Abstract
Signaling by the Ret receptor tyrosine kinase promotes cell movements in the Wolffian duct that give rise to the first ureteric bud tip, initiating kidney development. Although the ETS transcription factors Etv4 and Etv5 are known to be required for mouse kidney development and to act downstream of Ret, their specific functions are unclear. Here, we examine their role by analyzing the ability of Etv4 Etv5 compound mutant cells to contribute to chimeric kidneys. Etv4−/−;Etv5+/− cells show a limited distribution in the caudal Wolffian duct and ureteric bud, similar to Ret−/− cells, revealing a cell-autonomous role for Etv4 and Etv5 in the cell rearrangements promoted by Ret. By contrast, Etv4−/−;Etv5−/− cells display more severe developmental limitations, suggesting a broad role for Etv4 and Etv5 downstream of multiple signals, which are together important for Wolffian duct and ureteric bud morphogenesis.
Keywords: Kidney, Morphogenesis, Cell movement, Mouse, Wolffian duct
INTRODUCTION
One of the key signals that promote metanephric kidney development is glial cell-derived neurotrophic factor (Gdnf), which is secreted by metanephric mesenchyme (MM) cells and signals through the receptor tyrosine kinase (RTK) Ret and co-receptor Gfrα1 (Takahashi, 2001). Ret and Gfra1 are expressed in the Wolffian duct (WD) and then in the tips of the ureteric bud (UB), an outgrowth from the caudal WD that branches extensively to form the renal collecting system, while inducing MM progenitor cells to generate the nephron epithelia (Carroll and McMahon, 2003; Costantini, 2006; Dressler, 2009). Mice lacking Ret, Gfrα1 or Gdnf fail to make normal kidneys owing to defects in UB outgrowth or subsequent branching (Costantini and Shakya, 2006). Sprouty 1 (Spry1), a negative regulator of RTK signaling, is expressed in the UB and upregulated by Ret signaling to generate a crucial negative-feedback loop (Basson et al., 2005; Basson et al., 2006).
One of the main challenges in understanding how Gdnf promotes branching morphogenesis is to identify the specific cellular behaviors that are altered by Ret signaling. A fruitful approach has been to generate chimeras in which a subset of WD/UB cells lack Ret and examine their ability to contribute to a chimeric kidney (Chi et al., 2009b; Shakya et al., 2005). These studies have shown that an early role of Ret is to promote competitive cell rearrangements in the caudal WD, in which cells with the strongest Ret signaling preferentially form a localized epithelial domain, which emerges as the first UB tip. In Ret−/−↔wild-type chimeras (i.e. those generated by injecting Ret−/− cells into wild-type blastocyts), wild-type cells converge to form this ‘primary tip domain’, whereas Ret−/− cells are excluded. By contrast, in Spry1−/−↔wild-type chimeras, the Spry1−/− cells (with elevated Ret activity) preferentially form the primary tip domain, while excluding the wild-type cells (Chi et al., 2009b).
How does Ret signaling exert these effects on WD and UB cells? Several genes have been identified that are regulated by Ret signaling and are likely to contribute to altered cellular phenotypes and behaviors in response to Gdnf (Lu et al., 2009; Pepicelli et al., 1997). Among them are two closely related ETS transcription factors, Etv4 (Pea3) and Etv5 (Erm), which are expressed in the WD/UB lineage in a temporal and spatial pattern similar to that of Ret (as well as in the MM and nascent nephrons). Their expression in UB tips can be upregulated by exogenous Gdnf or Fgf10 and is greatly reduced in a hypomorphic Ret mutant (de Graaff et al., 2001; Lu et al., 2009). However, Etv4/Etv5 expression remains normal in Fgf10−/− or Fgfr2 mutant kidneys, despite reduced UB branching (Lu et al., 2009; Ohuchi et al., 2000; Zhao et al., 2004). Thus, whereas FGF signaling is a major effector of Etv4/Etv5 expression in several organs (Brent and Tabin, 2004; Firnberg and Neubuser, 2002; Liu et al., 2003; Mao et al., 2009), Gdnf is the main stimulus for their expression in the UB.
The roles of Etv4 and Etv5 in kidney development have been examined in mouse using an Etv4 knockout allele (Livet et al., 2002) and two Etv5 loss-of-function alleles, Etv5tm1Hass (Lu et al., 2009) and Etv5tm1Kmm (Chen et al., 2005). Etv5tm1Hass (which lacks the DNA-binding domain) is a stronger allele, with homozygotes dying at E8.5 (Lu et al., 2009), whereas homozygotes for Etv5tm1Kmm (exons 2-5 deleted) have normal kidneys (Chen et al., 2005). Etv5tm1Hass, the allele used in this study, will be referred to as ‘Etv5−’. Etv4−/− or Etv4+/−;Etv5+/− mice have low frequencies of renal agenesis or hypoplasia, whereas nearly all Etv4−/−;Etv5+/− compound mutants have renal agenesis or hypodysplasia, with greatly reduced UB branching (Lu et al., 2009). Etv4 Etv5 double homozygotes have been generated using the weaker Etv5tm1Kmm allele (to allow development past E8.5) and lack kidneys entirely (Lu et al., 2009). Thus, Etv4 and Etv5 are jointly required for kidney development.
