Abstract
BACKGROUND
The role of TP63 in cancer remains controversial since both oncogenic and tumor suppressive actions have been reported. p63 protein is found in the nuclei of basal cells of the normal prostate, yet it is absent in the vast majority of prostate cancer nuclei. Since a complex array of TP63 mRNA transcripts encode polypeptides with distinct functional properties, it is important to determine which forms are expressed in normal and prostate cancer tissue.
METHODS
We used real-time RT-PCR to distinguish TP63 mRNA isoforms in prostate cancer cell lines (n=7), samples from prostate cancer patients, and specimens from healthy subjects. We sequenced all TP63 exons from prostate carcinoma cell lines, patient tumor/normal pairs (n=48), and tumor xenografts (n=20).
RESULTS
TP63 mRNA isoforms were present in all tumors, albeit at levels lower than in normal prostate. We performed mutational analysis of TP63 in 20 primary tumors, 20 metastases, 28 tumor xenografts, and 7 prostate cancer cell lines. The most abundant N-terminal variant was ΔN; the most abundant C-terminal variant was the α form. Prostate tumor cell line CWR22Rv1 contained a single G to T substitution in exon 8 that is identical to a dominant-negative DNA binding inactivation mutation occurring in patients with a congenital TP63 deficiency syndrome. One patient tumor contained a somatic mutation in exon 11.
CONCLUSIONS
The pattern of TP63 mRNA expression in normal prostate tissue is retained in reduced amounts in prostate cancer, and a potentially functional TP63 mutation was identified in one prostate tumor. Thus it appears doubtful that TP63 causes prostate cancer to develop; if it is a prostate cancer gene it likely functions as a tumor suppressor. Further study of the role of TP63 isoforms in regulating stem cell functions of normal and neoplastic prostate epithelial cells is needed.
Keywords: TP63, prostate, cancer, mRNA, tumor suppressor gene, oncogene, p63
Introduction
A widely held view suggests that cancer cells arise from either long-lived tissue stem cells, or from their partially differentiated progeny (progenitor cells) that have co-opted the program for self renewal (1,2). Since these “cancer stem cells” are predicted to be largely resistant to standard chemotherapeutic regimens, new approaches are being sought to specifically eliminate them. In order to develop these approaches an improved understanding of the biological properties of tissue stem cells, and their progenitor cells, are required. Accordingly, to better understand the origins of prostate cancer, there is a great deal of work being done to identify and characterize tissue stem cells and to characterize cells with properties of prostate cancer stem cells. Much of this work involves the transcription factor encoded by the TP63 locus.
TP63 is a member of the TP53 and TP73 gene family (3-6). Unlike p53 protein, which is expressed at detectable levels only in response to environmental stress, p63 protein is constitutively expressed at high levels in a variety of epithelial tissues (3,7). Germ line mutations in the TP63 gene result in a group of autosomal dominant ectodermal dysplasia syndromes (including the ectrodactyly, ectodermal dysplasia, and facial clefts—or EEC—syndrome)(8,9). In the mouse, targeted disruption of both TP63 alleles results in epidermal defects as well as a lack of limbs, skin adnexa, hair, and teeth (10,11). While its exact role in stem cell function is still somewhat controversial, these results, together with in vitro experiments (12,13), have led to the concept that TP63 plays a critical role in the maintenance of epidermal stem cells.
TP63 contains 16 different exons coding for at least 6 different mRNA isoforms that share a common core DNA binding domain but exhibit varying 5' and 3' ends. Two alternate promoters produce two different N-terminal variants, termed TA and ΔN, and alternative splicing at the 3' end generates three different C-terminal variants, termed α, β, and γ. The functional role of the different isoforms has not been fully determined, although ΔNp63 in keratinocytes has been proposed to maintain the proliferative state, preventing terminal differentiation (14-16). In vitro experiments have shown that the TA-p63–containing isoforms appear to have some p53-like functions including transactivation of p53 target promoters and promotion of cell death, and that these functions can be counteracted by ΔNp63 isoforms (17).
A critical step in deciphering the function of TP63 in prostate stem cell biology is to characterize the pattern and relative levels of expression of the different mRNA variants encoded by the TP63 gene in human prostate tissues. Using quantitative PCR, Signoretti and colleagues found that the ΔNp63 and TAp63 mRNA isoforms were present in PrEC cells (human prostate epithelial cells characterized largely by a basal cell phenotype) and that ΔNp63 was the more abundant of the two. They also observed only very low levels of ΔNp63 mRNA in PC3 cells, undetectable levels of ΔNp63 in LNCaP and DU145 cells, and low levels of TAp63 in LNCaP and DU145 cells (18).
Takahashi and colleagues analyzed the expression of TP63 mRNAs in human prostate cancer specimens (19). While they reported downregulation compared to normal in 39% of cases, not all cancers were downregulated, and in fact 34% were upregulated. The finding of expression of TP63 mRNA in human clinical prostate cancer is somewhat surprising since the protein is usually absent and the prostate cancer cell lines examined were generally negative (18,20). Moreover, it is possible that contaminating normal basal cells might confound these studies, and there have been no studies on the relative levels of these isoforms, or the levels of the α, β, or γ variants from normal human prostate tissue and cancer tissue.
The role of TP63 as a tumor suppressor or oncogene in cancer remains enigmatic. In squamous cell cancers of the head and neck p63 has been found to be overexpressed, and the region encoding this gene amplified, suggesting an oncogenic role (21). However, in urothelial carcinoma, decreased p63 protein correlates with more aggressive disease (22). In chronic myelogenous leukemia in blast crisis, somatic mutations in the DNA binding domain of TP63 have been reported, suggesting a possible tumor suppressor role (23). It is important, therefore, to determine whether one mechanism of decreased levels of p63 protein in prostate cancer is by mutational inactivation. Although there has been one study of TP63 mutational analysis in primary prostate cancers using RT-PCR and sequencing, and no mutations were detected (19), there have been no comprehensive sequencing study of all exons across all stages of prostate clinical samples or cell lines/xenografts reported. Therefore, we sequenced all exons of TP63 from genomic DNA obtained from prostate cancer cell lines, prostate cancer xenografts, clinically localized primary cancers, and hormone refractory prostate cancers obtained at autopsy.
