization.
Materials and Methods
Animals
C57BL/6J breeders (4 wk of age) were obtained from The Jackson Laboratory (Bar Harbor, ME). The SIRT1+/− Tg mice on the 129/J background was a gift from Dr. Frederick W. Alt at the Howard Hughes Medical Institute, Children's Hospital, Center for Blood Research and Department of Genetics, Harvard University Medical School (Boston, MA) (15). SIRT1+/− Tg mice were backcrossed with C57BL/6 mice for five generations to obtain the C57BL/6 gene background. The heterozygous KO (SIRT1+/−) and wild-type (WT) littermates were used in the study. The mice were maintained at 23 ± 1 C with a 12-h light, 12-h dark cycle. Breeders were provided with corn-cob bedding and chow diet (5015, 11% fat in weight or 5001, 5% fat in weight). The experimental mice were housed with three to four mice per cage with free access to water and diet. All procedures were performed in accordance with National Institutes of Health guidelines and approved by the Institute Animal Care and Use Committee (IACUC) at the Pennington Biomedical Research Center.
Diets
The low-fat diet (LFD) is the regular chow diet (5% fat wt/wt; 5001 LabDiet). The mediate-fat diet (MFD) (11% fat wt/wt; 5015 LabDiet) and HFD (36% fat wt/wt, D12331, Research Diets) were used to test the response of the SIRT1+/− mice to dietary fat.
Nuclear magnetic resonance
Body composition was measured using quantitative nuclear magnetic resonance (NMR) as previously described (16). In the test, conscious and unrestrained mice were individually placed in small tubes and then inserted into a Brucker model mq10 NMR analyzer one at a time (Brucker, Milton, Ontario, Canada). The fat and lean mass were recorded within 1 min.
Genotyping
DNA was prepared from tails samples using the proteinase K protocol. The genotyping PCR was conducted in all of the mice. PCR was performed with the Peltier Thermal Cycler 100 (PTC-100) machine. The primers below were ordered from Sigma (St. Louis, MO):
SIRT1SKO-F: 5′-CTTGCACTTCAAGGGACCAA
SIRT1SKO-R1: 5′-GTATACCCACCACATCTGAG
SIRT1SKO-R2: 5′-CTACCACTCCTGGCTACCAA
Tissue collection
In the newborn pup, tissues were collected on the birth date after 4 h fast. In adult mice, the tissue collection was performed after 6 h fast. The samples include brown fat, white fat, skeletal muscle, and liver. The samples were either treated with 4% formaldehyde for histological analysis or frozen in liquid nitrogen for protein and mRNA analysis. The fixed samples were kept at room temperature. The frozen samples were then kept at −80 C.
Quantitative real-time RT-PCR
Tissues were collected after a 6-h fast and first kept in liquid nitrogen then stored at −80 C. Total RNA was extracted from frozen tissues using Tri-Reagent (T9424, Sigma) as described elsewhere (17). mRNA was quantified using Taqman RT-PCR. The primer and probe were from the Applied Biosystems (Foster City, CA). These include: glucose-6-phosphatase (G6Pase) (Mm00839363_m1), phosphoenolpyruvate carboxykinase 1 (PEPCK) (Mm00440636_m1), PPARγ (Mm00440945_m1), SREBP-1 (Mm00550338_m1), fatty acid synthase (FAS) (Mm00662319_m1), adipocyte-specific lipid-binding protein (Mm00445880_m1), hormone-sensitive lipase (HSL) (Mm00495359_ml), lipoprotein lipase (LPL) (Mm00434770-m1), fatty acid translocase (CD36) (Mm00432403_m1), stearoyl-coenzyme A desaturase 1 (SCD1) (Mm00772290_m1), PPARγ coactivator 1 (PGC1α) (Mm00447183_m1), uncoupling protein (UCP)-1 (Mm00494069_m1), carnitine palmitoyltransferase 1 (CPT1α) (Mm00550438_m1), CytoC (Mm01621048-m1), medium-chain acyl-coenzyme A dehydrogenase (MCAD) (Mm00431611_m1), adiponectin (Mm00456425_m1), leptin (Mm00434759_m1), preadipocyte factor (Pref) (Mm00494477_ m1), F4/80 antigen (a glycoprotein expressed by macrophages) (Mm00802530_m1), TNF-α (Mm00443258_m1), IL-1β (Mm00434228_m1), IL-6 (Mm00446190_m1), monocyte chemotactic protein-1 (MCP-1) (Mm00441242_m1), vascular endothelial growth factor (VEGF) (Mm00437304_m1), platelet-endothelial cell adhesion molecule 1, also known as PECAM1 (CD31) (Mm00476702_m1), VEGF receptor 2 (VEGFR2) (Mm00440099_m1), platelet-derived growth factor (PDGF) (Mm00440678_m1), Apelin (Mn00443562-m1), hepatocyte growth factor (HGF) (Mm01135177_m1), and TGFβ (Mm00441724_m1). Mouse ribosome 18S rRNA_s1 (without intron-exon junction) was used as an internal control. mRNA of liver genes in very low-density lipoprotein (VLDL) production was determined using the SYBR green method. The primer sequence is in Supplemental Fig. 1 published on The Endocrine Society's Journals Online web site at http://endo.endojournals. org. The quantitative RT-PCR (qRT-PCR) was conducted with 7900 HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA).
Energy metabolism
Energy expenditure, respiratory exchange ratio (RER), spontaneous physical movement, and food intake were measured simultaneously in each mouse with the Comprehensive Laboratory Animal Monitoring System (Columbus Instruments, Columbus, OH) as described previously (17). The calculations of carbohydrate and fat oxidation are based on stoichiometric equations reported by Watt (18). CHO oxidation = 4.585 × VCO2 − 3.226 × VO2; fat oxidation = 1.695 × VO2 − 1.701 × VCO2. The CO2 and O2 volume data from the metabolic chamber test were used in the calculation.
