Abstract
Orai1 and STIM1 are critical components of Ca2+ release-activated Ca2+ (CRAC) channels that mediate store-operated Ca2+ entry (SOCE) in immune cells. While Orai1 and STIM1 co-cluster and physically interact to mediate SOCE, the cytoplasmic machinery modulating these functions remains poorly understood. We sought to find modulators of Orai1 and STIM1 using affinity protein purification and identified a novel EF-hand protein, CRACR2A (CRAC regulator 2A, EFCAB4B, FLJ33805). We show that CRACR2A directly interacts with Orai1 and STIM1, forming a ternary complex that dissociates at elevated Ca2+ concentrations. Studies using siRNA-mediated knockdown and mutagenesis show that CRACR2A is important for clustering of Orai1 and STIM1 upon store depletion. Expression of an EF-hand mutant of CRACR2A enhanced STIM1 clustering, elevated cytoplasmic Ca2+ and induced cell death, suggesting its active interaction with CRAC channels. These observations implicate CRACR2A, a novel Ca2+ binding protein, highly expressed in T cells and conserved in vertebrates, as a key regulator of CRAC channel-mediated SOCE.
Ca2+ influx via CRAC channels is crucial for activation, proliferation and cytokine production in immune cells1–5. Recent studies have identified STIM1, a Ca2+-binding protein localized in the endoplasmic reticulum (ER) as an important component of store-operated Ca2+ entry (SOCE)6, 7. STIM1 is a single transmembrane (TM) segment-containing protein that detects ER Ca2+ via its N terminus and has a long C-terminal cytoplasmic region. Upon ER Ca2+ depletion, STIM1 oligomerizes and translocates to plasma membrane (PM)-proximal regions to activate SOCE6, 8, 9. Subsequent studies have identified Orai1 as a pore subunit of the CRAC channels10–16. Upon store depletion, Orai1 also clusters on the PM in the proximity of STIM1 clusters17, 18. Amplified CRAC currents have been observed upon co-expression of Orai1 and STIM1, suggesting that these are the limiting and essential components of CRAC channels15, 19–21. Several studies have identified that the cytoplasmic fragment of STIM1 directly interacts with Orai1 and is sufficient to activate CRAC currents when co-expressed with Orai122–28. However, the cellular machinery modulating Orai1-STIM1 interactions remains unexplored.
Recent studies showed that Orai1 exists in a macromolecular complex with 11–14 nm protrusion into the cytoplasm using chemically inducible bridge formation with linkers of variable lengths between the PM and ER membranes29. These results indicate the presence of additional components within the Orai1-STIM1 complex29. Using immunoaffinity purification of Orai1 after store depletion, we identified a macromolecular complex containing Orai1, STIM1 and putative interactors. An EF-hand containing protein, CRACR2A was validated as an important regulator of Orai1-STIM1 interaction. Our results show that CRACR2A directly interacts with the cytoplasmic regions of Orai1 and STIM1, forming a ternary complex. Interestingly, CRACR2A dissociates from Orai1 and STIM1 at higher Ca2+ concentrations ([Ca2+]). An EF-hand mutant of CRACR2A enhanced STIM1 clustering and elevated cytoplasmic [Ca2+] thereby causing cell death in T cells. These observations suggest a role of CRACR2A as a cytoplasmic Ca2+ sensor that modulates multiple steps of CRAC channel activation including translocation and clustering of Orai1 and STIM1 by direct protein interaction.
RESULTS
Orai1 and STIM1 exist in a macromolecular protein complex
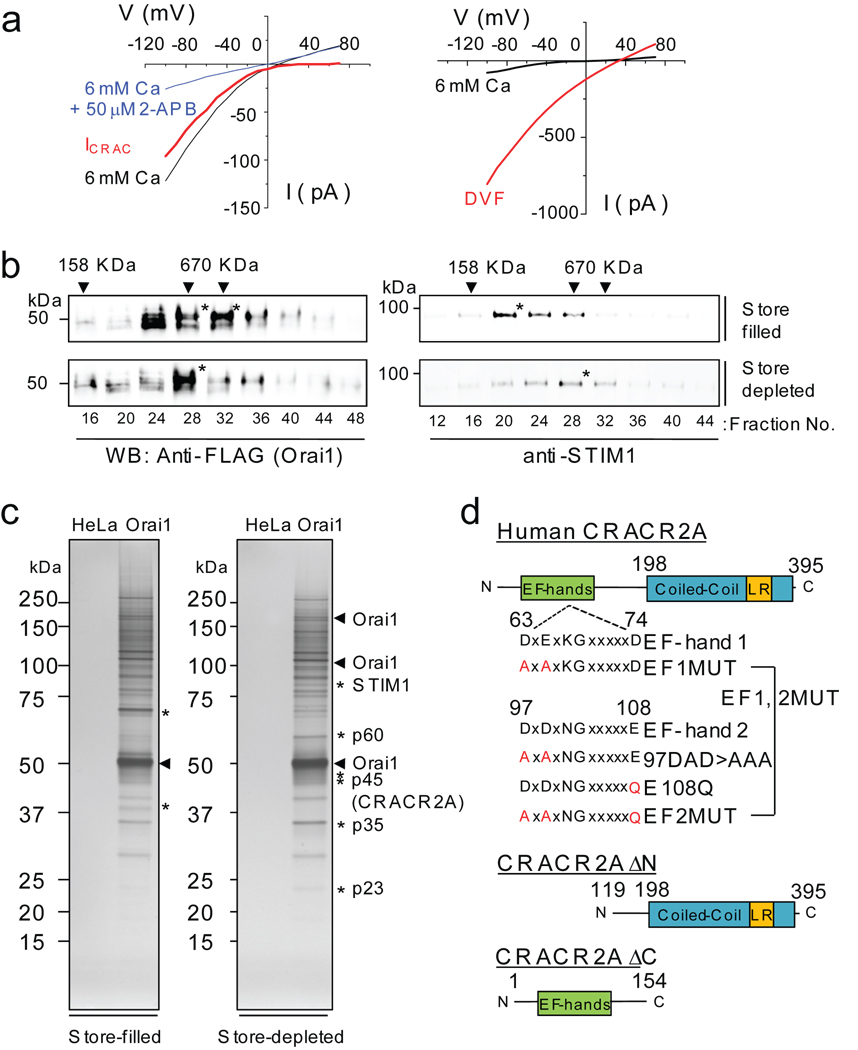
To identify novel regulators of the CRAC channel using Orai1 for affinity purification, we generated HeLa cells stably expressing Orai1 and STIM1 (HeLa O+S cells). Presence of an active CRAC channel complex was verified by detection of amplified CRAC currents (Fig. 1a, Supplementary Information, Fig. S1a)19–21. To capture Orai1 in its native complex, cells were treated with different concentrations of a membrane-permeable cross-linker, dithiobis succinimidyl propionate (DSP) and immunoblotted for Orai1. Upon treatment with 0.5 mM DSP, Orai1 and STIM1 were detected in high molecular weight complexes in non-reducing SDS-PAGE (Supplementary Information, Fig. S1b). These complexes were applied onto a 20–50% glycerol gradient to determine their size. The size of the Orai1 protein complex was estimated to be ~700 kDa under resting conditions and ~670 kDa after store depletion (Fig. 1b). Under resting conditions, STIM1 was primarily detected in a ~200 kDa protein complex (Fig. 1b, right), possibly as a dimer, whereas it co-migrated with Orai1 upon store depletion. These results are consistent with the observation that STIM1 self-associates at rest and forms a high molecular weight (MW) protein complex upon stimulation9, 30, 31.
Figure 1.
Identification of CRACR2A as a binding partner of Orai1 by large-scale affinity purification. (a) CRAC currents measured in HeLa cells stably expressing Orai1 and STIM1 (HeLa O+S cells). Left panel shows inwardly rectifying CRAC currents (red trace) obtained by subtracting 2-APB inhibited currents (blue trace) from whole-cell currents (black trace). The pipette solution contained 12 mM EGTA to deplete the intracellular Ca2+ stores and the external solution contained 6 mM CaCl2. Right panel shows current-voltage relationships of the currents in divalent free (DVF, red trace) or 6 mM CaCl2 (black) containing solution. (b) Glycerol gradient fractionation of DSP cross-linked HeLa O+S cells in store-filled (top two panels) and store-depleted (bottom two panels) conditions. Different fractions were separated on SDS-PAGE and immunoblotted for detection of Orai1 (left) and STIM1 (right). * represents fractions enriched in Orai1 or STIM1 proteins. Arrowheads denote the fractions in which MW markers were detected. For full scans see Supplementary Information, Fig. S12. (c) Affinity purification of Orai1 protein complex. The glycerol gradient fractions enriched in FLAG-Orai1 were pooled and immunoprecipitated with anti-FLAG resin. After elution with the FLAG peptide, fractions were separated on SDS-PAGE and visualized by silver staining. * indicates protein bands enriched in store-filled or store-depleted conditions. (d) Schematic showing the predicted domain structure of human CRACR2A protein. Human CRACR2A contains 395 amino acids with two predicted EF-hand motifs (SMART program) in its N terminus and a predicted coiled-coil domain with leucine rich (LR) sequence in its C terminus (Human Protein Reference Database and COILS). The mutants used in the current study are indicated.
Identification of CRACR2A as a binding partner of Orai1 and STIM1
To identify the binding partners of Orai1, fractions containing Orai1 were pooled, immunoprecipitated with anti-FLAG antibody coated resin and eluted with the FLAG peptide. Samples from HeLa cells showed very few non-specific bands, while those from HeLa O+S cells exhibited a number of specific bands (Fig. 1c). Enrichment of different size bands in resting or store-depleted conditions suggest presence of discrete binding partners of Orai1 (Fig. 1c, denoted by *). Specific bands after store depletion were excised and analyzed by mass spectrometry. Detection of STIM1 peptides served as a positive control (>17 peptides). In addition to STIM1, p45 renamed as CRACR2A (CRAC regulator 2A, EFCAB4B, FLJ33805, FLJ33046; amino acid sequence of the transcript NM_032680) was identified as a binding partner of Orai1. The N terminus of CRACR2A contains two predicted EF-hands while its C terminus has a predicted coiled-coil domain as well as a leucine-rich motif that is highly conserved among vertebrates (Fig. 1d and Supplementary Information, Fig. S2). Fluorescence protease protection assays32, 33 showed a predominantly cytoplasmic localization of CRACR2A (Supplementary Information, Fig. S3).