Although several downstream genes regulated by Etv4/Etv5 in the UB have been identified (Lu et al., 2009) (see below), how the activity of these transcription factors (and hence their target genes) alters the behavior of WD and UB cells, compromising UB outgrowth and branching, remains to be elucidated. To address this question, we have employed chimeric assays with Etv4/Etv5 mutant cells.
MATERIALS AND METHODS
Derivation and genotyping of ES cells
ES cell lines were derived from blastocysts from Hoxb7-myrVenus;Etv4−/− or Hoxb7-myrVenus;Etv4−/−;Etv5+/− females crossed to Hoxb7-myrVenus;Etv4+/−;Etv5+/− males, as described (Shakya et al., 2005). Etv4, Etv5 and Hoxb7-myrVenus alleles were genotyped as described (Chi et al., 2009a; Lu et al., 2009); wild-type Etv5 was amplified with primers 5′-GACCCCAGGCTGTACTTTGA-3′ and 5′-CAGTCCAGGCGATGAAGTG-3′.
Generation of chimeric embryos and genotyping of Ret-hypomorphic hosts
ES cell injections into wild-type and Rettm2(RET)Vpa/tm2(RET)Vpa hosts and genotyping of host embryos were performed as described (Chi et al., 2009b; Hogan et al., 1994; Shakya et al., 2005). To generate CFP+ host embryos, Hoxb7/Cre;R26RCFP/CFP males (Srinivas et al., 2001; Yu et al., 2002) were mated with R26RCFP/CFP females.
Kidney culture and antibody staining
Urogenital regions were dissected, cultured and imaged as described (Watanabe and Costantini, 2004). Antibodies against pan-cytokeratin (Sigma), calbindin (Santa Cruz), GFP (Molecular Probes) and phosphohistone H3 (Ser10) (6G3) (Cell Signaling) were used to stain whole-mount specimens (Kuure et al., 2005) and cryosections (Chi et al., 2009b).
Mitotic index
Total and phosphohistone H3+ cells were counted in 20-25 serial sections (per specimen) through the posterior ends of six Etv4+/− and five Etv4−/−;Etv5+/− E10.5 WDs and the UB of four Etv4+/− and four Etv4−/−;Etv5+/− E11.5 kidneys. P-value was calculated using Student's t-test (two-tailed, equal variance).
RESULTS AND DISCUSSION
To investigate the cellular defects that lead to failure of renal development in Etv4/Etv5 mouse mutants, we generated both Etv4−/−;Etv5+/− and Etv4−/−;Etv5−/− embryonic stem (ES) cells, injected them into wild-type blastocysts, and analyzed their ability to contribute to the WD and UB in the resulting chimeric kidneys. We compared their behavior with Ret−/− ES cells (Chi et al., 2009b; Shakya et al., 2005). All the mutant ES cell lines carried a transgene expressing a fluorescent protein in the WD and UB, either Hoxb7-GFP (Srinivas et al., 1999) or Hoxb7-myrVenus (Chi et al., 2009a), whereas the wild-type host embryos expressed cyan fluorescent protein (CFP) in the same cell lineage (see Materials and methods).