Materials and Methods
Cell Lines and Patient Samples
We used prostate tumor cell lines LNCaP, LNCaP C4-2B, PC3, LAPC4, DU145, MDAPCa2B, and CWR22Rv1; and hPrEC (epithelial cell origin); a cervical squamous cancer cell line, ME180; and tumor xenografts LUCaP 23.8, 23.12, 35, 35V, 49, 58, 69, 70, 73, 77, 78, 86.2, 92.1, 93, 96, 105 and LAPC 4AD, 9AD, 4cAI, 9cAI. All cell lines were maintained in RPMI 1640 with 10% fetal calf serum at 37°C with 5% C02 except hPrEB and ME180 cells, which were grown in McCoy's (Gibco, Carlsbad, CA, USA). hPrEB cells were derived from prostate cancer patients by immortalizing epithelial cells in culture with high-risk papilloma virus E6 and E7 (generously provided by Dr. Hyman Levitsky from Johns Hopkins). LNCaP, PC3, DU145, MDAPCa2B, and CWR22Rv1 cells were obtained from the American Type Culture Collection. C42B cells were obtained from the laboratory of William G. Nelson at Johns Hopkins. All xenograft DNA samples were obtained from Robert L. Vessella (University of Washington School of Medicine). Cells were harvested from a T175 flask at 80% confluence and stored as a pellet at -80°C for subsequent RNA extraction.
Radical prostatectomy specimens were obtained from surgery patients and metastatic tumors from autopsies at the Johns Hopkins Hospital in an IRB-approved protocol. Paired normal and tumor tissues were collected from each of the radical prostatectomy specimens by individually harvesting samples from presumptive normal and tumor regions and immediately snap-freezing them in liquid nitrogen. Hematoxylin-and eosin-stained sections provided histological confirmation of normal tissue and cancer tissue.
The snap-frozen tumor blocks were trimmed to provide sections with near homogeneous tumor populations containing minimum amounts of inflammatory cells, stroma and normal prostate cells. Fifty to one hundred sections, each between 5 and 10 μm thick, were transferred from each normal and each tumor block at -20°C to conical tubes and stored at -80°C for subsequent RNA extraction.
In situ hybridization
Non-radioactive in situ hybridization was performed using a probe corresponding to the DNA binding domain of the p63 gene in the following manner: a portion of the p63 gene was cloned using the primers FDN and RA from cDNA prepared from ME180 cells. The product (1472 bp) was gel purified (Qiagen QIAquick Gel Extraction Kit) and cloned using the Top TA Cloning Kit (Invitrogen, Vector PCR 2.1-Topo), and transformed into E. coli. The antisense and sense riboprobes were then generated by first producing a PCR product containing the T7 promoter in either the antisense or sense direction using primers amplifying exons 4 and 5 of the p63 cDNA (442 nucleotide products). For production of the antisense probe the following primers were used: antisense primer forward (T7 promoter in bold) – 5′-ctaatacgactcactataggggaattcacggctcagctcatg-3′. Antisense primer reverse-5′gtacacgaacctggggctcc- 3′. For construction of the sense probe, the following primers were used. Sense primer forward 5′-ctaatacgactcactactatagggcgaacctgggctcctgaac-3′. Sense primer reverse 5′- gaattcacggctcagctcatg-3′. RNA labeling was performed using T7 polymerase and digoxigenin-UTP using the DIG RNA labeling kit (Roche Molecular Biochemicals, Indianapolis, IN). In situ hybridization was carried out as follows. After deparaffinization and rehydration, slides were rinsed in DEPC water and incubated in 1% hydrogen peroxide and then in tris-buffered saline (TBS) (pH 7.4) for 5 min at room temperature. Slides were then treated with proteinase K (80 μg/ml) at 37°C for 40 min. After rinsing in TBS containing 0.05% Tween 20 (TBST), the RNA probe (either sense or antisense at 200 ng/μl) was applied and slides were hybridized overnight at 55°C. Slides were then washed in 2x SSC for 5 min at 45°C and then incubated with a 1:35 dilution of RNaseA/T1 cocktail (Ambion, Austin, TX, USA) in 2x SSC for 30 min at 37°C. Slides were washed twice by incubation at 60°C with 2x SSC containing 50% formamide for 30 min followed by a second wash for 20 min. Slides were washed again with 0.08 SSC for 20 min at 60°C, rinsed in TBS and then incubated with rabbit Ig fraction (Dako, Carpinteria, CA) for 15 min at room temperature. Then slides were incubated with rabbit HRP-anti-digoxigenin (Dako) diluted 1:100 for 30 min at room temperature. Slides were then washed 3 × 5 min each in TBST and were then treated with biotinylated-tryamide (Dako GenPoint kit) for 15 min in the dark at room temperature. After washing 3 × 5 min in TBST, slides were treated with the secondary streptavidin (Dako GenPoint kit) for 15 min at room temperature. Slides were washed and developed with DAB and counterstained with hematoxylin.
Laser Capture Microdissection
Laser capture microdissection (LCM) was performed to isolate areas of tumor from adjacent normal tissue as previously described (24).
RNA Extraction and Removal of Trace Genomic DNA
RNA was extracted from tissue and cell lines using the ToTALLY RNA Total RNA Isolation Kit according to the manufacturer's instructions (Ambion, Austin, TX, USA). Isolated RNA was treated with DNAse to remove traces of genomic DNA using the DNA-free Kit according to manufacturer instructions (Ambion, Austin, TX, USA). The purified RNA solution was then removed, quantified using UV spectrophotometry, and stored at -80°C.