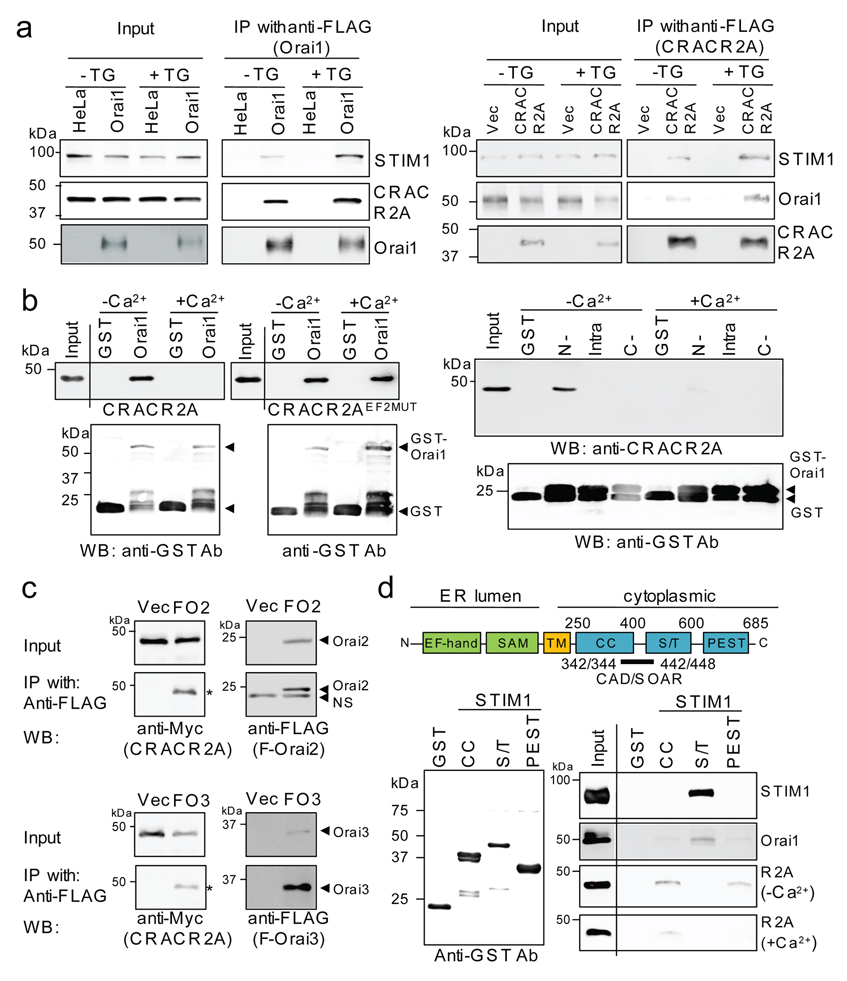
To examine whether CRACR2A binds Orai1, we carried out immunoprecipitation and pulldown analysis. For immunoprecipitation experiments, HeLa cells stably expressing either STIM1 alone or STIM1 and Orai1 proteins were transiently transfected with a plasmid encoding CRACR2A. As seen in Fig. 2a, both STIM1 and CRACR2A co-immunoprecipitated with Orai1 and showed enhanced binding upon store depletion. These results were further validated by reverse immunoprecipitation using FLAG-tagged CRACR2A (Fig. 2a, right). To test for a direct interaction between CRACR2A and Orai1, recombinant proteins purified from E. coli (Supplementary Information, Fig. S4) were used for in vitro binding assays. GST pulldown analysis showed a strong interaction between CRACR2A and Orai1 in the absence of Ca2+ while the binding was minimal in the presence of 2 mM Ca2+ (Fig. 2b, first panel). To confirm an inhibitory role of Ca2+, we used an EF-hand mutant of CRACR2A that abolishes Ca2+ binding (CRACR2AEF2MUT, Fig. 1d and see below). As shown in Fig. 2b, Ca2+ had no effect on Orai1-CRACR2AEF2MUT interaction, supporting its inhibitory role.
Figure 2.
CRACR2A directly interacts with Orai and STIM1. (a) CRACR2A co-immunoprecipitates with Orai1 and STIM1. Left two panels: HeLa cells stably expressing either STIM1 (HeLa), or STIM1 and Orai1 (Orai1) were transfected with a plasmid encoding CRACR2A. After immunoprecipitation with anti-FLAG resin (Orai1) and elution with the FLAG peptide, eluted fractions were immunoblotted using anti-STIM1, anti-CRACR2A, and anti-FLAG (Orai1) antibodies. Right two panels: HeLa cells stably expressing STIM1 were transfected with plasmids encoding Myc-Orai1 alone (Vec) or together with FLAG-CRACR2A (CRACR2A). After immunoprecipitation with anti-FLAG resin (CRACR2A), precipitates were immunoblotted with anti-STIM1, anti-Myc (Orai1), and anti-FLAG (CRACR2A) antibodies. (b) CRACR2A directly binds to the N terminus of Orai1. Left panels: GST pulldown analysis was performed between full-length GST-Orai1 and 6× His-tagged CRACR2A or CRACR2AEF2MUT proteins in the presence or absence of 2 mM CaCl2 (left panels). Lower panel shows input levels of GST-Orai1. Right panels: pulldown analysis with GST-fused fragments of the N terminus (N-, amino acids 64–91), intracellular loop (intra, amino acids 137–173) and the C terminus (C-, amino acids 254–301) of Orai1 and full-length 6× His-tagged CRACR2A. Lower panel shows input levels of Orai1 fragments. All the recombinant proteins were purified from E. coli. (c) CRACR2A interacts with Orai2 and Orai3 proteins. HeLa cells stably expressing STIM1 were transfected with plasmids encoding FLAG-Orai2 (FO2, top) or FLAG-Orai3 (FO3, bottom) together with Myc-CRACR2A. Lysates were immunprecipitated with anti-FLAG resin and immunoblotted for detection of CRACR2A (anti-Myc, left, indicated with *) and Orai2 or Orai3 (anti-FLAG, right). NS, non-specific band. (d) Interaction between the cytoplasmic fragments of STIM1 and CRACR2A. A schematic of STIM1 with cytoplasmic domains of coiled-coil (CC, amino-acid residues 250–400), serine/threonine rich sequence (S/T, residues 400–600), and PEST sequence (residues 600–685) is shown. The CAD/SOAR fragment is indicated. Purified GST-fused STIM1 fragments were immunoblotted with anti-GST antibodies (left). Their interactions with purified full-length Orai1, CRACR2A, or STIM1 proteins were assessed by pulldown analysis in the absence (−Ca2+) or presence (+Ca2+) of 2 mM CaCl2. STIM1 fragments and CRACR2A were purified from E. coli, while full-length Orai1 and STIM1 proteins were purified from baculovirus infected insect cells. For full scans see Supplementary Information, Fig. S12.
For domain interaction studies, we purified GST-fused fragments of the N, C termini and the intracellular loop of Orai1 (Fig. 2b, right and Supplementary Information, Fig. S4d). The N terminal fragment of Orai1 consisted of the amino acids 64–93, since truncation of first 63 residues did not alter CRACR2A binding (data not shown). These interaction studies showed that CRACR2A binds to the N terminus of Orai1 in a Ca2+-sensitive manner, with elevated Ca2+ favoring its dissociation from Orai1 (Fig. 2b, right panel). Since amino acids 64–93 of Orai1 are highly conserved among Orai family, we examined interaction of CRACR2A with Orai2 and Orai3. CRACR2A co-immunoprecipitated with Orai2 and Orai3 suggesting a conserved binding mechanism with all the Orai proteins (Fig. 2c). Since CRACR2A co-immunoprecipitated with Orai1 and STIM1, we hypothesized that Orai1, STIM1 and CRACR2A may form a ternary complex though direct interaction. To examine a direct interaction of STIM1 with CRACR2A, three different GST fusion proteins of the cytoplasmic domain of STIM1, corresponding to coiled-coil domain (CC), serine/threonine (S/T) rich sequence, and proline/lysine rich sequence (PEST) were examined (Fig. 2d). Full-length CRACR2A bound to the CC and PEST domains of STIM1 and the interaction was reduced in the presence of Ca2+. On the other hand, the S/T rich sequence, which did not bind to CRACR2A, was identified as a STIM1 multimerization and Orai1 interaction domain (Fig. 2d). This sequence partially overlaps with the recently identified CAD/SOAR domains (amino acids 342–448) of STIM1, which directly interact with Orai122–24. Together these data suggest that CRACR2A binds to both Orai1 and STIM1 in a Ca2+-sensitive manner.
Identification of a CRACR2A family member, CRACR2B
BLAST and Homologene analysis revealed a homologue of CRACR2A, renamed as CRACR2B (EFCAB4A, FLJ46033). NCBI depicts one transcript encoded by the human CRACR2B gene (NM_173584) while Ensembl shows four transcripts (ENST00000321883, 327417, 450448, 450828) with different intron-exon architecture. To characterize the mRNA sequence of human CRACR2B, we cloned its full-length cDNA from Jurkat T cells (registered as GenBank accession number, GU952799). Our sequencing analysis revealed similar intron-exon architecture between human and mouse CRACR2B (Supplementary Information, Fig. S5). Sequence alignment of human CRACR2A and CRACR2B proteins showed an overall identity of 36% (pair-wise BLAST analysis) with a conserved primary structure of predicted EF-hand motifs and a coiled-coil domain.