Western blot
Fresh liver tissue was collected and frozen immediately in liquid nitrogen. The whole cell lysate was prepared in a lysis buffer with sonication as described elsewhere (19). Antibodies include those against Tubulin (ab7291; Abcam, Cambridge, MA), SIRT1 (Sir2) (DAM1514081; Millipore, Billerica, MA), SREBP-1 (sc-13551; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and Sp1 (sc-59; Santa Cruz).
Hematoxylin and eosin (H&E) staining
Fresh tissues (liver and fat) were collected and fixed in 4% neutral buffered formalin solution (HT50-1-2; Sigma). The tissue slides were obtained through serial cross-section cutting at 8 μm thickness and processed with a standard procedure.
Plasma test
Alanine aminotransferase (ALT) was measured in the plasma according to standard enzymatic assay (Thermo Electron Corp., Waltham, MA). Insulin and IL-6 were determined in the plasma using the Mouse Serum Adipokine Multiplex Kit (MADPK-71K; LINCO Research, Billerica, MA). Triglyceride (TAG) and glycerol were determined using the Serum Triglyceride Determination Kit (TR0100; Sigma). Cholesterol was determined with the Cholesterol Reagent (80015; Raichem, San Diego, CA). Plasma free fatty acid (FFA) was determined using the Fatty Acid Detection Kit (catalog no. SFA-1; Zen-Bio, Inc., Research Triangle Park, NC).
Liver lipid test
Liver tissues were homogenized in PBS (1 g: 20 ml). The lipids were extracted from the liver tissue lysate using a chloroform/methanol (2:1) mixture (20). TAG and glycerol were determined using Serum Triglyceride Determination Kit (TR0100, Sigma). Cholesterol was determined with Cholesterol Reagent (80015; Raichem) according to instructions by the manufacturer.
Fatty acid oxidation
Fatty acid oxidation was determined in liver tissue using 14C-labled palmitic acid with a procedure modified from a published study (21). The oxidation reaction was conducted in Krebs-Ringer bicarbonate buffer containing 1 μCi/ml [1-14C] palmitic acid (GE Healthcare Bio-Sciences Corp.) and 0.5% BSA, which was aerated with 5% CO2 and 95% O2 gas for at least 30 min before use. Liver homogenates from 10 mg tissue were incubated with the reaction mixture for 1 h at 37 C. CO2 was collected using semidry filter paper saturated with 2 n NaOH. The radioactivity in CO2 was quantified and used to determine fatty acid oxidation.
Statistical analysis
In this study, the data were presented as the mean ± sem from multiple samples (n = 6–8 for each group in animal study). All of the in vitro experiments were conducted three times at least. Two-tailed, unpaired Student's t test was used in the statistical analysis with significance P ≤ 0.05.
Results
SIRT1+/− Tg mice have no hepatic steatosis on LFD
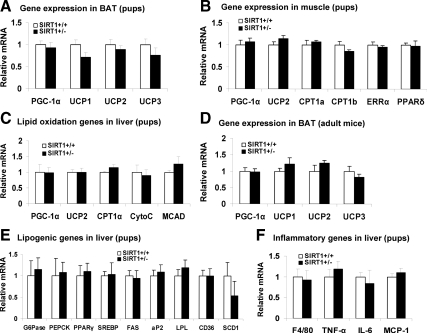
In this study, the SIRT+/− 129 mice were backcrossed with C57BL/6 mice for five generations to obtain the C57BL/6 gene background. Most of the SIRT1 null mice died within the first month after birth in the C57BL/6 background, and the surviving SIRT1 null mice were 50% smaller in body size (data not shown). Therefore, the null mice were not used in the metabolic phenotype study. The heterozygous SIRT1-KO (SIRT1+/−) mice were examined for metabolic phenotypes. On the regular chow diet (LFD, 5% fat wt/wt), the mice were normal in body weight, fat content, and lean body mass relative to their WT littermates in a 24-wk study (Supplemental Fig. 2A). Their livers did not exhibit a significant difference in weight, histology, or gene expression compared with those of WT control (Supplemental Fig. 2, B and C). We examined eight representative genes for glucose and lipid metabolism in the liver. However, no significant change was observed in them (Supplemental Fig. 2C). A macrophage marker (F4/80) and several inflammatory cytokines (TNF-α, IL-1, and MCP-1) were examined in the liver. No difference was observed between the SIRT1+/− and WT mice (Supplemental Fig. 2D). The data suggest that SIRT+/− mice are normal in metabolism on LFD.
Increased fat content and reduced energy expenditure in SIRT1+/− mice on the MFD
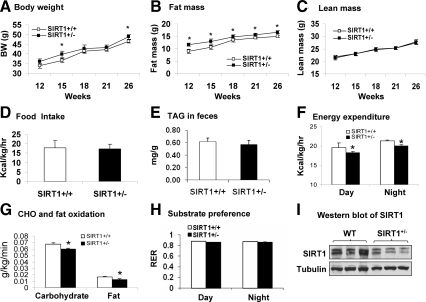
HFD is often used in the study of metabolic phenotype in Tg mice. In this study, we employed two HFDs with different fat content. The first diet with 11% fat (wt/wt) is designed as MFD. The second with 33% fat (wt/wt) is named for HFD. In the study, mouse embryonic fibroblast (MEF) cells were used in mice at 3 wk of age in a 26-wk study. The SIRT+/− mice did not exhibit any difference in the first 10 wk of age. They exhibited a modest increase in body weight and fat content after 10 wk (Fig. 1, A and B). The increased body weight occasionally reached statistical significance at 15 and 26 wk of age. The body fat content was increased consistently and became 30% more than the control (11.7 g vs. 9.0 g) at 12 wk of age (Fig. 1B). The increase remained for the rest of time (Fig. 1B). The lean body mass was not changed (Fig. 1C), suggesting that body fat may account for the gain in body weight. There was no difference in food intake between the Tg and WT mice (Fig. 1D). Fat digestion and absorption in the intestine were examined by determining fat content in the feces. The data do not suggest a defect in the gastrointestinal function in the SIRT1+/− mice (Fig. 1E).