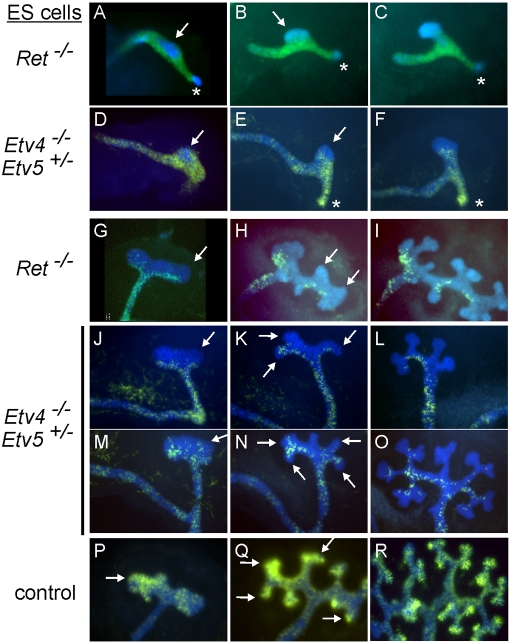
Fig. 1 shows whole-mount images of chimeras made with Etv4−/−;Etv5+/− cells, at stages from the initial WD swelling (E10.5) through early UB branching (E12.5), as compared with Ret−/− chimeras. As previously observed, the Ret−/− cells were specifically excluded from a dorsal domain of the E10.5 WD, termed the ‘primary tip domain’ because it gives rise to the first UB tip (Chi et al., 2009b) (Fig. 1A). As the UB emerged, Ret−/− cells were absent from the tip, but contributed to the trunk (Fig. 1B,C). In Etv4−/−;Etv5+/−↔wild-type chimeras, the mutant cells behaved very similarly to Ret−/− cells, as seen in whole-mount specimens (Fig. 1D-F) or in cross-sections (Fig. 2). During the first UB branching at E11.5, the Etv4−/−;Etv5+/− cells (Fig. 1J,M), like Ret−/− cells (Fig. 1G), contributed preferentially to the proximal side of the initial UB branches. During subsequent branching (E12.5), both types of mutant cells contributed to some of the trunks, but to few, if any, of the tips (Fig. 1H,I,K,L,N,O). By contrast, control (Etv4+/−) ES cells contributed without restriction throughout the WD (not shown) and UB epithelium (Fig. 1P-R). Thus, Etv4−/−;Etv5+/− mutant cells displayed essentially the same cell-autonomous defect as Ret−/− cells: an inability to contribute to the UB tip domain.
Fig. 1.
Etv4−/−;Etv5+/− cells, like Ret−/− cells, are excluded from the ureteric bud tip domain in mutant↔wild-type chimeric mouse kidneys. Wild-type or mutant ES cells, carrying Hoxb7-GFP or Hoxb7-myrVenus, were injected into host blastocysts carrying Hoxb7/Cre and R26R-CFP. In the chimeric embryos, host-derived Wolffian duct (WD) and ureteric bud (UB) cells express CFP (cyan), and ES-derived cells express GFP or myrVenus (green). (A) As previously reported (Chi et al., 2009b), Ret−/− ES cells contribute poorly to the primary UB tip domain (arrow) and common nephric duct (CND; asterisk) at E10.5. (B,C) At E11.0, the UB tip is composed mainly of wild-type cells. (D-F) Etv4−/−;Etv5+/− cells are mostly excluded from the UB tip domain (arrows) (n=11/13), similar to Ret−/− cells (Chi et al., 2009b). However, they contributed normally to the CND (asterisks) (n=14/16). (G-I) During initial UB branching, Ret−/− cells contribute mainly to the proximal side of branches (G), and during subsequent branching (H,I) to some trunks, but not tips (arrows) (Chi et al., 2009b; Shakya et al., 2005). (J-O) Two Etv4−/−;Etv5+/−↔wild-type chimeras, isolated at E11.5 (J,M) and cultured for 2 days (K,L,N,O). Like Ret−/− cells, Etv4−/−;Etv5+/− mutant cells contribute to UB trunks but not tips (arrows) (n=17/17). (P-R) Control ES cells (Etv4+/−) contribute throughout the trunks and tips (arrows).
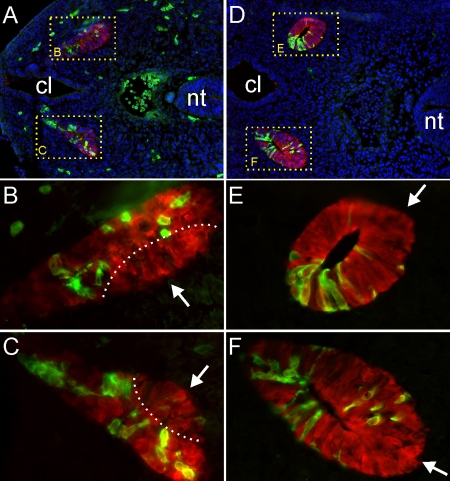
Fig. 2.
Distribution of Etv4−/−;Etv5+/− cells in chimeric Wolffian duct and ureteric bud epithelium. (A-F) Sections through Etv4−/−;Etv5+/−↔wild-type mouse chimeras at the 36-somite (A-C) and 39-somite (D-F) stages, stained with anti-GFP (green) to reveal mutant cells, with anti-calbindin (red) to label all WD and UB cells, and with Hoechst nuclear stain (blue; A,D). B and C are enlargements of the WDs in A; E and F are enlargements of the emerging UBs in D. The dorsal regions of the WD at the 36-somite stage (i.e. the primary UB tip domains; dashed lines) are devoid of mutant cells, as are the UB tips at the 39-somite stage (arrows). cl, cloaca; nt, neural tube.