CRACR2 proteins play an important role in SOCE
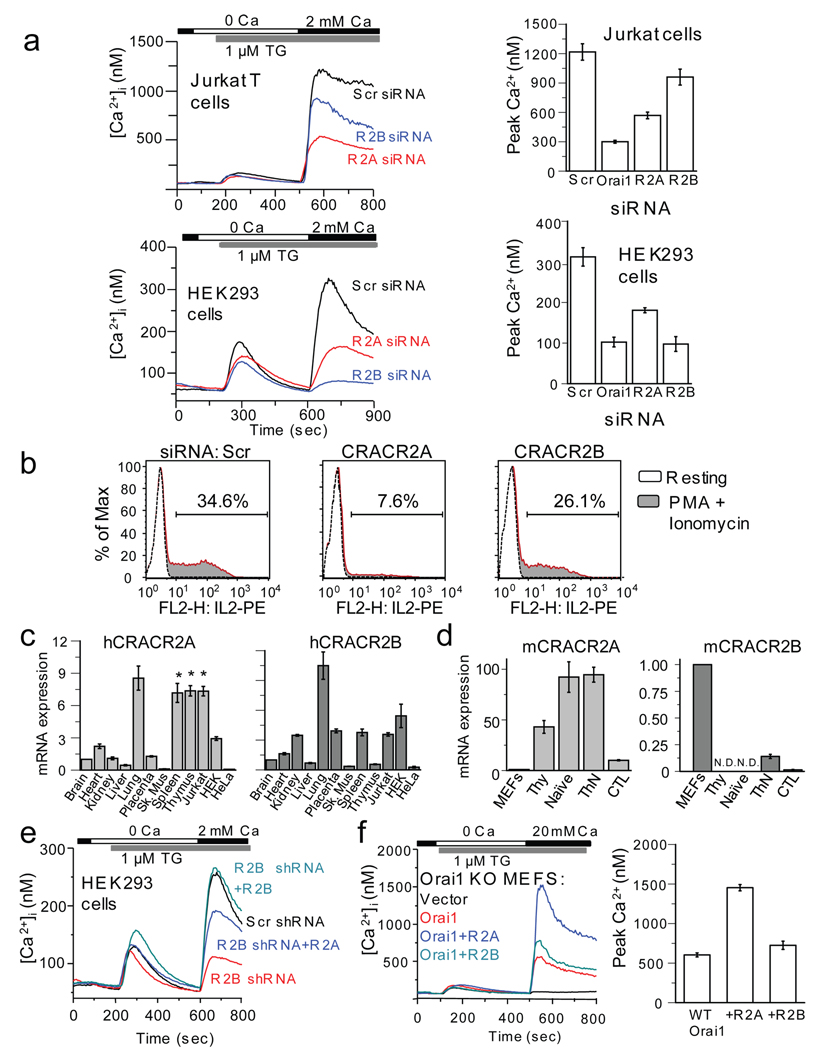
To examine the physiological role of CRACR2 proteins, we measured SOCE in Jurkat T cells upon siRNA-mediated depletion of CRACR2A or CRACR2B. As seen in Fig. 3a, depletion of CRACR2A decreased SOCE by 50% while that of CRACR2B had a mild effect. Consistent with a decrease in SOCE, depletion of CRACR2A resulted in a stronger impairment in IL-2 production by Jurkat T cells than CRACR2B knockdown (Fig. 3b). Surprisingly, in HEK293 cells, depletion of CRACR2B had a stronger effect on SOCE than that of CRACR2A (Fig. 3a, bottom). Specificity and efficiency of CRACR2 knockdown was examined by RT-PCR and immunoblotting (Supplementary Information, Fig. S6a–d). To determine the reason for the differences between Jurkat and HEK293 cells, we measured the expression levels of CRACR2 transcripts in various human tissues and cell lines. Quantitative RT-PCR analysis revealed abundant expression of CRACR2A transcripts in spleen and thymus distinct from CRACR2B (Fig. 3c). Among cell lines, CRACR2A transcripts were higher in Jurkat T cells when compared with HEK293 or HeLa cells, while those of CRACR2B were higher in HEK293 cells (Fig. 3c). Since CRACR2A transcripts were abundant in Jurkat T cells, we examined their expression in primary T cells. Murine thymocytes, naïve and effector CD4+ T cells showed high expression of CRACR2A transcripts when compared with murine embryonic fibroblasts (MEFs), while CRACR2B transcripts were very low (Fig. 3d). These data suggest that the relative contribution of CRACR2A and CRACR2B in SOCE in T cells versus HEK293 cells is partially due to differences in their expression levels.
Figure 3.
CRACR2A plays an important role in Orai1-mediated SOCE in T cells. (a) SOCE measurements in Jurkat and HEK293 cells depleted of CRACR2A and CRACR2B. Top panel, averaged responses from Jurkat T cells: scrambled (Scr, n = 75 cells), CRACR2A (R2A, n = 79), or CRACR2B (R2B, n = 70) siRNAs. Bottom, averaged responses from HEK293 cells: Scr (n = 41), R2A (n = 47) or R2B (n = 49). Bar graphs represent averaged peak [Ca2+]i ± s.e.m. from three independent experiments. (b) Measurement of IL-2 expression in Jurkat T cells transfected with siRNAs. A representative of three independent experiments is shown. (c) Real-time PCR analysis of human CRACR2A and CRACR2B transcripts from various tissues and cell lines. Normalized mRNA levels are plotted relative to those of brain tissue. Data represent average ± s.d. from 2 independent experiments performed in triplicate. * indicates tissues or cell-lines showing high expression of CRACR2A transcripts, distinct from CRACR2B. (d) Expression of CRACR2A and CRACR2B in murine primary cells. The mRNA levels of CRACR2A or CRACR2B were measured from mouse embryonic fibroblasts (MEFs), thymocytes (Thy), naïve CD4+ T cells (Naïve), effector CD4+ (ThN), or CD8+ (CTL) T cells. Normalized mRNA levels are plotted relative to those of MEFs. N.D., not detected. Data represent average ± s.d. from 2 independent experiments performed in triplicate. (e) Examination of functional redundancy between CRACR2 proteins. SOCE was measured in HEK293 cells stably expressing control (Scr, black trace, n = 46 cells) or CRACR2B shRNA (red, n = 49). CRACR2B-depleted cells with ectopic expression of CRACR2A (R2A, blue, n = 41) or CRACR2B (R2B, cyan, n = 43) were examined for SOCE. A representative of three independent experiments is shown. The plasmids contain IRES-GFP and GFP+ cells were selected for analysis. (f) Effect of CRACR2 protein expression on Orai1-mediated SOCE. SOCE was measured in Orai1-null MEFs expressing CRACR2A or CRACR2B together with Orai1. Each trace shows averaged responses from 25 (vector), 30 (Orai1), 35 (Orai1+R2A) or 33 (Orai1+R2B) MEFs. The bar graph shows averaged peak [Ca2+]i ± s.e.m. from three independent experiments.
To check for a possible functional redundancy between CRACR2 proteins, we used HEK293 cells stably depleted of CRACR2B by shRNA and ectopically expressed CRACR2A or CRACR2B (Supplementary Information, Fig. S6e). Expression of CRACR2B completely restored SOCE (Fig. 3e, cyan trace) validating the specificity of knockdown. Under these conditions, expression of CRACR2A significantly restored SOCE, indicating a functional redundancy between CRACR2 proteins (Fig. 3e, blue trace). To examine whether both CRACR2 proteins can modulate Orai1 activity, we expressed CRACR2A or CRACR2B in Orai1-null MEFs in the absence or presence of Orai1. Orai1-null MEFs show very low residual SOCE, which is restored to wild-type levels by exogenous expression of Orai1 (Fig. 3f, black and red traces)34. Expression of CRACR2A or CRACR2B alone did not restore SOCE in Orai1-null MEFs (Supplementary Information, Fig. S6f). However, co-expression of CRACR2A with Orai1 increased the peak [Ca2+]i by more than twofold compared with expression of Orai1 alone (Fig. 3f, bar graph). In contrast, co-expression of CRACR2B with Orai1 had a very mild effect on SOCE. These data suggest a specific role of CRACR2A in enhancement of Orai1-mediated SOCE that is distinct from CRACR2B.