Figure 1.
Increased fat content and reduced energy expenditure on MFD in SIRT1+/− mice. A, Time course of body weight gain. Body weight (BW) was determined every 3–5 wk after 12 wk of age. B, Fat mass. C, Lean mass. The fat and lean mass were determined using NMR. D, Food intake. Food intake was monitored daily for 3 d. Average daily food intake (g) was converted into kilocalories and normalized with body weight (kg) and time (hours). E, TAG in feces of MFD-fed mice. F, Energy expenditure in SIRT1+/− mice. Energy expenditure was examined in the mice at 13 wk of age using the metabolic chamber. The unit is kilocalories per kilogram of lean body mass. G, Carbohydrate (CHO) and fatty acid oxidation at night time normalized with the lean body mass. H, Substrate preference. It was expressed by RER, which is a volume ratio of oxygen consumed vs. CO2 exhaled. I, Western blot of SIRT1 protein in liver tissue. Values are the means ± se (n = 7). *, P ≤ 0.05.
The increased body fat was observed in the absence of elevated food intake, suggesting a reduction in energy expenditure. To test this possibility, we examined energy metabolism using a rodent metabolic chamber. The experiment was performed in mice at 13 wk of age. The data were collected for both day and night times, and normalized with lean body mass. In the Tg mice, the energy expenditure was reduced by 6% in both day and night times (Fig. 1F). The reduction was observed with decreased oxidation in carbohydrate (11%) and fatty acid (23%) (Fig. 1G). Carbohydrate is the main source of fuel in this model system as indicated by substrate oxidation rate and RER (>0.8) (Fig. 1, G and H). RER was not significantly altered in the SIRT1+/− mice, although fatty acid oxidation was reduced more than carbohydrate. These data suggest that substrate preference was not significantly changed on MFD. Locomotor activity was not reduced in the Tg mice (data not shown), suggesting that the basic metabolic rate was lower in the SIRT1+/− mice. The increased adiposity may be a result of reduced energy expenditure in the SIRT1+/− mice. The reduced SIRT1 activity was confirmed at protein level in the liver (Fig. 1I).
Hepatic steatosis in SIRT1+/− mice on MFD
SIRT1 activation by resveratrol was shown to attenuate fatty liver (11). The study suggests that if SIRT1 activity is reduced, lipid may accumulate in the liver. To test the possibility, we examined liver weight and fat content in the SIRT1+/− mice. As expected, the liver weight was increased by 42%, and liver to body ratio was increased in the Tg mice (Table 1). The plasma lipids including TAG, glycerol, and total cholesterol were examined. They were not elevated or decreased in the SIRT1+/− mice on MFD (Table 1). On HFD, TAG was reduced in the SIRT1+/− mice.
Table 1.
Association of liver and fat tissue weight with body weight
| Assay | MFD
|
HFD
|
||
|---|---|---|---|---|
| WT | SIRT1+/− | WT | SIRT1+/− | |
| Body weight (g) | 45.9 ± 1.1 | 47.2 ± 1.6a | 46.6 ± 1.9 | 49.0 ± 1.6a |
| Fat mass (g) | 14.9 ± 0.3 | 16.9 ± 0.4a | 18.5 ± 1.3 | 21.3 ± 0.5a |
| Lean mass (g) | 26.5 ± 0.9 | 26.0 ± 1.0 | 25.5 ± 0.3 | 25.7 ± 0.9 |
| Liver (g) | 1.75 ± 0.13 | 2.49 ± 0.15a | 1.64 ± 0.21 | 2.37 ± 0.28a |
| Liver/body weight ratio | 0.04 ± 0.003 | 0.05 ± 0.003a | 0.04 ± 0.005 | 0.05 ± 0.006a |
| Brown fat (g) | 0.20 ± 0.01 | 0.21 ± 0.01 | 0.21 ± 0.03 | 0.24 ± 0.03 |
| Epididymal fat (g) | 1.58 ± 0.15 | 1.37 ± 0.11 | 2.75 ± 0.09 | 2.45 ± 0.23 |
| Retroperitoneal fat (g) | 0.90 ± 0.04 | 1.05 ± 0.11 | 0.99 ± 0.10 | 1.10 ± 0.09 |
| Subcutaneous fat (g) | 12.42 ± 0.08 | 14.48 ± 0.28a | 14.76 ± 1.15 | 17.75 ± 0.53a |
| TAG (mg/dl) | 9.3 ± 0.5 | 8.6 ± 0.7 | 13.6 ± 2.2 | 10.7 ± 0.3a |
| Glycerol (mg/dl) | 7.2 ± 0.4 | 7.6 ± 1.2 | 8.7 ± 0.4 | 8.0 ± 0.7 |
| Cholesterol (mg/dl) | 134.9 ± 6.7 | 144.7 ± 10.0 | 263.4 ± 7.1 | 263.4 ± 11.3 |
Livers were collected from mice at 28 wk of age. In the MFD group, the mice were fed on the diet from 3 wk of age. In the HFD group, the mice were fed on LFD (regular chow) for 12 wk and then fed on HFD for 16 wk. Mice were euthanized using CO2. The sc fat is obtained from the total body fat by exclusion of epididymal and retroperitoneal fat. The total body fat contains fat in liver. Blood was collected first, and then liver, epididymal fat, retroperitoneal fat, and brown fat tissues were taken and weighed for the tissues to body weight ratio. Subcutaneous fat = (fat mass − epididymal fat − retroperitoneal fat). TAG, glycerol, and cholesterol were determined in plasma. Values are means ± se (n = 6 mice).