RT-PCR
Reverse transcription was performed using the RETROscript first-strand synthesis kit for RT-PCR (Ambion, Austin, TX, USA). Primers utilized for cDNA amplification are summarized in Table 1. Primers for the TA isotypes detected α, β, and γ splice variants but not the ΔN isotypes; conversely, primers for the ΔN isotypes detected α, β, and γ splice variants but not the TA isotypes. We used the same primers for the TA and ΔN oligonucleotides as those reported by Signoretti and colleagues, with the exception of the reverse primer for ΔN, which lacked an A at the 3′ end (18). The primers for the α, β and γ splice variants were designed based on known sequences from NCBI Genebank.
Table 1.
Oligonucleotide primers for RT-PCR.
| Gene Detected | Primer | Sequence | Product Size | Location | Notes |
|---|---|---|---|---|---|
| TP63 TA (exons 1-4, no 3') | F | TGTATCCGCATGCAGGACT | 127 | exon 3 | unique to TA |
| R | CTGTGTTATAGGGACTGGTGGAC | exon 4 | |||
| TP63 ΔN (exons3'-4, no 1-3) | F | GAAAACAATGCCCAGACTCAA | 125 | exon 3' | unique to ΔN |
| R | TGCGCGTGGTCTGTGTT | exon 4 | |||
| TP63 α (exons 10-14, no 15) | F | TCAGTTTCTTAGCGAGGTTG | 118 | overlaps exon 12 and 13 | unique for α |
| R | ATTTTCAGACTTGCCAGATC | exon 14 | |||
| TP63 β (exons 10-12,14, no 13 or 15) | F | AGCATTGTCAGGATCTGG | 113 | overlaps exon 12 and 14 | unique for β |
| R | GAGATGAGAAGGGGAGGA | ||||
| TP63 γ (exons 10 & 15) | F | AAACATCTCCTTTCAGCC | 115 | overlaps exon 10 and 15 | unique for γ |
| R | GGTACACTGATCGGTTTG | exon 15 | |||
| TBP | F | CACGAACCACGGCACTGATT | 89 | ||
| R | TTTTCTTGCTGCCAGTCTGGAC |
For both standard and quantitative PCR each reaction tube contained 1X PCR Gold buffer (15 mM Tris-HCl pH8, 50 mM KCl, Applied Biosystems, Foster City, CA, USA), 200 μM of dNTPs (LifeTechnologies, Rockville, MD, USA), 300 nM each of forward and reverse primer, 0.625 units of AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA, USA), 3 mM MgCl2, and (for quantitative PCR) 1/100,000 SYBR Green (Molecular Probes, Eugene, OR, USA), all held in DEPC-water. Reaction conditions were as follows: an initial 10 minutes at 95°C followed by 45 cycles of 5 seconds at 94°C and 60 seconds at 62°C. For quantitative PCR, each sample was amplified in triplicate.
After 45 cycles, the standard PCR reaction mixture underwent a final extension for 5 minutes at 72°C, while the quantitative PCR mixture underwent a melt curve to validate reaction product specificity. During PCR optimization all reactions were subjected to standard PCR and gel electrophoresis and all products were found to contain a single band at the correct size with no primer-dimer formation. For standard PCR, reaction products were detected using 3% (in 1X TBE) agarose gel electrophoresis in the presence of Gel Star (BioWhittaker Molecular Applications, Rockland, ME, USA). In all PCR reactions, a no-template reaction mixture was used as a negative control.
Quantitative PCR
Real-time quantitative RT-PCR was performed using iCycler IQ (BioRad, Hercules, CA, USA) and SYBR Green detection. All PCR reactions were run in triplicate. The relative expression of the various TP63 isoforms in the samples was determined using the comparative CT method as detailed in Applied Biosystems User Bulletin # 2 (http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocum ents/cms_040980.pdf). TBP, which codes for the TATA-binding protein, was used as an endogenous control reference standard for normalizing expression and quantifying levels of each mRNA isoform (25). The hPrEB cells were found to express all 6 TP63 mRNA isoforms and were used to assess the efficiency of the PCR reaction for each primer set as well as for TBP. All PCR reactions were approximately 95-100% efficient. An evaluation of relative expression was obtained first by normalizing the CT of the isoform to that of TBP mRNA (to normalize for amount of RNA) in the matched triplicate of the same sample. The fold increase of the isoform relative to TBP mRNA was obtained by equation 2ΔCT. This normalized level of isoform expression from a given patient was compared by dividing the normalized expression for the isoform for the normal prostate tissue by that of the tumor tissue from the same patient to obtain a relative fold difference. Statistical analysis comparing relative levels in tumor and paired normal was performed with the Wilcoxon signed-ranks test with Stata 8.1 (Stata Corporation, College Station, TX, USA).
TP63 Gene Sequencing
DNA Amplification and Purification
Intronic primers for each of the 16 exons of TP63 were employed as described (Table 2). (26). PCR amplification was performed by the following cycle: 95°C for 5 min, 95°C for 1 min, 57°C for 1 min, 72°C for 1 min (30 cycles), 72°C for 10 min. Verification of adequate, uncontaminated amplification was made by gel electrophoresis on 2% agarose (Gibco, Carlsbad, CA, USA). After amplification verification, DNA was purified with the Concert Rapid PCR Purification Kit (Life Technologies, Rockville, MD, USA).
Table 2.