CRACR2A mediates clustering of Orai1 and STIM1 at PM-ER junctions
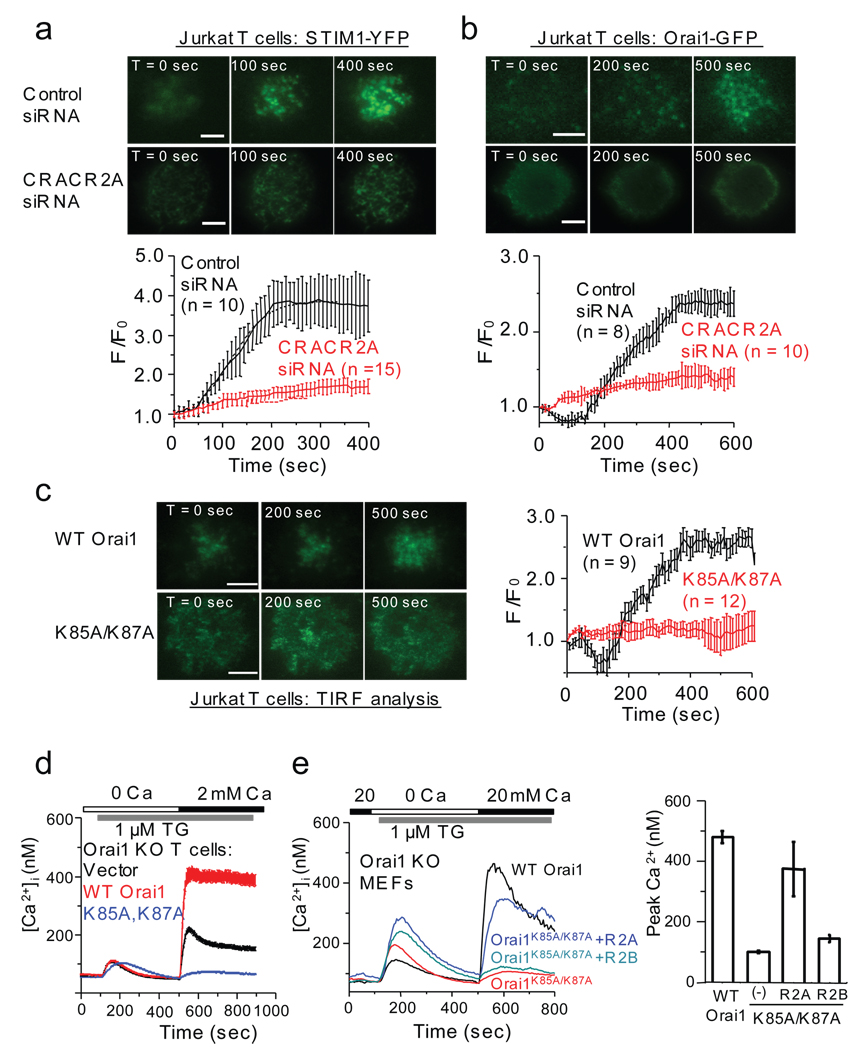
STIM1 is involved in clustering and gating of Orai1 and evokes large CRAC currents when co-expressed with Orai135–38. In contrast, CRACR2A did not induce large CRAC currents when co-expressed with Orai1 (data not shown), indicating that CRACR2A cannot substitute for STIM1. We hypothesized that binding of CRACR2A to Orai1 and STIM1 may stabilize their interaction in PM-proximal area, and consequently its depletion may impair clustering of Orai1 and STIM1. To examine the role of CRACR2A in STIM1 clustering, we transfected control or CRACR2A-depleted Jurkat T cells with STIM1-YFP and measured its accumulation into PM-proximal regions using total internal reflection fluorescence (TIRF) microscopy. Upon stimulation, STIM1-YFP clustered within ten minutes in control cells (Fig. 4a, top), but in CRACR2A-depleted cells clustering was severely reduced (Fig. 4a, bottom). To investigate the effects of CRACR2A on clustering of Orai1, we transfected control and CRACR2A-depleted Jurkat T cells with plasmids encoding Orai1-GFP and STIM1-mCherry in an equimolar ratio. Orai1 formed clusters upon thapsigargin treatment in control cells while CRACR2A depletion impaired this process (Fig. 4b). These data suggest that CRACR2A is important for clustering of Orai1 and STIM1.
Figure 4.
CRACR2A is essential for cluster formation of Orai1 and STIM1 in T cells. (a) TIRF microscopy images of STIM1-YFP in Jurkat T cells transfected with either control (top panel) or CRACR2A siRNA (bottom panel). Cells were imaged in Ca2+ free Ringer solution and ER Ca2+ was depleted with 1 µM of thapsigargin at the initial time point (T = 0). The graph below represents an average of normalized fluorescence intensity ± s.e.m. from measurements of 10 cells transfected with control siRNA (black) and 15 cells transfected with CRACR2A siRNA (red). Bar, 5 µm. (b) Clustering of Orai1-GFP in Jurkat T cells co-expressing STIM1-mCherry. Cells were transfected with control siRNA (top panel) or CRACR2A siRNA (bottom panel). Jurkat cells expressing both Orai1 and STIM1 proteins were imaged by TIRF microscopy. The graph below represents an average of normalized fluorescence intensity ± s.e.m. from measurements of 8 cells transfected with control siRNA and 10 cells transfected with CRACR2A siRNA. Bar, 5 µm. (c) TIRF microscopy analysis of Orai1K85A/K87A-GFP in Jurkat T cells. Jurkat T cells expressing STIM1-mCherry and wild type Orai1-GFP or Orai1K85A/K87A-GFP were imaged. The graph represents an average of normalized fluorescence intensity ± s.e.m. from 9 cells expressing Orai1-GFP and 12 cells expressing Orai1K85A/K87A-GFP. Bar, 5 µm. (d) Reconstitution of SOCE in Orai1-null CD4+ T cells by expression of WT Orai1 or Orai1K85A/K87A. T cells transduced with retroviral vectors expressing WT Orai1 or Orai1K85A/K87A together with GFP from an IRES site were examined for SOCE. Each trace shows average ± s.e.m. from 70 (vector), 77 (WT Orai1), or 79 (K85A, K87A) GFP+ cells. A representative of three independent experiments is shown here. (e) Recovery of defect in SOCE of Orai1K85A/K87A mutant by co-expression of CRACR2 proteins. Orai1-null MEFs were transduced with retroviruses encoding CRACR2A or CRACR2B together with Orai1K85A/K87A for measurement of SOCE. Each trace shows averaged responses from 35 (Orai1K85A/K87A), 32 (WT Orai1), 30 (Orai1K85A/K87A + R2A) or 39 (Orai1K85A/K87A + R2B) GFP+ MEFs. The bar graph shows averaged peak [Ca2+]i ± s.e.m. from three independent experiments.
To identify the residues in Orai1 N terminus necessary for CRACR2A binding, we generated mutants of amino-acids conserved in Orai family (Supplementary Information, Fig. S7a). Particularly, we focused on positively charged residues since CRACR2A contains many negatively charged residues (Supplementary Information, Fig. S2). GST pulldown assays revealed an important role of positively charged residues in binding to CRACR2A while mutations of conserved serine residues in Orai1 did not alter binding (Supplementary Information, Fig. S7b). The substitutions of lysines at positions 85 and 87 to alanine (K85A and K87A) significantly decreased interaction with CRACR2A. Pulldown and immunoprecipitation with a K85A/K87A double mutant of Orai1 showed very low residual binding to CRACR2A (Supplementary Information, Fig. S7b, c). To examine whether Orai1K85A/K87A showed impairment in clustering, we expressed Orai1K85A/87A-GFP with STIM1 in Jurkat T cells. While WT Orai1 readily clustered upon store depletion, Orai1K85A/K87A showed impairment (Fig. 4c). To assess the functional consequence of impaired clustering, we measured SOCE in Orai1-null T cells expressing Orai1K85A/K87A. Orai1K85A/K87A could not restore SOCE in Orai1-null CD4+ T cells (Fig. 4d). Moreover, it had a dominant negative effect over the residual SOCE, suggesting that the residual SOCE may be contributed by other members of the Orai family and hetero-multimerization with Orai1K85A/K87A may influence their function. Membrane insertion and expression levels of the Orai1K85A/K87A mutant were similar to WT (Supplementary Information, Fig. S7c and S8). To examine whether CRACR2A co-expression can overcome the inability of Orai1K85A/K87A to rescue SOCE, we generated Orai1-null MEFs stably co-expressing Orai1K85A/K87A with either CRACR2A or CRACR2B. Indeed, co-expression of CRACR2A with Orai1K85A/K87A showed significant recovery of SOCE, while that of CRACR2B had a marginal effect (Fig. 4e). These experiments suggest that CRACR2A physically interacts with the positively charged residues in the N terminus of Orai1 and this interaction is important for clustering and activity of Orai1.
The EF-hands of CRACR2A mediate Ca2+-sensitive interaction with Orai1 and STIM1
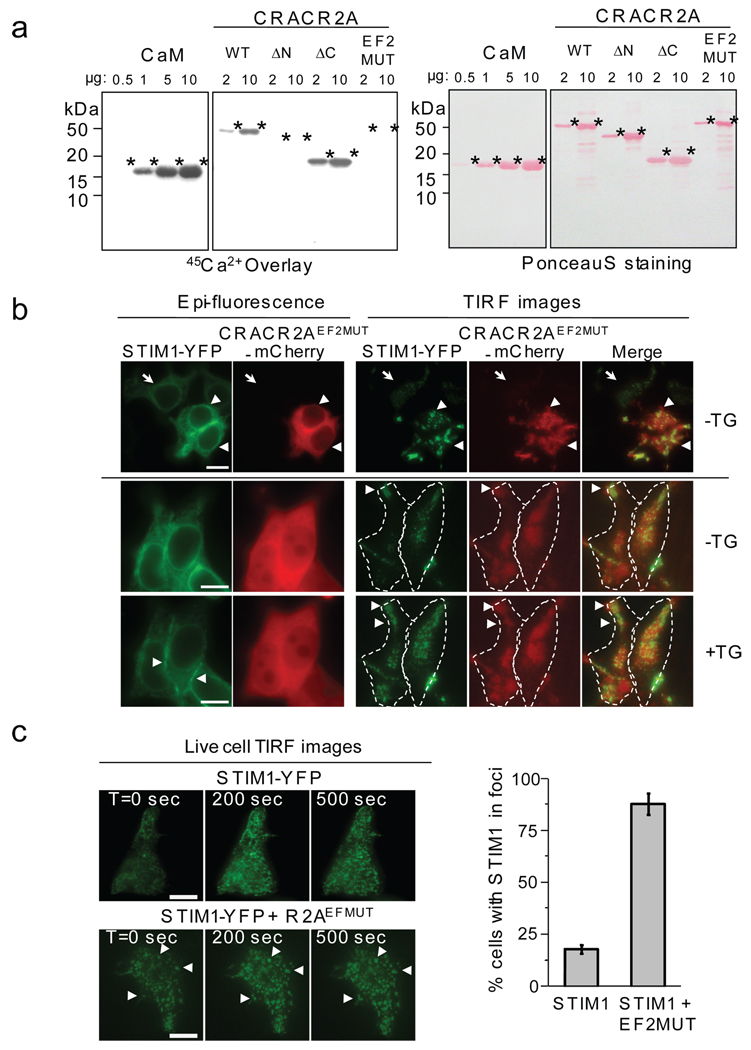
The EF-hands are highly conserved among CRACR2 family (Supplementary Information, Fig. S2 and S5). To test whether these predicted EF-hands bind Ca2+, 45Ca2+ overlay analysis was performed using recombinant full-length and various mutants of CRACR2A together with calmodulin (CaM) as a positive control. WT CRACR2A and ΔC mutant showed binding to Ca2+ while truncation of the N terminus or mutations in the second EF-hand abolished Ca2+ binding (Fig. 5a). These results together with those in Fig. 2 suggest that the EF-hand motifs of CRACR2A bind to Ca2+ and Ca2+ binding reduces its interaction with Orai1 and STIM1.