P ≤ 0.05.
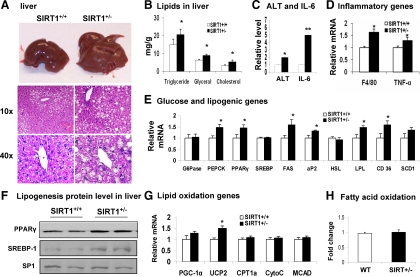
To characterize the hepatic steatosis, we examined morphology, histology, lipid content, and gene expression in the liver of SIRT1+/− mice. The SIRT1+/− liver exhibited white coloring (Fig. 2A), and a significant increase in the lipid droplets in hepatocytes (Fig. 2A). The lipid droplets appear as uncolored circles in the H&E-stained tissue slide. The size of lipid droplet was also increased and this was observed with high magnification (Fig. 2B, ×40). Consistently, TAG, glycerol, and cholesterol contents were increased by 38%, 45%, and 66% in the liver (Fig. 2B). The liver function was evaluated by the serum ALT and IL-6. A 100% increase was observed in ALT and 5-fold increases was found in IL-6 in the SIRT1+/− serum (Fig. 2C). Inflammation was increased in the liver as indicated by mRNA expression for macrophage marker (F4/80) and proinflammatory cytokine TNF-α (Fig. 2D). The data provide multiple lines of evidence for hepatic steatosis in the Tg mice.
Figure 2.
Hepatic steatosis in SIRT1+/− mice on MFD. Liver was examined in mice of 28 wk of age on MFD. A, Liver. Tissue was stained with H&E. Pictures were taken using a microscopy with ×10 or ×40 object lenses, respectively. B, TAG, glycerol, and cholesterol content in the liver of mice. C, ALT and IL-6 levels in the serum. D, Inflammatory gene mRNA in liver. E, Gluconeogenic and lipogenic gene expression in liver. F, Western blot of PPARγ and SREBP-1 protein in nucleus of liver. G, β-Oxidation and mitochondrial genes. H, Fatty acid oxidation in liver tissue. Fold change is used to express mRNA expression. Values are the means ± se (n = 7). *, P ≤ 0.05; **, P ≤ 0.001.
To understand the molecular basis of hepatic steatosis, we examined lipogenesis and fat oxidation in the liver by monitoring mRNA expressions of lipogenic genes and β-oxidation genes. Ten genes in lipogenesis (PPARγ, SREBP, FAS, PEPCK, G6Pase, SCD1, LPL, aP2, CD36, and HSL) were examined. An increase was observed in six of them (PPARγ, FAS, PEPCK, CD36, aP2, and LPL) in the Tg mice (Fig. 2E). PPARγ increase was confirmed in protein (Fig. 2F). The RT-PCR primer detects both SREBP1c and SREBP1a. Although the SREBP1 mRNA was not increased, the protein abundance was significantly augmented in the SIRT1+/− mice (Fig. 2F). Five β-oxidation-related genes (PGC-1α, UCP2, CPT1α, CytoC, and MCAD) were examined in the liver, and only UCP2 was increased modestly in the Tg mice (Fig. 2G). We further determined fatty acid oxidation potential in primary culture of liver tissues using radiolabeled fatty acid. No change was observed in the SIRT1+/− liver (Fig. 2H). These data suggest that lipogenesis is enhanced and β-oxidation is not altered in the liver of SIRT1+/− mice. Hepatic steatosis is likely a result of elevated lipogenesis.
Reduced energy expenditure in SIRT1+/− mice on HFD
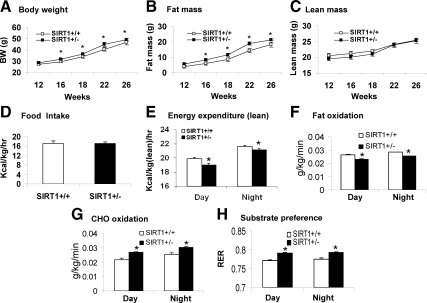
The metabolic phenotype of SIRT1+/− mice was examined on HFD (37% fat wt/wt). The Tg mice gained more body weight and body fat relative to the WT mice on HFD (Fig. 3, A and B). The lean body mass and food intake were not changed (Fig. 3, C and D). As expected, energy expenditure was significantly reduced in both day and night times (Fig. 3E). In substrate utilization, a decrease in fatty acid utilization was observed in the Tg mice (Fig. 3F). However, the carbohydrate utilization was higher in the SIRT1+/− mice (Fig. 3G). The increase led to a significant increase in RER (Fig. 3H), supporting that the fatty acid utilization is reduced in the SIRT1+/− mice. This group of results provides additional support to the reduction in energy expenditure and fatty acid metabolism in the SIRT1+/− mice.
Figure 3.
Reduced energy expenditure in SIRT1+/− mice on HFD. The mice were fed on HFD at 12 wk of age, and examined for energy expenditure after 3 wk on HFD. A, Time course of body weight gain. Body weight was weighed every 2–4 wk from 12 wk of age. B, Fat mass. C, Lean mass. D, Food intake. Food intake was monitored daily for 3 d. Average daily food intake (g) was converted into kilocalories and normalized with lean body mass (kg) and time (hours). E, Energy expenditure in SIRT1+/− mice. Energy expenditure was examined using the metabolic chamber and normalized with lean body mass. F, Fatty acid utilization normalized with lean body mass. G, Carbohydrate utilization normalized with lean body mass. H, RER is a volume ratio of oxygen consumed vs. CO2 exhaled. In this figure, values are the means ± se (n = 7). *, P ≤ 0.05.