Intronic primer sequences utilized for TP63 genomic DNA sequencing.
| Exon | Primer | Exon size (bp) | PCR fragment size (bp) | |
|---|---|---|---|---|
| Exon 1 | F | GACCCTATTGCTTTTAGCCTC | 62 | 235 |
| R | CATTCATAATACACAAGGCACTTC | |||
| Exon 2 | F | CCTGCATGGTTTTATAGATTCACTTG | 129 | 305 |
| R | GACCACCCACATTTACCTATTTAG | |||
| Exon 3 | F | CCCTTTCCATGCCTAACTCACT | 133 | 209 |
| R | CCAAAGACTGAAGAGAAAGCCTG | |||
| Exon 3' | F | GGCAAAATCCTGGAGCCAGAAG | 42 | 140 |
| R | AAAGCATCTCTAAATGGAGTGC | |||
| Exon 4 | F | GGCTTCAGCGGCTAATATTGGG | 255 | 353 |
| R | GTGAAGCCCATCCTTGGACTTG | |||
| Exon 5 | F | TCTCCTTCCTTTCTCCACTGGC | 187 | 284 |
| R | TGCCCACAGAATCTTGACCTTC | |||
| Exon 6 | F | GCCACCAACATCCTGTTCATGC | 116 | 259 |
| R | GTCTACTCAGTCCATAGAGGTGTTG | |||
| Exon 7 | F | GAAGGAACAACGTCAGTTTAAACCC | 110 | 245 |
| R | AAAGCAGCCACGATTTCACTTTGCC | |||
| Exon 8 | F | GTGGTAGATCTTCAGGGGACTTTC | 137 | 263 |
| R | CCAACATCTGGAGAAGATTCAACC | |||
| Exon 9 | F | GTGTTCCCAGGATGAAACTTGC | 83 | 153 |
| R | GAAGCAACCATGAACACCCAAGG | |||
| Exon 10 | F | CCACACTTCTAACAGTTCTACAGC | 137 | 278 |
| R | TCATCAATCACCCTATTGCTGATCC | |||
| Exon 11 | F | CGTGCTCACCATCATTTCCATG | 158 | 239 |
| R | CTAGCCTGTTCATCCTTCAGCC | |||
| Exon 12 | F | CACTGGGATGCTGGTACATGATG | 145 | 228 |
| R | GGGCACATGCTGTGGACTC | |||
| Exon 13 | F | CTGTTTCCTCCTTCCTCTTCC | 94 | 212 |
| R | CAGGGATTGAACTACAAGGC | |||
| Exon 14 | F | CCCTGTTTTCATTCTCCATGACAC | 297 | 366 |
| R | GGGATAGGAAGAGCTCACATGG | |||
| Exon 15 | F | CATGAAGGTATAAGGAGTGTGTTTCTG | 115 | 240 |
| R | ACACACACTTAAAATATAGAGATAGGGC |
DNA Sequencing
Purified DNA at a concentration of 1 μM primer was combined with primer. Sequencing analysis was performed using the Applied Biosystems 3730 DNA analyzer, ABI PRISMã DNA Sequencing Analysis Software, and Sequencher for Windows (Gene Codes, Ann Arbor, MI, USA). All sequencing was performed in both the sense and antisense directions and all mutations were verified by re-sequencing of both strands after a repeated PCR reaction.
Results
Expression of TP63 mRNA in Cell Lines
Levels of TP63 mRNA variants in the cervical squamous cell carcinoma cell line ME-180 were analyzed as a positive control for quantitative real-time RT-PCR and all isoforms examined were expressed. Normal prostate epithelial cells (hPrEC), which are known to express a phenotype most consistent with basal cells or intermediate basal cells, also expressed all isoforms with a similar pattern of relative expression levels to that seen in ME-180 (Table 3). The most abundant variants detected were the 5' promoter variant ΔN and the 3' splice variant α, which suggests that the predominant mRNA isoform in both ME-180 and PrEC cells is ΔNp63α. The relative levels of mRNA isoforms containing both TA and beta were higher in ME-180 than PrEC cells. All 7 prostate cancer cell lines were negative for TP63 mRNA isoforms.
Table 3.
Expression of TP63 mRNA isoforms in cell lines.
| Cell Line | Fold increase of TA over TBP | Fold increase of ΔN over TBP | Fold increase of α over TBP | Fold increase of β over TBP | Fold increase of γ over TBP |
|---|---|---|---|---|---|
| hPrEC | 0.02 | 37 | 28 | 1.4 | 0.06 |
| C4-2B | ND | ND | ND | ND | ND |
| CWR22rV1 | 0.01 | ND | ND | 0.01 | ND |
| hPrEB | 0.24 | 14 | 7.2 | 0.58 | 0.09 |
| PC3 | 0.05 | 0.03 | ND | 0.06 | ND |
| LNCAP | 0.01 | ND | ND | ND | ND |
| DU145 | 0.01 | ND | ND | ND | ND |
| LAPC4 | ND | ND | ND | ND | ND |
| ME180 | 1.2 | 31 | 22 | 3.8 | 0.41 |
ND = nondetectable.
Expression of TP63 mRNA in Normal Prostate and Adenocarcinoma in Radical Prostatectomy Specimens
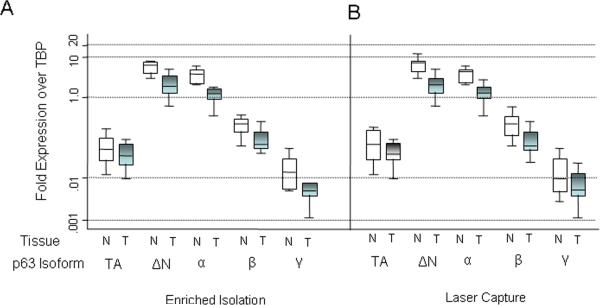
Normal prostate tissue obtained from frozen sections of manually enriched tissue blocks (without laser capture microdissection) in 12 of 12 radical prostatectomy specimens from patients with localized prostate cancer also expressed all 5 TP63 mRNA variants (Fig. 1). The most abundant variants contained ΔN and α, which suggests that the predominant mRNA isoform in normal prostate is also ΔNp63α.