Figure 5.
An EF-hand mutant of CRACR2A causes spontaneous clustering of STIM1. (a) CRACR2A binds Ca2+ via its EF-hands. 45Ca2+ overlay experiments were performed with purified full-length CRACR2A, CRACR2A truncated in its N terminus (ΔN, amino acids 119–395), C terminus (ΔC, amino acids 1–154), or mutated in its EF-hand2 (EF2MUT). 2 or 10 µg of each protein were used to examine Ca2+ binding. Similar amount of purified calmodulin (CaM) was run (left panel) to compare Ca2+ binding affinity. Compare Ca2+ binding of CaM with ΔC mutant of CRACR2A for similar molar concentrations of each protein. Panels on the right show Ponceau S staining of the same blots to compare protein amounts. (b) Live cell epifluorescence and TIRF microscopy analysis of HEK293 cells expressing STIM1-YFP and CRACR2AEF2MUT-mCherry in the absence or presence of store depletion. Left panels show epifluorescence images from the middle of the cell and right panels show TIRF images from the footprint of the cell. Arrowheads in the top panel show clustering of STIM1-YFP in cells co-expressing CRACR2AEF2MUT. An arrow in the same panel depicts minimal clustering in a cell expressing only STIM1-YFP. Arrowheads in the bottom TIRF panels show co-accumulation of CRACR2AEF2MUT-mCherry with STIM1-YFP at sites of newly formed clusters after store depletion (+TG). Bar, 10 µm. (c) TIRF microscopy analysis of HEK293 cells expressing STIM1-YFP in the absence (top) or presence (bottom) of CRACR2AEF2MUT. Cells co-expressing mCherry (top) or CRACR2AEF2MUT-mCherry (bottom) were selected. CRACR2AEF2MUT induced clustering of STIM1-YFP without ER Ca2+ depletion (compare images at T = 0 s in the top and bottom panels). Thapsigargin was added at T = 0 s and images of STIM1 clustering are shown at 200 and 500 s. Arrowheads represent preformed clusters of STIM1 that expand upon store depletion. Bar graph depicts averaged number of HEK293 cells (± s.d.) showing STIM1 clustering in the presence (n = 112 cells) or absence (n = 106) of CRACR2AEF2MUT. Cells moderately expressing STIM1-YFP were examined. Bar, 10 µm.
Our previous data showed CRACR2A depletion reduced clustering of STIM1 and Orai1 (Fig. 4a, b). To examine whether co-expression of WT or Ca2+-insensitive CRACR2A (CRACR2AEF2MUT) influences STIM1 clustering, we examined STIM1 localization in HEK293 cells co-expressing CRACR2AEF2MUT-mCherry using TIRF microscopy. Cells expressing STIM1-YFP showed minimal clustering under resting conditions (Fig. 5b, top, arrow). However, in cells co-expressing CRACR2AEF2MUT-mCherry, STIM1 was clustered to varying degrees even without store depletion (see two separate examples in the top and middle panels of Fig. 5b). Upon store depletion, clusters of STIM1 expanded to become larger aggregates (Fig. 5b and c). As mentioned previously (Supplementary Information, Fig. S3), CRACR2A is a predominantly cytoplasmic protein and was detected everywhere in the footprint of the cell (Fig. 5b). Interestingly, we could detect preferential accumulation of CRACR2AEF2MUT at sites of STIM1 clustering upon store depletion (Fig. 5b, middle and bottom panels of TIRF images, arrowheads). Visual examination of cells expressing STIM1 and CRACR2AEF2MUT revealed more than 80% cells with STIM1 in clusters (Fig. 5c). These results were confirmed independently by using live cell confocal microscopy (Supplementary Information, Fig. S9). In addition, biochemical analysis also showed STIM1 oligomerization mediated by CRACR2A overexpression. Upon co-expression of CRACR2A, both STIM1 and CRACR2A could readily be detected in high MW complexes without store depletion (Supplementary Information, Fig. S10a, b). To investigate whether the spontaneous clusters of STIM1 upon co-expression of CRACR2A mediate Ca2+ entry, we measured [Ca2+]i in HEK293 cells co-expressing STIM1 and various mutants of CRACR2A in the presence of 0.1 or 10 mM extracellular [Ca2+]. Co-expression of CRACR2A with STIM1 increased [Ca2+]i and the increase was higher in cells expressing CRACR2AEF2MUT (Supplementary Information, Fig. S10c), suggesting that the clustered STIM1 is functionally active.
CRACR2A regulates CRAC channel-mediated Ca2+ entry and cell death in T cells
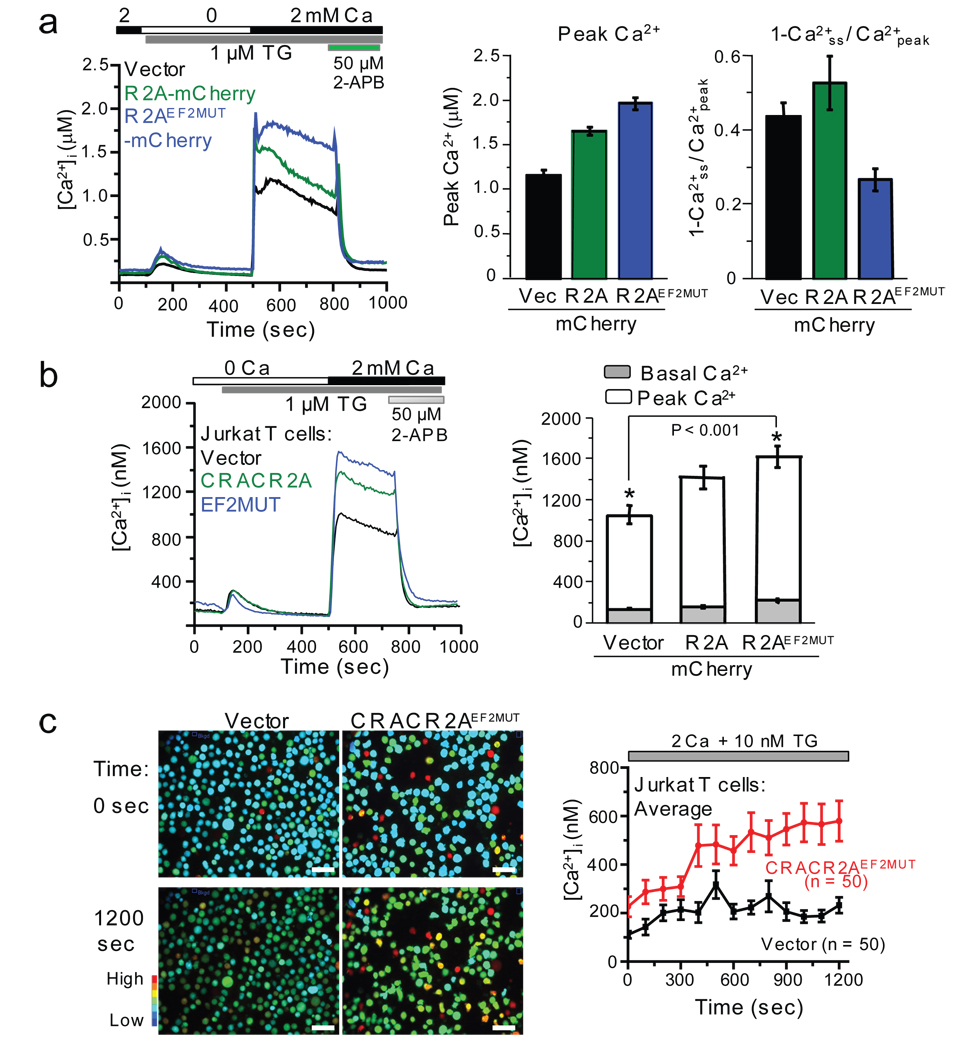
To examine how CRACR2A influences Orai1-mediated SOCE, we measured SOCE in HeLa O+S cells expressing mCherry, CRACR2A-mCherry, or CRACR2AEF2MUT-mCherry. CRACR2A expression significantly increased the peak [Ca2+]i, while CRACR2AEF2MUT-mCherry increased both peak and sustained [Ca2+]i (Fig. 6a). To examine its effect on endogenous CRAC channel function, we measured SOCE in Jurkat T cells expressing WT or mutant CRACR2A. Expression of CRACR2A as well as CRACR2AEF2MUT enhanced SOCE in Jurkat T cells, with the latter having a stronger effect (Fig. 6b). We also observed a significant increase in resting [Ca2+]i in Jurkat cells expressing CRACR2AEF2MUT (Fig. 6b, right panel and Fig. 6c), supporting our observations in HEK293 cells.
Figure 6.
CRACR2A regulates SOCE and CRAC channel-mediated Ca2+ oscillations in T cells. (a) Measurement of SOCE in HeLa O+S cells expressing CRACR2A. Averaged responses from HeLa O+S cells expressing mCherry (n = 33 cells), CRACR2A-mCherry (n = 36), or CRACR2AEF2MUT-mCherry (n = 39) are shown. Bar graph shows peak [Ca2+]i values immediately upon addition of 2 mM Ca2+ (Left). A decrease in sustained [Ca2+]i is plotted as 1-Ca2+ss / Ca2+peak, where Ca2+ss represents steady state [Ca2+]i at 800 sec and Ca2+peak represents the peak [Ca2+]i (right). Bar graphs show average ± s.e.m. from three independent experiments. (b) Measurement of SOCE in Jurkat T cells expressing CRACR2A. Averaged responses from cells expressing mCherry (n = 55 cells), CRACR2A-mCherry (n = 50), or CRACR2AEF2MUT-mCherry (n = 60) are shown. Bar graph depicts average ± s.e.m. of [Ca2+]i before (basal, gray bars) and after (peak, open bars) store depletion from three independent experiments. * represents statistically significant differences in resting and stimulated [Ca2+]i (P < 0.001 by t-test). (c) Expression of CRACR2AEF2MUT in Jurkat T cells disrupts normal Ca2+ oscillations induced by thapsigargin. Jurkat T cells expressing mCherry or CRACR2AEF2MUT-mCherry were treated with 10 nM thapsigargin in 2 mM Ca2+ containing Ringer solution to induce asynchronous [Ca2+]i oscillations. Left panel shows pseudocolored images of transfected cells for [Ca2+]i (>70% transfection efficiency in each case) and right panel shows the traces of [Ca2+]i oscillations averaged from 50 cells. Data are representative of three independent experiments. Bar, 50 µm.