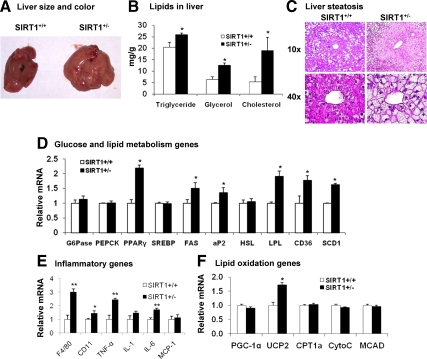
Hepatic steatosis was accelerated by HFD
The increased dietary fat may accelerate liver steatosis in the Tg mice on HFD. To test the possibility, we examined the hepatic steatosis at 16 wk on HFD (28 wk of age). As expected, the Tg livers were significantly larger and heavier than those of WT (Fig. 4A). The liver-to-body ratio was significantly higher (Table 1). The increase in TAG and glycerol content was observed (Fig. 4B). The cholesterol content was increased by 250% on HFD (66% on MFD) (Fig. 4B). The liver tissue exhibited many more lipid droplets under the H&E staining (Fig. 4C). The hepatocytes were filled with lipid droplets, and the cytoplasm was less stained (Fig. 4C). The cell morphology change was visible with the ×10 objective lens, and a striking difference was observed with the ×40 objective lens. In the SIRT1+/− liver, expression of lipogenic genes (PPARγ, LPL, FAS, CD36, and SCD1) was significantly elevated (Fig. 4D), supporting the increased lipogenesis. Inflammation was also increased in the liver as indicated by expression of macrophage markers (F4/80 and CD11b) and inflammatory cytokines (TNF-α and IL-6) (Fig. 4E), suggesting that the steatosis is associated with inflammation. These data suggest that liver steatosis was augmented by HFD in the SIRT1+/− mice.
Figure 4.
Hepatic steatosis was accelerated by HFD in SIRT1+/− mice. The liver was examined in the SIRT1+/− mice on HFD for 16 wk (28 wk of age). A, Increased liver weight and size in the SIRT1+/− Tg mice. B, TAG, glycerol, and cholesterol content in liver. C, H&E staining of liver. Pictures were taken under a microscopy with object lenses of ×10 or ×40, respectively. D, Gluconeogenic and lipogenic gene mRNA in liver. E, Inflammatory gene mRNA in liver. F, mRNA in liver. Values are the means ± se (n = 7). *, P ≤ 0.05; **, P ≤ 0.001.
β-Oxidation related gene was not changed in the Tg mice
SIRT1 was reported to induce gene expression for fatty acid oxidation in skeletal muscle (22) and liver (23). If this is the primary activity of SIRT1, we expect to see a reduced expression in these genes in the SIRT1+/− mice. To test the possibility, we selected five most important mitochondrial genes and examined their expression in the liver of SIRT+/− mice on MFD (Fig. 2G). However, we did not obtain expected result, but observed an increase in UCP-2 (Fig. 2G). To exclude the impact of environment, we examined new born mice. The assay was conducted in brown fat (PGC-1α, UCP1, UCP2, and UCP3), skeletal muscle (PGC-1α, UCP2, CPT1a, CPT1b, estrogen-related receptor α, and PPARδ), and liver (PGC-1α, UCP2, CPT1a, CytoC, and MCAD). The ten different genes related to fatty acid oxidation were examined. No change was detected in any of them in the SIRT1+/− mice (Fig. 5, A–C). In adult mice, no significant change was observed in brown fat (Fig. 5D). Genes in lipogenesis and inflammation were also examined in the liver of pups. No significant change was observed (Fig. 5, E and F). The data suggest that the reduction of SIRT1 activity does not change gene expression directly. The gene expression may be changed in response to dietary fat challenges.
Figure 5.
Energy expenditure genes in brown fat, muscle, and liver. In this figure, mRNA expression was determined for fatty acid oxidation-related genes in different tissues of new born mice except for panel D. A, BAT. B, Muscle. C, Liver. D, BAT of adult mice. E, Lipogenic genes in liver. F, Inflammatory genes. Values are the means ± se (n = 6–8). ERRα, Estrogen-related receptor-α.
Inflammation in adipose tissue of SIRT1+/− mice
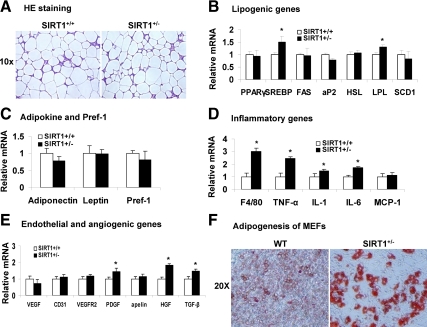
We examined fat pad mass to determine fat distribution in the body. The adipose tissues such as epididymal and retroperitoneal fat pads were weighted in mice on both MFD and HFD. Interestingly, the fat mass was not increased in the Tg mice (Table 1). The fat tissues were analyzed in histology and gene expression. There was no difference in adipocyte size, cell density, and the extracellular matrix between Tg and WT mice on MFD (Fig. 6A), suggesting that the adipose tissue has a normal structure and cell size in the SIRT1+/− mice. Expression of most lipogenic genes (PPARγ, FAS, aP2, HSL, and SCD1) were not changed in the adipose tissue of SIRT1+/− mice except SREBP and LPL whose expression was increased (Fig. 6B). Adipokines (adiponectin and leptin) and Pref-1 were examined, and they were not altered (Fig. 6C). These data suggest that the mature adipocytes and preadipocytes both were normal in the SIRT1+/− mice.