Figure 1. Relative levels of TP63 mRNA isoforms in primary prostate cancer and matched benign tissues.
A) Tissue obtained by macrodissection (Enriched Isolation) of frozen normal and tumor pairs. Box represents distribution of data from n = 12 patients each. N = normal tissue. T = tumor tissue. B) Relative levels of TP63 mRNA isoforms obtained after laser capture microdissection (n = 9).
Tumor tissue in 12 of 12 radical prostatectomy specimens obtained from frozen sections of manually enriched tissue blocks (without laser capture microdissection) also expressed all 5 mRNA variants (Fig. 1). As in the normal prostate tissue, the most abundant variants were ΔN and α. The levels of expression in carcinoma, however, were markedly reduced (approximately 5- to 10-fold on average) compared to those in the matched normal tissues (Fig. 1). The difference in expression between tumor and normal tissue was highly statistically significant for the most common isoforms, the ΔN and α variants (P=.008). Differences in the other isoforms were not statically significant between tumor and normal, perhaps as a result of the low level of expression of these forms in both types of tissue.
Since this pattern of mRNA expression was similar to that of normal tissue, we sought to rule out the possibility the TP63 mRNA results might represent contamination by intermixed normal basal cells. Although our tissues obtained by manual dissection of the frozen blocks were approximately 75-80% pure tumor, contamination with normal basal cells was plausible since prostate cancer cells often permeate in and around benign glands. Therefore, we repeated the analysis using tumor tissue harvested with LCM (n=9), where our purity was raised to approximately 95-99% tumor cells. Somewhat surprisingly, we observed the same pattern of mRNA expression (Fig. 1). Like the non-LCM samples, decreased expression was most pronounced for the most common isoforms, the ΔN and α variants (P=.008) (Fig. 1). Differences in expression between normal tissue and tumor for the other isoforms were not significant.
In situ Hybridization
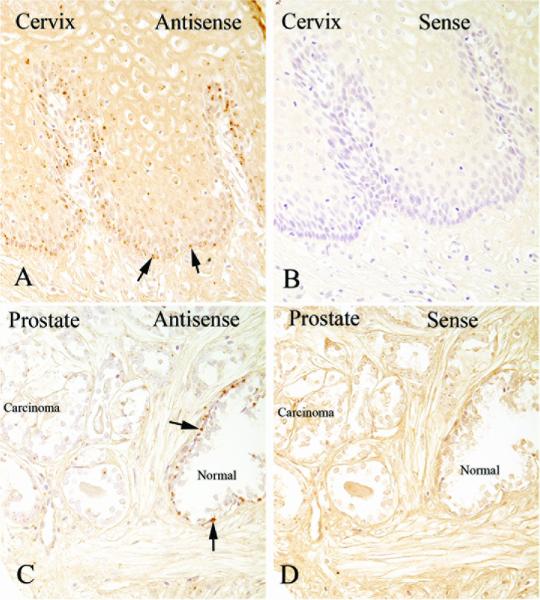
In order to attempt to determine whether the reduced, albeit present, levels of p63 mRNA that we identified in tumor samples were detectable by in situ hybridization we used a non-radioactive technique. As a positive control sections of human squamous epithelium, which express high levels of p63, were subjected to the in situ hybridization procedure. A strong signal was achieved using the antisense probe, but no signal was detected with the sense probe. There was a clear gradient of staining with cells towards the basal layer staining much stronger than those near the surface. As a negative control, sections of human colon were also subjected to the in situ protocol and no staining was detected with either the sense or antisense probes. In prostate tissues, a strong in situ hybridization signal for p63 mRNA was detected exclusively in the basal cells in normal epithelium using a prostate cancer tissue microarray containing sampled from 20 patients. By contract, p63 mRNA was undetectable in normal prostate luminal cells or in adenocarcinoma cells by in situ hybridization in the same specimens (Fig. 2). Since the RT-PCR results were clearly positive in cancer cells for all isoforms after the LCM procedure, and we were careful during the LCM procedure not to collect any benign prostate epithelial cells, we conclude that while our in situ assay is highly specific, we were not able to achieve a sufficiently sensitive assay to detect it in prostate cancer cells using this non-radioactive approach. Since we have not laser captured luminal vs. basal cells in the prostate, we have not determined whether luminal cells in normal acini are expressing any p63 mRNA.
Figure 2.
In situ hybridization against p63 mRNA shows strong signals in normal prostate basal cells. (A) Positive control tissue consisting of uterine cervix showing strong signals in the basal and para-basal cell regions when the antisense probe was used. (B) Adjacent tissue section in which the sense probe was used. (C) Prostate normal and carcinoma showing strong signals in basal cells in the normal gland that is present but absent signals in luminal cells and carcinoma cells. (D) Sense probe shows absence of signals in the prostate. All images x200 original magnification.