In T cells, asynchronous [Ca2+]i oscillations evoked by T cell receptor signaling are important for activation of NFAT and NF-κB pathways39, 40. These oscillations can also be induced by low concentrations of thapsigargin41. Consistent with previous observations41, control Jurkat T cells showed asynchronous oscillations with an average frequency of 4–6 mHz (Supplementary Information, Fig. S11a) and a marginal increase in average responses from 50 cells (Fig. 6c). Interestingly, cells expressing CRACR2AEF2MUT showed higher basal [Ca2+]i which increased progressively upon treatment with 10 nM thapsigargin (Fig. 6c). Population average responses were threefold higher in the presence of CRACR2AEF2MUT when compared with control cells. The oscillations in control cells and cells expressing CRACR2AEF2MUT were blocked by 2-APB, implying a role of CRAC channel in this function as previously reported41 (Supplementary Information, Fig. S11b and data not shown).
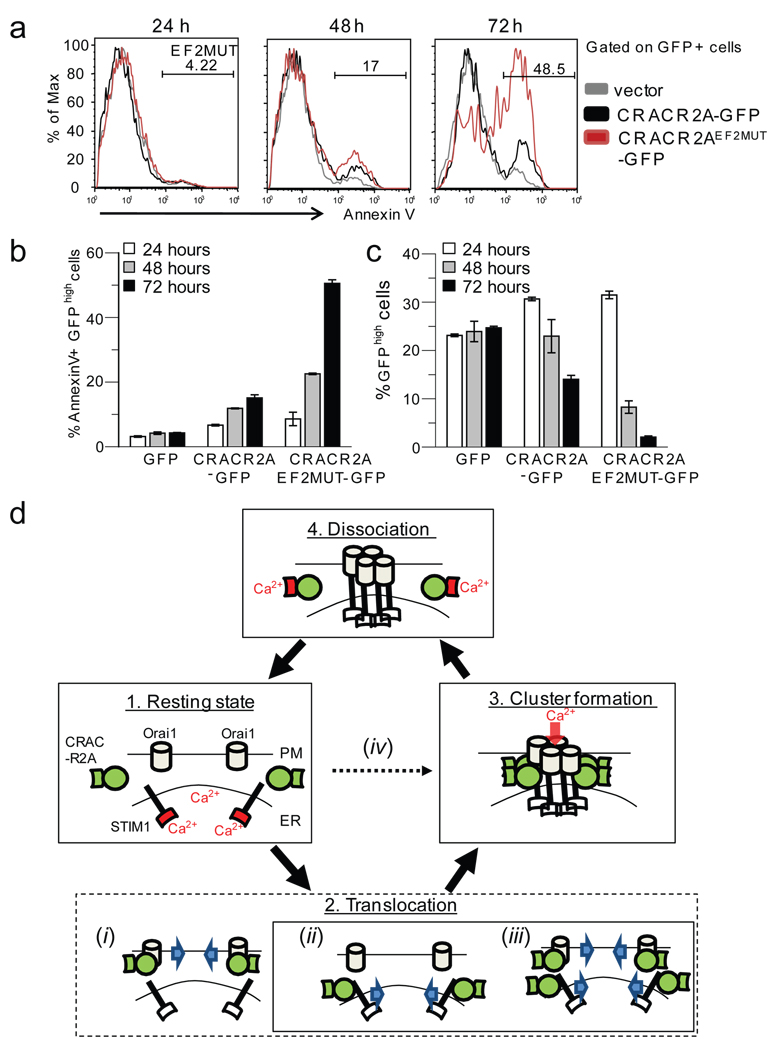
Since overexpression of CRACR2AEF2MUT increased basal [Ca2+]i, we measured its effect on cell death in T cells. Jurkat T cells expressing CRACR2A-GFP or CRACR2AEF2MUT-GFP showed twofold and fivefold increase in apoptotic cell population respectively, when compared with GFP-expressing cells at 72 h post transfection (Fig. 7a, b). As a result, population of cells expressing CRACR2A-GFP or CRACR2AEF2MUT-GFP drastically decreased by 72 h while that of control cells remained unchanged (Fig. 7c). These results suggest that CRACR2A plays a pivotal role in regulation of [Ca2+]i and an uncontrolled increase in [Ca2+]i upon expression of CRACR2AEF2MUT can result in cell death.
Figure 7.
Expression of EF-hand mutant of CRACR2A induces cell death in Jurkat T cells. (a) Cell death induced by CRACR2A overexpression in T cells. Jurkat T cells expressing GFP, CRACR2A-GFP or CRACR2AEF2MUT-GFP were examined for cell death by Annexin V staining at 24, 48, and 72 h after transfection. One representative from three independent experiments is shown. (b) Quantification of the data shown in (a). Bar graphs represent average ± s.e.m. from three independent experiments. (c) Live cell population is reduced in Jurkat cells expressing CRACR2AEF2MUT-GFP. Jurkat T cells expressing GFP, CRACR2A-GFP or CRACR2AEF2MUT-GFP were assessed for percent of live, GFP-positive populations at 24, 48 and 72 h after transfection. Cells with high expression of GFP (GFPhigh) were analyzed by flow cytometry. Data represents average ± s.e.m. from three independent experiments. (d) A proposed model showing possible role(s) of CRACR2A in CRAC channel function. Under resting conditions, Orai1 and STIM1 are distributed at the PM and ER membranes, respectively (1. Resting state) while CRACR2A (green) localizes in the cytoplasm. Upon store depletion, Orai1 and STIM1 translocate to form clusters at the junctional regions between PM and ER (State 2). CRACR2A may either be actively involved in translocation of Orai1 (i), STIM1 (ii) or both (iii). It is also possible that CRACR2A passively interacts with Orai1 and STIM1 at sites of clustering (iv). Based on our data, we propose that CRACR2A is important for stabilization of Orai1- STIM1 complex via direct protein interaction under physiological conditions where amounts of Orai1 and STIM1 proteins are limiting (State 3). Upon increase of cytoplasmic [Ca2+] via opening of CRAC channels, the EF hands of CRACR2A bind Ca2+ ions, resulting in its dissociation from Orai1 and STIM1 (State 4), thereby destabilizing the Orai1-STIM1 complex. The EF-hands of STIM1 and CRACR2A are indicated and their Ca2+-bound status is colored in red. The schematic does not represent molecular stoichiometry of Orai1, STIM, and CRACR2A proteins.
DISCUSSION
We have identified CRACR2A as an important component of the Orai1-STIM1 complex. CRACR2A directly interacts with the N terminus of Orai1; however, its co-expression with Orai1 is not sufficient to amplify CRAC currents (data not shown). Instead, CRACR2A seems to act as a stabilizer for the interaction between Orai1 and STIM1. Several experimental data support this hypothesis. First, the biochemical analyses showed that CRACR2A directly interacts with both Orai1 and STIM1 and their interaction increases upon store depletion. Second, depletion of CRACR2A decreases CRAC channel-mediated SOCE due to reduced clustering of Orai1 and STIM1. Third, CRACR2A facilitates clustering of STIM1 and Orai1 at PM-ER junctions and potentiates SOCE. Based on these observations, we propose the following model (Fig. 7d). Under resting conditions, Orai1 and STIM1 are dispersed in the PM and ER membranes respectively, while CRACR2A is localized in the cytoplasm (Fig. 7d, state 1). Upon store depletion, STIM1 and Orai1 translocate and cluster at PM-ER junctions. Whether CRACR2A plays an active role in translocation of Orai1 and/or STIM1 (State 2, i-iii) or is recruited into junctions after clustering (iv) awaits further studies. Since Orai1 translocation is dependent on STIM1, CRACR2A is likely to be involved in translocation of STIM1 or both of Orai1 and STIM1 (ii and iii) rather than Orai1 alone (i). In all the scenarios, our results suggest the presence of a ternary complex of Orai1, STIM1, and CRACR2A (State 3) that dissociates upon elevation of [Ca2+]i (State 4). CRACR2A may play an important role in a physiological context where the amount of Orai1 and STIM1 is limiting, unlike transient overexpression of Orai1 and STIM1. Recently, CaM binding to the N terminus of Orai1 has been shown to evoke inactivation of CRAC channels42. CaM binding sites on Orai1 and STIM1 partially overlap with those of CRACR2A42, 43. In addition, CRACR2A and CaM have comparable affinity for Ca2+ (Fig. 5a). Interestingly, CaM co-immunoprecipitates with Orai1 only in the presence of Ca2+, contrary to CRACR2A, which dissociates from Orai1 and STIM1 at elevated [Ca2+]. These results suggest a scenario where CRACR2A binding favors an active CRAC channel complex upon store depletion (at low [Ca2+]i) while CaM facilitates inactivation of CRAC channels at high [Ca2+]i. Whether CRACR2A plays any role in modulating electrophysiological properties of CRAC channels awaits future investigations. In summary, we have identified a novel family of SOC channel regulators that modulate protein interactions in a [Ca2+]i-dependent manner.