One significant difference was observed between Ret−/− and Etv4−/−;Etv5+/− cells in the formation of the common nephric duct (CND), the caudal-most segment of the WD that transiently connects the WD and UB to the urogenital sinus. Between E11.5 and E12.5 the CND is remodeled, allowing the ureter to detach from the WD and move to the bladder (Batourina et al., 2005). Whereas Ret−/− cells are excluded from the CND (Chi et al., 2009b), as confirmed here (Fig. 1A-C, asterisks), Etv4−/−;Etv5+/− mutant cells contributed extensively to the CND (Fig. 1E,F, asterisks). Thus, the roles of Ret in the formation of the UB tip domain and the CND, two different WD derivatives with distinct fates, appear to be mediated through different downstream pathways: contribution to the UB tip, but not the CND, depends on Etv4/Etv5. This is consistent with the finding that although the ureter often fails to connect to the bladder (a process that is dependent on the proper formation and remodeling of the CND) in Ret−/− mice (Batourina et al., 2002), it apparently connects to the bladder in Etv4−/−;Etv5+/− mice (Lu et al., 2009) (our unpublished data).
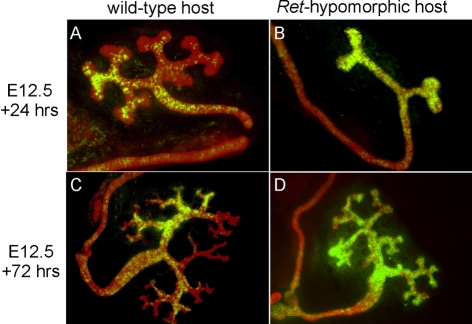
Rettm2(RET)Vpa is a hypomorphic allele with reduced signaling, causing renal hypoplasia with defective UB branching (de Graaff et al., 2001). Unlike in wild-type hosts, Ret−/− cells contribute extensively to the tips in Ret-hypomorphic host embryos (Chi et al., 2009b). Together with the behavior of Spry1−/− cells in Spry1−/−↔wild-type chimeras, this has suggested that WD cells compete with each other, based on the level of Ret signaling, to contribute to the UB tip domain (Chi et al., 2009b). When Etv4−/−;Etv5+/− ES cells were injected into Ret-hypomorphic hosts they contributed extensively to the UB tips as well as the trunks (Fig. 3). Their unrestricted contribution was similar to that of control Etv4+/− cells in wild-type hosts (Fig. 1P-R). Thus, Etv4−/−;Etv5+/− cells are not inherently unable to contribute to UB tips: they fail to compete successfully with wild-type cells, but compete effectively with cells in which Ret signaling is reduced. This supports the hypothesis that Ret signaling and transcriptional regulation by Etv4/Etv5 control similar cellular behaviors.
Fig. 3.
In Ret-hypomorphic host embryos, Etv4−/−;Etv5+/− ES cells contribute strongly to ureteric bud tips. (A-D) Etv4−/−;Etv5+/− ES cells were injected into wild-type (A,C) or Ret-hypomorphic (B,D) mouse blastocysts. Kidneys were cultured from E12.5 for 24 (A,B) or 72 (C,D) hours, then stained with anti-pan-cytokeratin (red), which stains all UB cells, and with anti-GFP (green), which stains the Etv4−/−;Etv5+/− cells. In wild-type hosts, the Etv4−/−;Etv5+/− ES cells contribute to UB trunks, but not (or weakly) to the tips (n=8/8 kidneys). However, in Ret-hypomorphic hosts, the Etv4−/−;Etv5+/− cells usually contribute strongly to the entire UB epithelium, including the tips (n=4/5 kidneys).
Ret signaling has been implicated in both cell proliferation (Pepicelli et al., 1997) and cell movement (Chi et al., 2009b; Tang et al., 1998) in the WD/UB lineage. As such, the failure of Etv4−/−;Etv5+/− cells to contribute to the UB tip domain in wild-type hosts might reflect a competitive disadvantage in cell movement or reduced proliferation in the primary UB tip domain. To examine proliferation, we counted phosphohistone H3+ (pH3+) cells in Etv4−/−;Etv5+/− mutant embryos and Etv4+/− controls, in the E10.5 caudal WD and E11.5 UB (see Fig. S1 in the supplementary material). In the UB, pH3+ cells were 3.5-fold less abundant in the mutant (0.6% versus 2.1%, P<0.005); thus, the severely reduced growth of Etv4−/−;Etv5+/− UBs (Lu et al., 2009) can be attributed, at least in part, to reduced cell proliferation. However, in the caudal WD at E10.5, before UB outgrowth, mutants and controls had the same percentage of pH3+ cells (2.6% versus 2.3%, P=0.487). Therefore, the failure of Etv4−/−;Etv5+/− cells to contribute to the UB tip is not due to a proliferative defect in the WD. In Ret−/−↔wild-type chimeras, time-lapse imaging of organ cultures showed that Ret−/− cells were excluded from the primary UB tip domain by wild-type cells because the mutant cells failed to participate in cell rearrangements that generate this localized domain (Chi et al., 2009b). The nearly identical distributions of Etv4−/−;Etv5+/− and Ret−/− cells in wild-type chimeras, and the lack of a proliferative disadvantage, indicate that these cells share the same deficiency in cell rearrangement.