Figure 6.
Inflammation in adipose tissue of SIRT1+/− mice on MFD. Epididymal fat was collected from mice at 28 wk of age and used in the analysis. A, H&E staining of epididymal fat. B, Lipogenic genes in epididymal fat. C, mRNA for adipokines and Pref-1. D, mRNA for inflammatory genes. E, mRNA for endothelial and angiogenic genes. F, Oil-Red O staining in differentiated MEFs. Values are the means ± se (n = 7). *, P ≤ 0.05. Apelin, an angiogenic factor produce by adipocytes.
The disassociation of adipose tissue mass with body weight suggests something wrong in the SIRT1+/− mice. To understand the mechanism, we examined inflammation and angiogenesis in the epididymal fat pads since adipose tissue growth is controlled by these factors (24,25,26). In the SIRT1+/− mice on MFD, macrophage infiltration and expression of inflammatory genes were enhanced. This is indicated by expression of F4/80, TNF-α, IL-1, and IL-6 (Fig. 6D). The angiogenic activity was examined by expression of endothelial cell markers (CD31 and VEGFR2) and angiogenic genes (VEGF, PDGF, Alpine, HGF, and TGF-β). The result suggests that angiogenesis was not reduced in the SIRT1+/− mice (Fig. 6E). Instead, angiogenic factors such as PDGF, HGF, and TGF-β were increased (Fig. 6E). The increase is consistent with the enhanced macrophage infiltration as macrophages express these angiogenic factors (27). We examined adipogenesis in the SIRT1+/− mice by examining differentiation of MEFs in vitro. The SIRT1+/− cells exhibited an enhanced potential in adipogenesis (Fig. 6F). The lipid accumulation was increased in the SIRT1+/− cells after differentiation as indicated by oil red O staining. This differentiation potential may be inhibited in the adipose tissue by the elevated inflammation. This possibility may explain the disassociation of adipose tissue mass with body weight in the SIRT1+/− mice.
TAG export in liver of SIRT1+/− mice
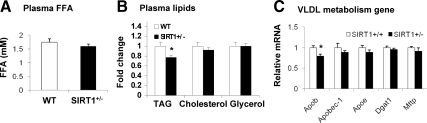
Liver delivers TAG to peripheral tissues by production of VLDL. If the production is impaired, TAG will accumulate in hepatocytes to induce steatosis. To test this possibility, total FFA and TAG were examined in the plasma. Genes in the control of VLDL synthesis were examined in the liver. FFA was not significantly different between WT and SIRT1+/− mice (Fig. 7A). The total TAG was reduced in the SIRT1+/− mice (Fig. 7B). Five genes (Apob, Apobec1, Apoe, Dgat1, and Mttp) were examined in the VLDL pathway. Four genes were normal, and one gene (Apob) was reduced (Fig. 7C). The data suggest that liver is able to keep plasma FFA in the normal range in the SIRT1+/− mice. However, fat export by liver may be impaired by the reduced Apob expression. This defect is reflected by the reduced plasma TAG in mice.
Figure 7.
TAG export in liver. Plasma lipids and liver gene expression were determined in mice on HFD. A, FFA level in plasma. B, Total TAG, glycerol, and cholesterol in plasma. C, Expression of genes in VLDL production pathway. Apob, Apolipoprotein B; Apobec1, apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1; Apoe, apolipoprotein E; Dgat1, diacylglycerol acyltransferase 1; Mttp, microsomal TAG transfer protein. Results are means ± sem (n = 7). *, P < 0.05.
Discussion
Liver steatosis and energy metabolism in response to dietary fat
The metabolic phenotype was characterized in the SIRT1 heterozygous knockout mice (SIRT1+/−) in the C57BL/6 gene background. The body fat content was increased and energy expenditure was reduced in the mice on either MFD or HFD but not on the LFD. Liver fat accounts for 30%, and sc fat accounts for 70% of the increased fat (Table 1). Visceral fat mass (Table 1) and skeletal muscle fat (Supplemental Fig. 3A) were not increased in the SIRT+/− mice. The liver steatosis was enhanced in the mice as indicated by liver weight, liver-to-body ratio, and liver lipids parameters (TAG, glycerol, and cholesterol). The liver function was impaired as indicated by plasma ALT. This liver phenotype is consistent with those from SIRT1 knockdown or overexpression. The liver fat content was increased after transient knockdown of SIRT1 in liver (28) and decreased with SIRT1 overexpression in liver (29). However, the mechanism by which SIRT1 regulates the liver lipid was not extensively investigated in the two studies. Our data suggest that the hepatic steatosis is a result of reduced lipid mobilization in the absence of normal SIRT1 activity.
Lipogenic genes in the liver of SIRT1+/− mice
SIRT1 was reported to inhibit lipid accumulation and promote lipid mobilization in adipocytes through inhibition of the ligand-dependent PPARγ activity (7). It is not clear if SIRT1 acts in the same way in hepatocytes. Data in the current study strongly support the SIRT1 activity in hepatocytes. However, there is difference in mechanism. Our data suggest that SIRT1 may control PPARγ expression. The PPARγ mRNA was increased in the liver of SIRT1+/− mice and this change was dependent on the dietary fat. The PPARγ function was enhanced as the PPARγ target genes (FAS and CD36) was elevated. During preparation of the manuscript, a study of liver-specific SIRT1 knockout was reported (23). The tissue-specific KO increased the hepatic steatosis in mice on HFD (40% calorie in fat) in an 11-wk study. The study enforces the role of SIRT1 in the pathogenesis of liver steatosis. However, a different mechanism is proposed for the steatosis mechanism. The study concluded that a reduction in fatty acid oxidation contributes to the hepatic steatosis. This is based on reduced expression in fatty acid oxidation-related gene, such as PGC-1a, CPT1a, CytoC, and MCAD. The same genes were examined in our study and the reduction was not observed although we conducted an extensive investigation under several conditions. We examined these genes in newborn mice and adult mice. We examined liver, brown fat, and skeletal muscle. In adult mice, these genes were examined in liver in response to three different diets. We also examined fat oxidation in the primary liver tissues using a radiolabeled fatty acid. All the data suggest that mitochondrial function was not reduced in fatty acid oxidation in the liver of SIRT1+/− mice. Although the discrepancy exists in fat oxidation, both studies consistently suggest that lipogenesis are increased in the liver in response to SIRT1 inhibition. Lipogenic genes such as FAS and SREBP were up-regulated in both studies. The data support that the primary biological activity of SIRT1 is inhibition of lipogenesis (7).