METHODS
Reagents and antibodies
Fura 2-AM, Fura 4F-AM, and Lipofectamine 2000 were obtained from Invitrogen (Carlsbad, CA). Anti-FLAG antibody (F3165) from Sigma-Aldrich (St. Louis, MO) was used at 1:10,000 dilution. STIM1 was detected using monoclonal antibody (No. 610954, BD Biosciences, San Jose, CA) at 1:2,000 dilution. Anti-actin (sc-1616) and anti-Myc (sc-47694) antibodies were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA) (1:2,000 dilution). Polyclonal rabbit antibody for detection of CRACR2A was generated using purified human CRACR2A protein (Open Biosystems, Huntsville, AL) and used at 1:10,000 dilution.
Plasmids and cells
Full-length cDNAs of human Orai family and STIM1 were subcloned into pMSCV-CITE-eGFP-PGK-Puro16. For generation of STIM1-YFP and STIM1-mCherry, cDNA was cloned into pEYFP-N1 or pmCherry-N1 plasmids (Clontech, Mountain View, CA). Orai1K85A/K87A mutant was generated with an extracellular FLAG-tag in pMSCV-CITE-eGFP-PGK-Puro vector and with C-terminal GFP in pEGFP-N1 vector. Full-length cDNAs for human CRACR2A (clone ID 3531511) and mouse CRACR2B (clone ID 5256663) were purchased from Open Biosystems and subcloned into pMSCV-CITE-eGFP-PGK-Puro vector with a FLAG-tag. A plasmid encoding Myc-CRACR2A was generated by subcloning into pcDNA3.1mychis (Invitrogen). WT CRACR2A or CRACR2AEF2MUT cDNA was subcloned into pEGFP-N1, pEGFP-C1 and pmCherry-N1 plasmids, respectively. CRACR2B cDNA was subcloned into pcDNA3.1mychis and pMSCV-CITE-eGFP-PGK-Puro vectors. Primers used are described in Suppl. Table 1. HEK293, HeLa and Jurkat T cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA) center. HeLa O+S cells were generated by transducing with viruses encoding FLAG-tagged Orai1 and untagged STIM1 (1:3 ratio) together with hCD2544–45 followed by selection using hCD25 antibody-coated magnetic beads (Invitrogen).
Glycerol gradient analysis, affinity protein purification and mass spectrometry
2 × 1010 HeLa O+S cells were left untreated or treated with 1 µM thapsigargin for 10 min in PBS and cross-linked with 0.5 mM dithiobis succinimidyl propionate (DSP) for 1 h on ice, followed by quenching with 20 mM Tris-Cl, pH 7.5. Cells were lysed in cell lysis buffer (20 mM Tris-Cl, 2 mM EDTA, 100 mM NaCl, 10% glycerol, 0.5% Igepal CA-630, protease inhibitor cocktail [Roche]). Precleared lysates were applied onto a 20–50% glycerol cushion and centrifuged at 288,000g for 16 h. 56 fractions were collected from the top and analyzed by immunoblotting. Orai1 containing fractions were pooled and immunoprecipitated overnight at 4°C with anti-FLAG antibody agarose, washed five times in lysis buffer with high salt (200 mM NaCl), five times with low salt buffer (100 mM NaCl), and eluted with the FLAG-peptide (0.1 mg/ml). The eluates were run on a SDS-PAGE, and analyzed by silver staining or Colloidal Blue Staining Kit (Invitrogen) for mass spectrometry. In-gel digestion with trypsin, nanoLC-MSMS and automated database searching were performed essentially as described previously46.
Immunoprecipitation and GST pulldown analysis
2 × 107 transfected HEK293 or HeLa cells were harvested in PBS and DSP cross-linked. Cells were lysed in lysis buffer (20 mM Tris-Cl, 2 mM EDTA, 100 mM NaCl, 10% glycerol, 0.5% Igepal CA-630, protease inhibitor cocktail [Roche]) and centrifuged at 100,000g for 1 h before preclearing with protein G-Sepharose. Lysates were immunoprecipitated overnight with anti-FLAG antibody agarose. Immunoprecipitates were washed in lysis buffer and analyzed by immunoblotting. For pulldown analysis, the GST-fused N terminus of Orai1, STIM1 fragments and 6× His-tagged full-length CRACR2A were incubated with glutathione sepharose 4B beads for 6 h in binding buffer (0.1% Igepal CA-630, 20 mM Tris-HCl, 100 mM NaCl, 2 mM EDTA, protease inhibitors, 10% glycerol, and 100 µg/ml of bovine serum albumin) with or without 2 mM CaCl2. After extensive washing, proteins bound to beads were analyzed by immunoblotting.
Purification of recombinant proteins from insect cells and E. coli
Full-length FLAG-tagged human Orai1 and STIM1 cDNAs were integrated into baculoviruses, expressed in Sf9 cells, and purified following manufacturer’s instructions (Bac-to-Bac Baculovirus Expression Systems, Invitrogen). Orai1, STIM1 fragments and full-length CRACR2A proteins were subcloned into pGEX4T-1 and pET21A plasmids. GST fusion protein expressing transformants were grown in liquid cultures and induced with isopropyl-1-thio-β-D-galactopyranoside (IPTG, 0.2 mM) at 18°C overnight. Subsequently, cells were harvested and resuspended in lysis buffer (50 mM NaH2PO4, 500 mM NaCl, 10% glycerol, pH 8.0) containing protease inhibitors and 0.5% Triton X-100. The lysates were incubated with glutathione sepharose 4B beads for 2 h. The recombinant proteins were eluted using glutathione. For 6× His-tagged proteins, cells were harvested and resuspended in lysis buffer (50 mM NaH2PO4, 500 mM NaCl, 10% glycerol, 0.5% Triton X-100, and 10 mM Imidazole, pH 8.0). After sonication and clearing, the supernatant was incubated with Ni-NTA agarose (Qiagen). The recombinant proteins were eluted in buffer containing 250 mM imidazole.
siRNA/shRNA-mediated depletion and quantitative PCR
0.5 × 106 HEK293 cells were transfected with siRNAs (Dharmacon) using lipofectamine reagent. Cells were re-transfected after 24 h and harvested for RT- PCR or [Ca2+]i measurement 3 days after transfection. Jurkat cells were electroporated with 100 nM of siRNA and used for experiments 3 days after transfection. The sequences of siRNAs are: hOrai1, UCACUGGUUAGCCAUAAGA; hCRACR2A, GCUCAGCAGCAGUUGGAAA; hCRACR2B, GAACAUGCAGAAAGAGAAA. Knockdown efficiency was quantified by RT-PCR analysis. Purified RNA was oligo(dT)-primed for first-strand cDNA synthesis (Superscript III kit; Invitrogen). Quantitative SYBR green real-time PCR was performed using an iCycler IQ5 (Bio-Rad). The cDNA levels were normalized to GAPDH. Sequences of the primers are mentioned in Suppl. Table 1. For shRNA expression, the corresponding sense and antisense siRNA sequences were subcloned into pRetro-Super-puro (OligoEngine). For assessment of expression of CRACR2A and CRACR2B in various human tissues, cDNA made from commercially available purified total RNA (Applied Biosystems, Foster City, CA) was used for PCR with primers (Suppl. Table 1).
Single-cell Ca2+ imaging, TIRF and confocal microscopy
T Cells were loaded at 1 × 106 cells ml−1 with 1 µM Fura 2-AM for 30 min at 25°C and attached to poly-L-lysine–coated coverslips. Fibroblasts, HeLa and HEK293 cells were grown directly on UV-sterilized coverslips and loaded with 2 µM Fura 2-AM for 45 min. For SOCE measurements in Fig. 6a and 6b, cells were loaded with 2 µM Fura 4F-AM for 45 min. Intracellular [Ca2+]i measurements were performed using essentially the same methods as recently described47. TIRF microscopy was performed using an Olympus IX2 illumination system mounted on an Olympus IX51 inverted microscope using recently described methods47. For quantification of TIRF intensity across different cells, individual regions of interest were selected and data were analyzed as the ratio of fluorescence intensity at each time-point (F) to the fluorescence intensity at the start of the experiment (F0). For confocal analysis, at 6 h after transfection, HEK293 cells were plated onto sterilized coverslips and maintained in complete DMEM for 18 h before imaging in Ringer’s solution containing (in mM): 155 NaCl, 4.5 KCl, 2 CaCl2, 1 MgCl2, 10 D-glucose, and 5 Na-HEPES (pH 7.4). For depletion of stores, cells were treated with 1 µM thapsigargin in Ca2+-free Ringer’s solution (prepared by substitution of CaCl2 with 2 mM MgCl2 and 0.2 mM EGTA) for 5 min. eGFP and mCherry were excited simultaneously at 488 and 594 nm on a Leica SP2 AOBS inverted confocal microscope equipped with a PL APO 60×/NA 1.4 oil immersion objective. Fluorescence emission was collected at 615–700 nm (mCherry) and 510–570 nm (eGFP). Images were processed for enhancement of brightness or contrast using LCSlite (Leica) software.
T cell purification, cytokine measurement, Annexin V staining and immunocytochemistry
T cell purification, activation and transduction were carried out as previously described47. Cells were harvested and used for measurement of Ca2+ entry or RNA extraction after 5–6 days of stimulation. Jurkat T cells were electroporated with 20 µg of plasmids. For intracellular cytokine staining, Jurkat T cells were stimulated with 10 nM phorbol 12-myristate 13-acetate (PMA) and 1 µM ionomycin for 16 h. Cells were fixed with 4% paraformaldehyde in PBS, and permeabilized in saponin buffer (PBS, 0.5% saponin, 1% bovine serum albumin, and 0.1% sodium azide). Cells were stained with Phycoerythrin-conjugated human IL-2 antibody (eBioscience) for 20 min, washed and analyzed. For apoptosis assay, transfected Jurkat cells were stained using Annexin V staining kit (BD Biosciences). Cells were examined on a FACSCalibur flow cytometer (BD Biosciences). Immunocytochemistry and flow cytometry on HEK293 cells expressing WT and mutant Orai1 constructs were performed as described47.
Measurement of CRAC currents by whole cell patch clamp recording
CRAC current measurements were carried out as previously described47.