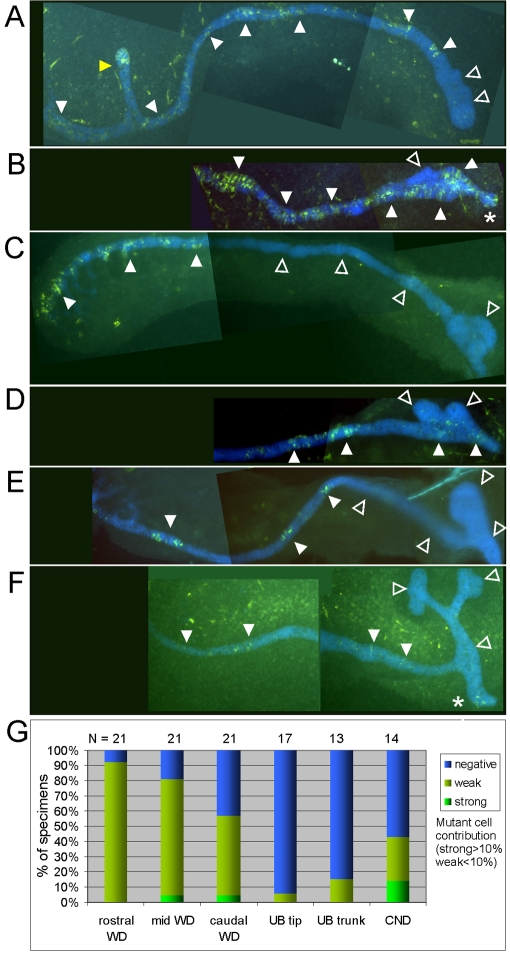
To examine the effects of complete loss of Etv4 and Etv5 we generated chimeras with Etv4−/−;Etv5−/− cells. Etv4−/−;Etv5−/−↔wild-type chimeras developed past the stage when Etv4−/−;Etv5−/− embryos die (E8.5), and remained grossly normal at E11.5-12.5. The Etv4−/−;Etv5−/− cells were able to contribute (albeit usually very weakly) to the WD (Fig. 4), but were very rarely found in the UB, even in the trunks (Fig. 4C-G). They often contributed weakly to the rostral and mid-WD and less often to the caudal WD (e.g. Fig. 4G). This graded distribution suggests a possible defect in cell movement or proliferation during WD elongation in cells lacking both Etv4 and Etv5. By contrast, Etv4−/−;Etv5+/− or Ret−/− cells extensively populated the entire WD and showed no proliferative defect in the WD, but failed to contribute to the UB tip domain. In those chimeras in which Etv4−/−;Etv5−/− cells were found in the caudal WD, the mutant cells were able to contribute to the CND (Fig. 4B,F,G), like Etv4−/−;Etv5+/− cells (Fig. 1E,F), further supporting the conclusion that Etv4 and Etv5 are not required for CND formation. The nature of the defect that restricts Ret−/−, but not Etv4−/−;Etv5−/−, cells from contributing to the CND remains to be identified.
Fig. 4.
Etv4−/−;Etv5−/− cells contribute weakly to the Wolffian duct but not to the ureteric bud in chimeric mouse embryos. (A-F) Etv4−/−;Etv5−/−↔wild-type chimeras analyzed at E10.5-12.0. Rostral, left; caudal, right. Mutant cells are green (Venus+); wild-type cells are blue (CFP+). Six specimens are shown arranged from top to bottom by developmental stage, illustrating the range of mutant cell contributions. B has the most extensive contribution observed in the WD (white arrowheads indicate some of the areas with Venus+ mutant cells), but the forming UB tip domain is devoid of mutant cells (open arrowhead). A,D,F show mutant cells at different positions along the WD (white arrowheads) and in a mesonephric tubule (A, yellow arrowhead), but not in the extreme caudal WD (A) or in UBs (D,F) (open arrowheads). In C and E, a longer segment of caudal WD (as well as the UB) is devoid of mutant cells. Asterisks in B and F indicate mutant cells in the CND. The Etv4−/−;Etv5−/− cells contribute to the mesonephric tubules, primitive nephrons that form near the rostral end of the WD (A), as do Etv4−/−;Etv5+/− cells and Ret−/− cells (data not shown). (G) The percentage of specimens showing strong, weak or no contribution to different regions. N indicates the number of informative specimens.