Liver TAG export in SIRT+/− mice
Lipid accumulation in liver is regulated by multiple events, such as lipogenesis, fatty acid oxidation, and fat export activity. Liver delivers TAG to peripheral tissues through production of very low-density lipoprotein (VLDL), which contains a high content of TAG. A reduction in VLDL production will lead to impairment in TAG export by the liver. In consequence, TAG will accumulate in hepatocytes for steatosis and the total TAG will decrease in the plasma. To test the TAG export activity, plasma TAG and genes in the control of VLDL production were examined in the SIRT1+/− mice. The reduction in Apob expression and plasma TAG suggests that liver export of TAG is reduced in the SIRT1+/− mice. This defect provides a second mechanism for the TAG accumulation in liver. It may also explain the reduced fatty acid oxidation and decreased energy expenditure in the SIRT1+/− mice on HFD. These data suggest that fatty acid mobilization may be a key to understand the hepatic steatosis and low energy expenditure in the SIRT1+/− mice. The conclusion is consistent with that in the WT mice, SIRT1 promotes fatty acid mobilization leading to fatty acid oxidation in skeletal muscle (22) and liver (23). Boily et al. (30) reported that energy expenditure was higher in the SIRT1 null mice. The data were obtained using a similar approach to the current study. However, the body size effect might influence the conclusion. The SIRT1 null mice are 50% smaller than the SIRT1+/− mice in body size (15). The metabolic rate is usually higher in animal with small body size. This body size effect is excluded in the current study because the heterozygous SIRT1 KO mice are not small relative to the WT mice. In addition, the metabolic rate was normalized with lean body mass in the current study.
Inflammation in adipose tissue and liver may promote liver steatosis in the SIRT1+/− mice
Our data suggest that inflammation is elevated in the epididymal fat tissue and liver of SIRT1+/− mice on MFD and HFD. Expression of macrophage markers (F4/80, CD11b) and proinflammatory cytokines (TNF-α and IL-6 and similar cytokines) were increased (Figs. 2, 4, and 6). SIRT1 was reported to inhibit the transcriptional activity of nuclear factor κB (NF-κB) (31,32). NF-κB will contribute to the inflammation through transcriptional regulation of gene expression when the SIRT1 activity is reduced. In adipose tissue, this possibility explains elevated inflammatory cytokines, and macrophage infiltrations. IL-6 induces lipolysis in adipocytes, and TNF-α inhibits lipid accumulation in adipocytes (33). These events will limit growth of epididymal fat although the SIRT1+/− cells have a strong potential in adipogenesis (Fig. 6F). The epididymal fat pads did not show an increased mass in the SIRT1+/− mice. The inflammation in adipose tissue together with inflammation in liver provides the third mechanism for the liver steatosis in the SIRT1+/− mice.
In summary, our findings suggest that energy expenditure is reduced and adiposity is increased in the SIRT1+/− mice. A reduction in SIRT1 activity increased a risk for fatty liver in SIRT1+/− Tg mice. The increase in lipogenesis and reduction in fat export may contribute to the hepatic steatosis. Inflammation in epididymal adipose tissue and liver are also likely involved in the pathogenesis of fatty liver. In response to the SIRT1 deficiency, an increase in PPARγ and NF-κB activities may play a critical role in the molecular mechanisms of energy metabolism and hepatic steatosis in the SIRT1+/− mice.
Supplementary Material
Acknowledgments
We deeply thank Dr. Can Pang and Ms. Tianyi Tang for their excellent technical support.
Footnotes
This work was supported by National Institutes of Health (NIH) Grant DK068036 and ADA Research Award 7-07-RA-189 (to J.Ye) and American Diabetes Association Award 1-09-JF-17 (to Z.G.). F.X. is supported by a scholarship from the China Scholarship Council. The qRT-PCR test, metabolic phenotyping, and imaging studies were conducted in the genomic core, phenotyping core, and imaging core that are supported by NIH Grants 1P30-DK072476 and P20-RR021945.
Disclosure Summary: Authors have no conflict of interest to declare.
First Published Online March 25, 2010
Abbreviations: ALT, Alanine aminotransferase; CD31, platelet-endothelial cell adhesion molecule 1, also known as PECAM1; CD36, fatty acid translocase; CPT1a, carnitine palmitoyltransferase 1; CytoC, cytochrome c; FAS, fatty acid synthase; F4/80 antigen, a glycoprotein expressed by macrophages; FFA, free fatty acid; G6Pase, glucose-6-phosphatase; HDAC, histone deacetylase; H&E, hematoxylin and eosin; HFD, high-fat diet; HGF, hepatocyte growth factor; HSL, hormone-sensitive lipase; LFD, low-fat diet; LPL, lipoprotein lipase; MCAD, medium-chain acyl-coenzyme A dehydrogenase; MCP-1, monocyte chemotactic protein-1; MEF, mouse embryonic fibroblast; MFD, mediate-fat diet; NF-κB, nuclear factor κB; qRT-PCR, quantitative RT-PCR; PDGF, platelet-derived growth factor; PEPCK, phosphoenolpyruvate carboxykinase 1; PGC1α, PPAR-γ coactivator 1; PPARγ, peroxisome proliferator-activated receptor-γ; Pref, preadipocyte factor; RER, respiratory exchange ratio; SCD1, stearoyl-coenzyme A desaturase 1; SIRT1, mammalian sirtuin 1; SREBP, sterol regulatory element-binding protein; TAG, triglyceride; Tg, transgenic; UCP, uncoupling protein; VEGF, vascular endothelial growth factor; VEGFR2, VEGF receptor 2; VLDL, very low-density lipoprotein; WT, wild type.