Fluorescence protease protection (FPP) assays
FPP assays were performed using recently described protocol33. Briefly, HEK293 cells expressing GFP-CRACR2A or CRACR2A-GFP on UV-sterilized coverslips were perfused with KHM buffer (110 mM potassium acetate, 20 mM HEPES, and 2 mM MgCl2). For permeabilization, 75 µM of digitonin was used. Subsequently 50 µg ml−1 of proteinase K containing KHM buffer was perfused. GFP images were acquired at every 5-s interval after background subtraction. For each experiment, 50–100 individual HEK293 cells were analyzed using OriginPro (Originlab).
Detection of Ca2+ binding by using 45Ca2+ Overlay method
45CaCl2 (1 mCi, 37 MBq) was purchased from NEN. The protocol was modified from a previously described method48. Purified 6× His-fusion proteins and CaM (EMD Biosciences) were transferred to a nylon membrane after separating on a 12% SDS PAGE. The blot was extensively washed for 3 × 30 min with binding buffer (in mM: 60 KCl, 5 MgCl2, and 10 imidazole-HCl, pH 6.8). The overlay assay was performed by incubating the blot with 8.8 µM 45CaCl2 at 25°C for 1 h, followed by 3 × 5 min washes in dH2O. After autoradiography, the blot was stained with Ponceau S solution for detection of proteins.
Database accession numbers
Genbank accession numbers for CRACR2A proteins are; Homo sapiens (NP_116069.1), Pan troglodytes (XP_001156341), Mus musculus (NP_001028636), Danio rerio (XP_001922837.1). Genbank accession numbers of CRACR2B proteins are: Mus musculus (NP_001020274), Homo sapiens (registered as GenBank accession number, GU952799).
Statistical analysis
Statistical analysis was carried out using Student’s t-test.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kenneth Philipson for critical reading of this manuscript and kind suggestions on Ca2+ overlay experiments. We thank Bernard Ribalet for help with patch clamp experiments, Baljit Khakh for sharing equipments and Earl Homsher for helpful discussions. This work was supported by National Institute of Health grant AI-083432, Stein Oppenheimer Endowment Award, Cancer Research Coordinating Committee Award (Y.G.), and a fellowship from the American Heart Association (S.S.).
Footnotes
AUTHOR CONTRIBUTIONS
S.S. and Y.G. designed research. S.S. and Y.G. carried out experiments with technical help from H.J.J.. K.D.K. performed and analyzed Ca2+ binding assays and P.S. and J.W. carried out mass spectrometry. S.S. and Y.G. analyzed data and wrote the manuscript. Y.G. supervised the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Feske S. Calcium signalling in lymphocyte activation and disease. Nat Rev Immunol. 2007;7:690–702. doi: 10.1038/nri2152. [DOI] [PubMed] [Google Scholar]
- 2.Vig M, Kinet JP. Calcium signaling in immune cells. Nat Immunol. 2009;10:21–27. doi: 10.1038/ni.f.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gwack Y, Feske S, Srikanth S, Hogan PG, Rao A. Signalling to transcription: store-operated Ca2+ entry and NFAT activation in lymphocytes. Cell Calcium. 2007;42:145–156. doi: 10.1016/j.ceca.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Lewis RS. Calcium signaling mechanisms in T lymphocytes. Annu Rev Immunol. 2001;19:497–521. doi: 10.1146/annurev.immunol.19.1.497. [DOI] [PubMed] [Google Scholar]
- 5.Cahalan MD, Chandy KG. The functional network of ion channels in T lymphocytes. Immunol Rev. 2009;231:59–87. doi: 10.1111/j.1600-065X.2009.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liou J, et al. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roos J, et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang SL, et al. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luik RM, Wang B, Prakriya M, Wu MM, Lewis RS. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 2008;454:538–542. doi: 10.1038/nature07065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feske S, et al. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 11.Prakriya M, et al. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- 12.Vig M, et al. CRACM1 multimers form the ion-selective pore of the CRAC channel. Curr Biol. 2006;16:2073–2079. doi: 10.1016/j.cub.2006.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vig M, et al. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeromin AV, et al. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature. 2006;443:226–229. doi: 10.1038/nature05108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang SL, et al. Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proc Natl Acad Sci U S A. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gwack Y, et al. Biochemical and functional characterization of Orai proteins. J Biol Chem. 2007;282:16232–16243. doi: 10.1074/jbc.M609630200. [DOI] [PubMed] [Google Scholar]
- 17.Luik RM, Wu MM, Buchanan J, Lewis RS. The elementary unit of store-operated Ca2+ entry: local activation of CRAC channels by STIM1 at ER-plasma membrane junctions. J Cell Biol. 2006;174:815–825. doi: 10.1083/jcb.200604015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu P, et al. Aggregation of STIM1 underneath the plasma membrane induces clustering of Orai1. Biochem Biophys Res Commun. 2006;350:969–976. doi: 10.1016/j.bbrc.2006.09.134. [DOI] [PubMed] [Google Scholar]
- 19.Mercer JC, et al. Large store-operated calcium selective currents due to co-expression of Orai1 or Orai2 with the intracellular calcium sensor, Stim1. J Biol Chem. 2006;281:24979–24990. doi: 10.1074/jbc.M604589200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peinelt C, et al. Amplification of CRAC current by STIM1 and CRACM1 (Orai1) Nat Cell Biol. 2006;8:771–773. doi: 10.1038/ncb1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soboloff J, et al. Orai1 and STIM reconstitute store-operated calcium channel function. J Biol Chem. 2006;281:20661–20665. doi: 10.1074/jbc.C600126200. [DOI] [PubMed] [Google Scholar]
- 22.Muik M, et al. A Cytosolic Homomerization and a Modulatory Domain within STIM1 C Terminus Determine Coupling to ORAI1 Channels. J Biol Chem. 2009;284:8421–8426. doi: 10.1074/jbc.C800229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park CY, et al. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan JP, et al. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muik M, et al. Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. J Biol Chem. 2008;283:8014–8022. doi: 10.1074/jbc.M708898200. [DOI] [PubMed] [Google Scholar]
- 26.Navarro-Borelly L, et al. STIM1-Orai1 interactions and Orai1 conformational changes revealed by live-cell FRET microscopy. J Physiol. 2008;586:5383–5401. doi: 10.1113/jphysiol.2008.162503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, et al. STIM1 gates the store-operated calcium channel ORAI1 in vitro. Nat Struct Mol Biol. 2010;17:112–116. doi: 10.1038/nsmb.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, et al. Mapping the interacting domains of STIM1 and Orai1 in Ca2+ release-activated Ca2+ channel activation. J Biol Chem. 2007;282:29448–29456. doi: 10.1074/jbc.M703573200. [DOI] [PubMed] [Google Scholar]
- 29.Varnai P, Toth B, Toth DJ, Hunyady L, Balla T. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 Complex. J Biol Chem. 2007;282:29678–29690. doi: 10.1074/jbc.M704339200. [DOI] [PubMed] [Google Scholar]
- 30.Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci U S A. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baba Y, et al. Coupling of STIM1 to store-operated Ca2+ entry through its constitutive and inducible movement in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2006;103:16704–16709. doi: 10.1073/pnas.0608358103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorenz H, Hailey DW, Lippincott-Schwartz J. Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat Methods. 2006;3:205–210. doi: 10.1038/nmeth857. [DOI] [PubMed] [Google Scholar]
- 33.Lorenz H, Hailey DW, Wunder C, Lippincott-Schwartz J. The fluorescence protease protection (FPP) assay to determine protein localization and membrane topology. Nat Protoc. 2006;1:276–279. doi: 10.1038/nprot.2006.42. [DOI] [PubMed] [Google Scholar]
- 34.Gwack Y, et al. Hair loss and defective T- and B-cell function in mice lacking ORAI1. Mol Cell Biol. 2008;28:5209–5222. doi: 10.1128/MCB.00360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cahalan MD. STIMulating store-operated Ca(2+) entry. Nat Cell Biol. 2009;11:669–677. doi: 10.1038/ncb0609-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Putney JW., Jr New molecular players in capacitative Ca2+ entry. J Cell Sci. 2007;120:1959–1965. doi: 10.1242/jcs.03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis RS. The molecular choreography of a store-operated calcium channel. Nature. 2007;446:284–287. doi: 10.1038/nature05637. [DOI] [PubMed] [Google Scholar]
- 38.Baba Y, Kurosaki T. Physiological function and molecular basis of STIM1-mediated calcium entry in immune cells. Immunol Rev. 2009;231:174–188. doi: 10.1111/j.1600-065X.2009.00813.x. [DOI] [PubMed] [Google Scholar]
- 39.Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 40.Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 41.Dolmetsch RE, Lewis RS. Signaling between intracellular Ca2+ stores and depletion-activated Ca2+ channels generates [Ca2+]i oscillations in T lymphocytes. J Gen Physiol. 1994;103:365–388. doi: 10.1085/jgp.103.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mullins FM, Park CY, Dolmetsch RE, Lewis RS. STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0906781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bauer MC, O'Connell D, Cahill DJ, Linse S. Calmodulin binding to the polybasic C-termini of STIM proteins involved in store-operated calcium entry. Biochemistry. 2008;47:6089–6091. doi: 10.1021/bi800496a. [DOI] [PubMed] [Google Scholar]
- 44.Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- 45.Shi Y, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 46.Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007;581:3545–3549. doi: 10.1016/j.febslet.2007.06.052. [DOI] [PubMed] [Google Scholar]
- 47.Srikanth S, Jung HJ, Ribalet B, Gwack Y. The intracellular loop of Orai1 plays a central role in fast inactivation of Ca2+ release-activated Ca2+ channels. J Biol Chem. 2010;285:5066–5075. doi: 10.1074/jbc.M109.072736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maruyama K, Mikawa T, Ebashi S. Detection of calcium binding proteins by 45Ca autoradiography on nitrocellulose membrane after sodium dodecyl sulfate gel electrophoresis. J Biochem. 1984;95:511–519. doi: 10.1093/oxfordjournals.jbchem.a134633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.