Mutational Analysis of TP63 in Prostate Cancer Cell Lines
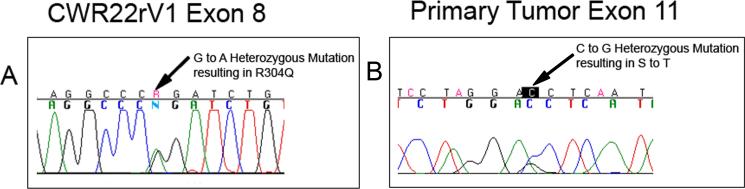
Using intronic primers, we performed mutational analysis of TP63 in 7 prostate cancer cell lines. Cell lines DU145, ME180, LNCaP, LNCaP C4-2B, MDAPCa2B, and PC3 contained wild-type TP63 sequences. Cell line CWR22Rv1 contained a single G to A mutation in exon 8, predicting an amino acid change of arginine (CGG) to glutamine (CAG) within the DNA-binding domain (Fig. 3). This mutation can be assumed to be functional since it is the exact change found in the germ line of one of the families with EEC syndrome (9). CWR22Rv1 is a hormone refractory prostate cancer cell line that was derived from its parental CWR22 xenograft, which does not grow in tissue culture. In order to determine whether the mutation we detected occurred during the development of the hormone refractory state, tissue was also obtained from the nonhormone refractory xenograft and subjected to DNA sequencing. Since the same mutation was identified in CWR22, we conclude that the mutation did not arise during the development of hormone refractory behavior. By immunohistochemical staining, the CWR22Rv1 cell line was negative for p63 (data not shown).
Figure 3.
Chromatogram traces of heterozygous mutations detected in CWR22Rv1 (A) and a single primary tumor (B).
Mutational Analysis of TP63 in Tumor/Normal Pairs in Radical Prostatectomy Specimens
Nineteen of twenty patients who underwent radical prostatectomy for localized prostate cancer contained wild-type sequences in tumor and matched normal tissue. One patient had a heterozygous single G to C mutation in exon 11 in the tumor tissue, predicting an amino acid change of serine (AGC) to threonine (ACC) (Fig. 3). It is not known whether this change is functional. The matched normal tissue in this patient was wild type, revealing a somatic mutation occurring in the tumor. Immunohistochemical staining of this tumor specimen revealed positive, albeit weak, staining in the nuclei of most tumor cells (data not shown).
Mutational Analysis of TP63 in Hormone Refractory Metastatic Tumors
To determine whether the development of hormone refractory lethal prostate cancer was related to TP63 mutations, multiple metastatic sites from 20 patients who underwent autopsy for prostate cancer were also examined. All tissues from all 20 patients’ metastatic tumors collected at autopsy contained wild-type TP63 sequences.
Mutational Analysis of TP63 in Tumor Xenografts
Since the only prostate cancer xenograft that we analyzed contained a point mutation in TP63 DNA-binding domain, we also analyzed a number of other prostate cancer xenografts, many of which contained both hormone naïve and hormone refractory counterparts. All 28 tumor xenografts contained wild-type TP63 sequences.
Discussion
We report that all well-characterized TP63 mRNA isoforms are expressed in the normal human prostate and that the most abundant isoforms contain ΔN and α, implying that the major isoform is TP63 ΔNα. Surprisingly, all isoforms were also expressed in a similar relative distribution to each other in primary prostate cancers, albeit the levels of expression were markedly reduced. To rule out the possibility that TP63 mRNA from adjacent normal tissue may have contaminated areas of tumor, we employed LCM to enhance the purity of the tumor cell isolation. The fact that we found similar levels of TP63 mRNA for all isoforms, even after LCM, indicates that primary prostate cancers indeed express TP63 mRNA.
The majority of prostate cancer cell lines and patient tumors we examined did not contain TP63 mutations, which suggests that somatic mutations are not the cause of decreased stable TP63 mRNA in the majority of prostate cancers. Still, the mutation in CWR22Rv1 is interesting because it is identical to a functional dominant negative TP63 mutation present in patients with a congenital TP63 defect that manifest EEC syndrome (9). Thus we infer that in CWR22Rv1 cells, TP63 is functionally inactivated through mutation. It has not been shown whether EEC is associated with an increased risk for cancer, and it is possible that the mutation in CWR22Rv1 is a germline mutation that has no role in prostate cancer.
Although we have not shown whether the mutation in the single primary tumor that showed a mutation is functionally important, the fact that the mutation was somatic, and apparently clonal, raises the possibility that it was indeed selected for during neoplastic transformation or progression. Of interest this patient's tumor did show some weak nuclear staining in tumor cells for p63. Taken together, our mutational data raise the possibility that a rare subset of human prostate cancers indeed contain TP63 mutations as part of their repertoire of important genetic changes and suggest that if TP63 is functioning as a cancer gene in the prostate, it appears to act more as a tumor suppressor than an oncogene.
Although it is clear that p63 protein is absent in the vast majority of human prostate cancer specimens, as determined by immunohistochemistry (18,20), the molecular mechanisms for absent expression of p63 protein in prostate cancer remain to be elucidated. On the one hand, since p63 is localized specifically to basal cells in the prostate, and basal cells are absent in prostate cancer, it is perhaps not unexpected that p63 protein is absent in prostate cancer. Yet other markers—including keratin 5, bcl-2, and c-met—that are more highly expressed in basal cells relative to luminal cells in normal prostate tissue have been identified in at least some prostate cancers and are increased in metastatic and/or hormone refractory disease (27-30). Although we have reported low-level p63 protein expression in the nuclei of a minority of high-grade prostate cancers (31), and others have reported much more rare cases with strong nuclear staining (32), the vast majority of human primary prostate cancers do not express detectable levels of p63 protein. Whether this can be explained at least in part by the fact that a microRNA (miR-203) is responsible for downregulating expression of p63 protein in the mouse epidermis as cells differentiate (15,16) is currently unknown.