References
- Imai S, Armstrong CM, Kaeberlein M, Guarente L 2000 Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795–800 [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD 2000 A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA 97:6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L 2000 Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126–2128 [DOI] [PubMed] [Google Scholar]
- Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW 2005 DNA repair, genome stability, and aging. Cell 120:497–512 [DOI] [PubMed] [Google Scholar]
- Longo VD, Kennedy BK 2006 Sirtuins in aging and age-related disease. Cell 126:257–268 [DOI] [PubMed] [Google Scholar]
- Schwer B, Verdin E 2008 Conserved metabolic regulatory functions of sirtuins. Cell Metab 7:104–112 [DOI] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L 2004 Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 429:771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J 2006 Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell 127:1109–1122 [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P 2005 Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434:113–118 [DOI] [PubMed] [Google Scholar]
- Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L 2007 SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell 6:759–767 [DOI] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA 2006 Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444:337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajmo JM, Liang X, Rogers CQ, Pennock B, You M 2008 Resveratrol alleviates alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol 295:G833–G842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks AS, Kon N, Knight C, Matsumoto M, Gutiérrez-Juárez R, Rossetti L, Gu W, Accili D 2008 SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metabolism 8:333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M 2003 The mammalian SIR2α protein has a role in embryogenesis and gametogenesis. Mol Cell Biol 23:38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF 2003 Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA 100:10794–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Wang Z, Zhang X, Butler AA, Zuberi A, Gawronska-Kozak B, Lefevre M, York D, Ravussin E, Berthoud HR, McGuinness O, Cefalu WT, Ye J 2007 Inactivation of PKCθ leads to increased susceptibility to obesity and dietary insulin resistance in mice. Am J Physiol Endocrinol Metab 292:E84–E91 [DOI] [PubMed] [Google Scholar]
- Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, Cefalu WT, Ye J 2009 Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 58:1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt MJ, Heigenhauser GJ, O'Neill M, Spriet LL 2003 Hormone-sensitive lipase activity and fatty acyl-CoA content in human skeletal muscle during prolonged exercise. J Appl Physiol 95:314–321 [DOI] [PubMed] [Google Scholar]
- Gao Z, Yin J, Zhang J, He Q, McGuinness OP, Ye J 2009 Inactivation of NF-κB P50 leads to insulin sensitization in liver through post-translational inhibition of p70S6K. J Biol Chem 284:18368–18376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Yang R, Tarr PT, Wu PH, Handschin C, Li S, Yang W, Pei L, Uldry M, Tontonoz P, Newgard CB, Spiegelman BM 2005 Hyperlipidemic effects of dietary saturated fats mediated through PGC-1β coactivation of SREBP. Cell 120:261–273 [DOI] [PubMed] [Google Scholar]
- Muoio DM, Dohm GL, Tapscott EB, Coleman RA 1999 Leptin opposes insulin's effects on fatty acid partitioning in muscles isolated from obese ob/ob mice. Am J Physiol 276:E913–E921 [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P 2007 Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J 26:1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X 2009 Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab 9:327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho CH, Koh YJ, Han J, Sung HK, Jong Lee H, Morisada T, Schwendener RA, Brekken RA, Kang G, Oike Y, Choi TS, Suda T, Yoo OJ, Koh GY 2007 Angiogenic role of LYVE-1-positive macrophages in adipose tissue. Circ Res 100:e47–e57 [DOI] [PubMed] [Google Scholar]
- Nishimura S, Manabe I, Nagasaki M, Hosoya Y, Yamashita H, Fujita H, Ohsugi M, Tobe K, Kadowaki T, Nagai R, Sugiura S 2007 Adipogenesis in obesity requires close interplay between differentiating adipocytes, stromal cells, and blood vessels. Diabetes 56:1517–1526 [DOI] [PubMed] [Google Scholar]
- Pang C, Gao Z, Yin J, Zhang J, Jia W, Ye J 2008 Macrophage infiltration into adipose tissue may promote angiogenesis for adipose tissue remodeling in obesity. Am J Physiol Endocrinol Metab 295:E313–E322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JR, Harris RA, Lee SR, Craigon MH, Binley K, Price T, Beard GL, Mundy CR, Naylor S 2004 Genetic amplification of the transcriptional response to hypoxia as a novel means of identifying regulators of angiogenesis. Genomics 83:1–8 [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Puigserver P 2007 Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci USA 104:12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschöp MH 2008 Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA 105:9793–9798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, Xuan J, Evans M, Harper ME, McBurney MW 2008 SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One 3:e1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW 2004 Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J 23:2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhou Y, Mueller-Steiner S, Chen LF, Kwon H, Yi S, Mucke L, Gan L 2005 SIRT1 protects against microglia-dependent amyloid-β toxicity through inhibiting NF-κB signaling. J Biol Chem 280:40364–40374 [DOI] [PubMed] [Google Scholar]
- Ye J 2008 Regulation of PPARγ function by TNF-α. Biochem Biophys Res Commun 374:405–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.