Ureter and kidney development are affected very similarly in Ret−/− and Etv4/Etv5 mutant embryos, and expression of Etv4 and Etv5 requires normal Ret signaling. This has suggested that the effects of Ret mutations on the developing WD/UB are caused, at least in part, by loss of Etv4 and Etv5 expression (Lu et al., 2009). However, the conclusions that could previously be drawn from a comparison of Ret and Etv4/Etv5 mutant phenotypes were limited, and the present study provides additional insight into the role of Etv4 and Etv5. One issue is that these genes are expressed in the MM as well as in the WD and UB (Lu et al., 2009), and signals from the mesenchyme are crucial for UB morphogenesis (Airik and Kispert, 2007; Dressler, 2006; Schedl, 2007). Therefore, the branching defects in Etv4 Etv5 compound mutants could be due, at least in part, to a failure of induction by the mutant MM. In the chimera studies, however, Etv4−/−;Etv5+/− compound mutant WD and UB cells showed cell-autonomous defects that were very similar to those displayed by Ret−/− cells, which strongly supports a direct role of Etv4/Etv5 in UB epithelial cells, downstream of Ret.
Furthermore, Ret and Etv4/Etv5 do not function in a linear pathway, but in a more complex signaling network. For example, several genes regulated by Ret (e.g. Wnt11, Crlf1, Dusp6) are expressed normally in Etv4−/−;Etv5+/− mutant kidneys, suggesting that their regulation uses different downstream pathways (Lu et al., 2009). In addition, Etv4 and Etv5 function downstream of multiple RTKs, including FGF receptors (Brent and Tabin, 2004; Firnberg and Neubuser, 2002; Liu et al., 2003), that signal to promote UB branching morphogenesis (Maeshima et al., 2007; Michos et al., 2010; Ohuchi et al., 2000; Zhao et al., 2004), so not all the effects of Etv4/Etv5 mutations can be attributed to a block in Ret signaling. As such, it could not be predicted to what extent the behavior of Etv4/Etv5 mutant cells would resemble that of Ret−/− cells in chimeric embryos.
We observed that Etv4−/−;Etv5+/− cells behave similarly to Ret−/− cells, revealing a role of Etv4 and Etv5 in the WD cell rearrangements that are promoted by Ret signaling (Chi et al., 2009b). Etv4 and Etv5 have been implicated in cell migration in other contexts, such as in the radial migration of cortical neurons and in invasive cancer cells (Firlej et al., 2008; Hasegawa et al., 2004). Furthermore, several genes regulated by Etv4 or Etv5 (or other ETS transcription factors) in other cell types, and implicated in cell migration, are also strongly downregulated in the Etv4−/−;Etv5+/− mutant UB (Lu et al., 2009). These include chemokine receptor Cxcr4, Met [the RTK for hepatocyte growth factor (Hgf)], and the matrix metalloprotease Mmp14 (Belien et al., 1999; Peschard and Park, 2007; Schier, 2003). Future studies will seek to identify the full set of genes regulated by Etv4 and Etv5 in the WD and UB.
In contrast to Etv4−/−;Etv5+/− cells, Etv4−/−;Etv5−/− cells were more severely compromised than Ret−/− cells. WD/UB morphogenesis is promoted not only by Gdnf, but also by the combined effects of many factors, including several that signal via RTKs, such as FGFs, Hgf and epidermal growth factor (Egf) (Ishibe et al., 2009; Michos et al., 2010; Tufro et al., 2007; Zhao et al., 2004). The fact that the defect is more severe in Etv4−/−;Etv5−/− than Ret−/− cells supports the conclusion that Etv4 and Etv5 not only function downstream of Ret in kidney development, but also have a broader role downstream of multiple RTKs (Lu et al., 2009; Michos et al., 2010).