In the prostate, it is not yet entirely clear whether p63-positive cells represent the true tissue stem cells, although mounting evidence in favor of this, at least in the mouse, has been generated recently. Tp63 null mice die within 1 day of birth and are born lacking a prostate (33). After isolation of the urogenital sinus from Tp63-/- mouse embryos and transplantation under the renal capsule of a wild type recipient, however, branching morphogenesis takes place and a structure with features of a prostate can develop (34). In this situation there are no basal cells and the luminal epithelial cells that form are abnormal in that while some contain androgen receptor and Nkx3.1 protein, most of them contain abundant mucin reminiscent of goblet cells in the intestine—cells that are not present in the normal prostate. By contrast, in intact mouse embryonic rescue experiments in which Tp63-/- embryos were injected with embryonic stem cells containing wild-type Tp63 and constitutively active β-galactosidase, p63 appeared to be absolutely required for normal prostate development including the appearance of both basal and luminal epithelial cells (35). This suggests that Tp63 is critical for the development and maintenance of normal prostatic epithelium (3,18). Several other studies in mice have indicated that cells with properties of prostate epithelial stem cells likely reside in the basal compartment (36,37), and a very recent study has shown that indeed clonally derived mouse prostate stem cells likely express p63 (38). In the human prostate, recent data indicate that cells with surface expression of CD133 possess stem cell–like features (39,40). While some studies have indicated that these cells are p63 positive (39), a recent study has suggested that these cells are actually p63 negative but quickly appear to give rise to cells that are p63 positive (40).
Conclusions
In summary we have shown that all major isoforms of TP63 mRNA are present in human prostate cancer and that a somatic mutation in TP63, while rare, has occurred in prostate cancer. Additional studies are required to determine why p63 protein is not expressed in most prostate cancers and the functional role, if any, that the different TP63 mRNA isoforms play in prostate cancer.
Acknowledgement
The authors would like to thank the John T. Isaacs lab for providing the CWR22 xenograft tissue.
Funded by Public Health Services NIH/NCI #R01CA084997, and NIH/NCI Specialized Program in Research Excellence (SPORE) in Prostate Cancer #P50CA58236 (Johns Hopkins). AMD is the Beth W. and A. Ross Myers Scholar supported through the Patrick C. Walsh Prostate Cancer Research Fund
References
- 1.Boman BM, Wicha MS. Cancer stem cells: a step toward the cure. J Clin Oncol. 2008;26(17):2795–2799. doi: 10.1200/JCO.2008.17.7436. [DOI] [PubMed] [Google Scholar]
- 2.Pierce GB. The cancer cell and its control by the embryo. Rous-Whipple Award lecture. American Journal of Pathology. 1983;113(1):117–124. [PMC free article] [PubMed] [Google Scholar]
- 3.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2(3):305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 4.Trink B, Okami K, Wu L, Sriuranpong V, Jen J, Sidransky D. A new human p53 homologue. Nat Med. 1998;4(7):747–748. doi: 10.1038/nm0798-747. [DOI] [PubMed] [Google Scholar]
- 5.Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M, Ikawa S. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med. 1998;4(7):839–843. doi: 10.1038/nm0798-839. [DOI] [PubMed] [Google Scholar]
- 6.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90(4):809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 7.Di Como CJ, Urist MJ, Babayan I, Drobnjak M, Hedvat CV, Teruya-Feldstein J, Pohar K, Hoos A, Cordon-Cardo C. p63 expression profiles in human normal and tumor tissues. Clin Cancer Res. 2002;8(2):494–501. [PubMed] [Google Scholar]
- 8.Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, Newbury-Ecob R, Hennekam RC, Van Buggenhout G, van Haeringen A, Woods CG, van Essen AJ, de Waal R, Vriend G, Haber DA, Yang A, McKeon F, Brunner HG, van Bokhoven H. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell. 1999;99(2):143–153. doi: 10.1016/s0092-8674(00)81646-3. [DOI] [PubMed] [Google Scholar]
- 9.Brunner HG, Hamel BC, Van Bokhoven H. The p63 gene in EEC and other syndromes. J Med Genet. 2002;39(6):377–381. doi: 10.1136/jmg.39.6.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398(6729):714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 11.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 12.Pellegrini G, Dellambra E, Golisano O, Martinelli E, Fantozzi I, Bondanza S, Ponzin D, McKeon F, De Luca M. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci U S A. 2001;98(6):3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129(3):523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 14.King KE, Ponnamperuma RM, Yamashita T, Tokino T, Lee LA, Young MF, Weinberg WC. deltaNp63alpha functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene. 2003;22(23):3635–3644. doi: 10.1038/sj.onc.1206536. [DOI] [PubMed] [Google Scholar]
- 15.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452(7184):225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lena AM, Shalom-Feuerstein R, di Val Cervo PR, Aberdam D, Knight RA, Melino G, Candi E. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15(7):1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 17.Yang A, Kaghad M, Caput D, McKeon F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 2002;18(2):90–95. doi: 10.1016/s0168-9525(02)02595-7. [DOI] [PubMed] [Google Scholar]