Supplementary Material
Acknowledgements
We thank Zaiqi Wu and Linda Williams for expert technical assistance. This work was supported by NIH grants 1R01DK075578 and 1R01DK083289 (F.C.), and fellowships from the National Kidney Foundation, Sigrid Juselius Foundation and Finnish Culture Foundation (S.K.) and the American Heart Association (X.C.). Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.051656/-/DC1
References
- Airik R., Kispert A. (2007). Down the tube of obstructive nephropathies: the importance of tissue interactions during ureter development. Kidney Int. 72, 1459-1467 [DOI] [PubMed] [Google Scholar]
- Basson M. A., Akbulut S., Watson-Johnson J., Simon R., Carroll T. J., Shakya R., Gross I., Martin G. R., Lufkin T., McMahon A. P., et al. (2005). Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev. Cell 8, 229-239 [DOI] [PubMed] [Google Scholar]
- Basson M. A., Watson-Johnson J., Shakya R., Akbulut S., Hyink D., Costantini F. D., Wilson P. D., Mason I. J., Licht J. D. (2006). Branching morphogenesis of the ureteric epithelium during kidney development is coordinated by the opposing functions of GDNF and Sprouty1. Dev. Biol. 299, 466-477 [DOI] [PubMed] [Google Scholar]
- Batourina E., Choi C., Paragas N., Bello N., Hensle T., Costantini F. D., Schuchardt A., Bacallao R. L., Mendelsohn C. L. (2002). Distal ureter morphogenesis depends on epithelial cell remodeling mediated by vitamin A and Ret. Nat. Genet. 32, 109-115 [DOI] [PubMed] [Google Scholar]
- Batourina E., Tsai S., Lambert S., Sprenkle P., Viana R., Dutta S., Hensle T., Wang F., Niederreither K., McMahon A. P., et al. (2005). Apoptosis induced by vitamin A signaling is crucial for connecting the ureters to the bladder. Nat. Genet. 37, 1082-1089 [DOI] [PubMed] [Google Scholar]
- Belien A. T., Paganetti P. A., Schwab M. E. (1999). Membrane-type 1 matrix metalloprotease (MT1-MMP) enables invasive migration of glioma cells in central nervous system white matter. J. Cell Biol. 144, 373-384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent A. E., Tabin C. J. (2004). FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development 131, 3885-3896 [DOI] [PubMed] [Google Scholar]
- Carroll T. J., McMahon A. P. (2003). Overview: the molecular basis of kidney development. In The Kidney. From Normal Development to Congenital Disease (ed. Vize P. D., Woolf A. S., Bard J. B. L.), pp. 343-376 Amsterdam: Academic Press; [Google Scholar]
- Chen C., Ouyang W., Grigura V., Zhou Q., Carnes K., Lim H., Zhao G. Q., Arber S., Kurpios N., Murphy T. L., et al. (2005). ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 436, 1030-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Hadjantonakis A. K., Wu Z., Hyink D., Costantini F. (2009a). A transgenic mouse that reveals cell shape and arrangement during ureteric bud branching. Genesis 47, 61-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Michos O., Shakya R., Riccio P., Enomoto H., Licht J. D., Asai N., Takahashi M., Ohgami N., Kato M., et al. (2009b). Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev. Cell 17, 199-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F. (2006). Renal branching morphogenesis: concepts, questions, and recent advances. Differentiation 74, 402-421 [DOI] [PubMed] [Google Scholar]
- Costantini F., Shakya R. (2006). GDNF/Ret signaling and the development of the kidney. BioEssays 28, 117-127 [DOI] [PubMed] [Google Scholar]
- de Graaff E., Srinivas S., Kilkenny C., D'Agati V., Mankoo B. S., Costantini F., Pachnis V. (2001). Differential activities of the RET tyrosine kinase receptor isoforms during mammalian embryogenesis. Genes Dev. 15, 2433-2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler G. R. (2006). The cellular basis of kidney development. Annu. Rev. Cell Dev. Biol. 22, 509-529 [DOI] [PubMed] [Google Scholar]
- Dressler G. R. (2009). Advances in early kidney specification, development and patterning. Development 136, 3863-3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firlej V., Ladam F., Brysbaert G., Dumont P., Fuks F., de Launoit Y., Benecke A., Chotteau-Lelievre A. (2008). Reduced tumorigenesis in mouse mammary cancer cells following inhibition of Pea3- or Erm-dependent transcription. J. Cell Sci. 121, 3393-3402 [DOI] [PubMed] [Google Scholar]
- Firnberg N., Neubuser A. (2002). FGF signaling regulates expression of Tbx2, Erm, Pea3, and Pax3 in the early nasal region. Dev. Biol. 247, 237-250 [DOI] [PubMed] [Google Scholar]
- Hasegawa H., Ashigaki S., Takamatsu M., Suzuki-Migishima R., Ohbayashi N., Itoh N., Takada S., Tanabe Y. (2004). Laminar patterning in the developing neocortex by temporally coordinated fibroblast growth factor signaling. J. Neurosci. 24, 8711-8719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B., Bedington R., Costantini F., Lacy E. (1994). Manipulating the Mouse Embryo: a Laboratory Manual Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Ishibe S., Karihaloo A., Ma H., Zhang J., Marlier A., Mitobe M., Togawa A., Schmitt R., Czyczk J., Kashgarian M., et al. (2009). Met and the epidermal growth factor receptor act cooperatively to regulate final nephron number and maintain collecting duct morphology. Development 136, 337-345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuure S., Sainio K., Vuolteenaho R., Ilves M., Wartiovaara K., Immonen T., Kvist J., Vainio S., Sariola H. (2005). Crosstalk between Jagged1 and GDNF/Ret/GFRalpha1 signalling regulates ureteric budding and branching. Mech. Dev. 122, 765-780 [DOI] [PubMed] [Google Scholar]
- Liu Y., Jiang H., Crawford H. C., Hogan B. L. (2003). Role for ETS domain transcription factors Pea3/Erm in mouse lung development. Dev. Biol. 261, 10-24 [DOI] [PubMed] [Google Scholar]
- Livet J., Sigrist M., Stroebel S., De Paola V., Price S. R., Henderson C. E., Jessell T. M., Arber S. (2002). ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron 35, 877-892 [DOI] [PubMed] [Google Scholar]
- Lu B. C., Cebrian C., Chi X., Kuure S., Kuo R., Bates C. M., Arber S., Hassell J., MacNeil L., Hoshi M., et al. (2009). Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat. Genet. 41, 1295-1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima A., Sakurai H., Choi Y., Kitamura S., Vaughn D. A., Tee J. B., Nigam S. K. (2007). Glial cell-derived neurotrophic factor independent ureteric bud outgrowth from the Wolffian duct. J. Am. Soc. Nephrol. 18, 3147-3155 [DOI] [PubMed] [Google Scholar]
- Mao J., McGlinn E., Huang P., Tabin C. J., McMahon A. P. (2009). Fgf-dependent Etv4/5 activity is required for posterior restriction of Sonic Hedgehog and promoting outgrowth of the vertebrate limb. Dev. Cell 16, 600-606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michos O., Cebrian C., Hyink D., Grieshammer U., Williams L., D'Agati V., Licht J. D., Martin G. R., Costantini F. (2010). Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet 6, e1000809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi H., Hori Y., Yamasaki M., Harada H., Sekine K., Kato S., Itoh N. (2000). FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem. Biophys. Res. Commun. 277, 643-649 [DOI] [PubMed] [Google Scholar]
- Pepicelli C. V., Kispert A., Rowitch D. H., McMahon A. P. (1997). GDNF induces branching and increased cell proliferation in the ureter of the mouse. Dev. Biol. 192, 193-198 [DOI] [PubMed] [Google Scholar]
- Peschard P., Park M. (2007). From Tpr-Met to Met, tumorigenesis and tubes. Oncogene 26, 1276-1285 [DOI] [PubMed] [Google Scholar]
- Schedl A. (2007). Renal abnormalities and their developmental origin. Nat. Rev. Genet. 8, 791-802 [DOI] [PubMed] [Google Scholar]
- Schier A. F. (2003). Chemokine signaling: rules of attraction. Curr. Biol. 13, R192-R194 [DOI] [PubMed] [Google Scholar]
- Shakya R., Watanabe T., Costantini F. (2005). The role of GDNF/Ret signaling in ureteric bud cell fate and branching morphogenesis. Dev. Cell 8, 65-74 [DOI] [PubMed] [Google Scholar]
- Srinivas S., Goldberg M. R., Watanabe T., D'Agati V., al-Awqati Q., Costantini F. (1999). Expression of green fluorescent protein in the ureteric bud of transgenic mice: a new tool for the analysis of ureteric bud morphogenesis. Dev. Genet. 24, 241-251 [DOI] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C. S., William C. M., Tanabe Y., Jessell T. M., Costantini F. (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M. (2001). The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev. 12, 361-373 [DOI] [PubMed] [Google Scholar]
- Tang M. J., Worley D., Sanicola M., Dressler G. R. (1998). The RET-glial cell-derived neurotrophic factor (GDNF) pathway stimulates migration and chemoattraction of epithelial cells. J. Cell Biol. 142, 1337-1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufro A., Teichman J., Banu N., Villegas G. (2007). Crosstalk between VEGF-A/VEGFR2 and GDNF/RET signaling pathways. Biochem. Biophys. Res. Commun. 358, 410-416 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Costantini F. (2004). Real-time analysis of ureteric bud branching morphogenesis in vitro. Dev. Biol. 271, 98-108 [DOI] [PubMed] [Google Scholar]
- Yu J., Carroll T. J., McMahon A. P. (2002). Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development 129, 5301-5312 [DOI] [PubMed] [Google Scholar]
- Zhao H., Kegg H., Grady S., Truong H. T., Robinson M. L., Baum M., Bates C. M. (2004). Role of fibroblast growth factor receptors 1 and 2 in the ureteric bud. Dev. Biol. 276, 403-415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.