- 18.Signoretti S, Waltregny D, Dilks J, Isaac B, Lin D, Garraway L, Yang A, Montironi R, McKeon F, Loda M. p63 is a prostate basal cell marker and is required for prostate development. Am J Pathol. 2000;157(6):1769–1775. doi: 10.1016/S0002-9440(10)64814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi H, Fukutome K, Watanabe M, Furusato M, Shiraishi T, Ito H, Suzuki H, Ikawa S, Hano H. Mutation analysis of the p51 gene and correlation between p53, p73, and p51 expressions in prostatic carcinoma. Prostate. 2001;47(2):85–90. doi: 10.1002/pros.1050. [DOI] [PubMed] [Google Scholar]
- 20.Parsons JK, Gage WR, Nelson WG, De Marzo AM. p63 protein expression is rare in prostate adenocarcinoma: implications for cancer diagnosis and carcinogenesis. Urology. 2001;58(4):619–624. doi: 10.1016/s0090-4295(01)01311-5. [DOI] [PubMed] [Google Scholar]
- 21.Hibi K, Trink B, Patturajan M, Westra WH, Caballero OL, Hill DE, Ratovitski EA, Jen J, Sidransky D. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci U S A. 2000;97(10):5462–5467. doi: 10.1073/pnas.97.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urist MJ, Di Como CJ, Lu ML, Charytonowicz E, Verbel D, Crum CP, Ince TA, McKeon FD, Cordon-Cardo C. Loss of p63 expression is associated with tumor progression in bladder cancer. Am J Pathol. 2002;161(4):1199–1206. doi: 10.1016/S0002-9440(10)64396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi H, Inokuchi K, Sakuma Y, Dan K. Mutation of the p51/p63 gene is associated with blastic crisis in chronic myelogenous leukemia. Leukemia. 2001;15(11):1729–1734. doi: 10.1038/sj.leu.2402265. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama M, Bennett CJ, Hicks JL, Epstein JI, Platz EA, Nelson WG, De Marzo AM. Hypermethylation of the human glutathione S-transferase-pi gene (GSTP1) CpG island is present in a subset of proliferative inflammatory atrophy lesions but not in normal or hyperplastic epithelium of the prostate: a detailed study using laser-capture microdissection. Am J Pathol. 2003;163(3):923–933. doi: 10.1016/s0002-9440(10)63452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latil A, Vidaud D, Valeri A, Fournier G, Vidaud M, Lidereau R, Cussenot O, Biache I. htert expression correlates with MYC over-expression in human prostate cancer. Int J Cancer. 2000;89(2):172–176. [PubMed] [Google Scholar]
- 26.Hagiwara K, McMenamin MG, Miura K, Harris CC. Mutational analysis of the p63/p73L/p51/p40/CUSP/KET gene in human cancer cell lines using intronic primers. Cancer Res. 1999;59(17):4165–4169. [PubMed] [Google Scholar]
- 27.Shi XB, Ma AH, Tepper CG, Xia L, Gregg JP, Gandour-Edwards R, Mack PC, Kung HJ, deVere White RW. Molecular alterations associated with LNCaP cell progression to androgen independence. Prostate. 2004;60(3):257–271. doi: 10.1002/pros.20039. [DOI] [PubMed] [Google Scholar]
- 28.Pisters LL, Troncoso P, Zhau HE, Li W, von Eschenbach AC, Chung LW. c-met proto-oncogene expression in benign and malignant human prostate tissues. J Urol. 1995;154(1):293–298. [PubMed] [Google Scholar]
- 29.Colombel M, Symmans F, Gil S, O'Toole KM, Chopin D, Benson M, Olsson CA, Korsmeyer S, Buttyan R. Detection of the apoptosis-suppressing oncoprotein bc1-2 in hormone-refractory human prostate cancers. Am J Pathol. 1993;143(2):390–400. [PMC free article] [PubMed] [Google Scholar]
- 30.McDonnell TJ, Troncoso P, Brisbay SM, Logothetis C, Chung LW, Hsieh JT, Tu SM, Campbell ML. Expression of the protooncogene bcl-2 in the prostate and its association with emergence of androgen-independent prostate cancer. Cancer Res. 1992;52(24):6940–6944. [PubMed] [Google Scholar]
- 31.Parsons JK, Gage WR, Nelson WG, De Marzo AM. p63 protein expression is rare in prostate adenocarcinoma: implications for cancer diagnosis and carcinogenesis. Urology. 2001;58(4):619–624. doi: 10.1016/s0090-4295(01)01311-5. [DOI] [PubMed] [Google Scholar]
- 32.Osunkoya AO, Hansel DE, Sun X, Netto GJ, Epstein JI. Aberrant diffuse expression of p63 in adenocarcinoma of the prostate on needle biopsy and radical prostatectomy: report of 21 cases. Am J Surg Pathol. 2008;32(3):461–467. doi: 10.1097/PAS.0b013e318157020e. [DOI] [PubMed] [Google Scholar]
- 33.Signoretti S, Waltregny D, Dilks J, Isaac B, Lin D, Garraway L, Yang A, Montironi R, McKeon F, Loda M. p63 is a prostate basal cell marker and is required for prostate development. Am J Pathol. 2000;157(6):1769–1775. doi: 10.1016/S0002-9440(10)64814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurita T, Medina RT, Mills AA, Cunha GR. Role of p63 and basal cells in the prostate. Development. 2004;131(20):4955–4964. doi: 10.1242/dev.01384. [DOI] [PubMed] [Google Scholar]
- 35.Signoretti S, Pires MM, Lindauer M, Horner JW, Grisanzio C, Dhar S, Majumder P, McKeon F, Kantoff PW, Sellers WR, Loda M. p63 regulates commitment to the prostate cell lineage. Proc Natl Acad Sci U S A. 2005;102(32):11355–11360. doi: 10.1073/pnas.0500165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burger PE, Xiong X, Coetzee S, Salm SN, Moscatelli D, Goto K, Wilson EL. Sca-1 expression identifies stem cells in the proximal region of prostatic ducts with high capacity to reconstitute prostatic tissue. Proc Natl Acad Sci U S A. 2005;102(20):7180–7185. doi: 10.1073/pnas.0502761102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci U S A. 2007;104(1):181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barclay WW, Axanova LS, Chen W, Romero L, Maund SL, Soker S, Lees CJ, Cramer SD. Characterization of adult prostatic progenitor/stem cells exhibiting self-renewal and multilineage differentiation. Stem Cells. 2008;26(3):600–610. doi: 10.1634/stemcells.2007-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson GD, Robson CN, Lang SH, Neal DE, Maitland NJ, Collins AT. CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci. 2004;117(Pt 16):3539–3545. doi: 10.1242/jcs.01222. [DOI] [PubMed] [Google Scholar]
- 40.Litvinov IV, Vander Griend DJ, Xu Y, Antony L, Dalrymple SL, Isaacs JT. Low-calcium serum-free defined medium selects for growth of normal prostatic epithelial stem cells. Cancer Res. 2006;66(17):8598–8607. doi: 10.1158/0008-5472.CAN-06-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]