Abstract
Population genotyping (PG) can underestimate resistance if resistance-containing low abundance variants go undetected. PG and clonal analysis (CA) results were compared in virologic failures (VFs) from a 48-week clinical trial that evaluated once-daily fosamprenavir/ritonavir (FPV/r) 1400 mg/100 mg or atazanavir/ritonavir (ATV/r) 300 mg/100 mg, each combined with tenofovir/emtricitabine, in antiretroviral-naive patients. VF was defined as confirmed HIV-1 RNA ≥400 copies/ml at ≥24 weeks or viral rebound >400 copies/ml any time following viral suppression. All patients had baseline PG. One hundred and six patients enrolled (53/arm). Baseline resistance mutations were more prevalent in patients receiving FPV/r (10/53) than ATV/r (3/53). Seven patients (7%) were VFs-four on FPV/r and three on ATV/r. In the four FPV/r-treated VFs, baseline HIV TAMs combinations and/or PI mutations were detected in one by PG at VF (RT: L210W + T215C; PR: M46I + L76V) and three others by CA alone (RT: L210W + T215Y; RT: M41L; RT: K65R + K70R; PR: I47V); all four had study drug-associated mutations (CA detecting more HIV-1 resistance mutations than PG). In the three ATV/r VFs, no baseline drug-associated mutations were detected by PG; for one patient CA detected RT: K65R; PR: I84V. Phylogenetic analysis revealed tight clustering for FPV/r-treated VFs with highly related clones, whereas HIV-1 from ATV/r-treated VFs had no outgrowth from baseline of low abundance resistance-containing variants. In conclusion, low-abundance HIV resistance-containing variants were detected in baseline samples from patients with VF. The archived viruses that reemerged under selection pressure and acquired additional mutations were found primarily in patients in the FPV/r arm. Despite this and a baseline resistance imbalance between the two arms, FPV/r and ATV/r provided similar virologic suppression through 48 weeks; however, these findings highlight the necessity for the development of quick and inexpensive methods for detection of minority species to better guide therapy selection.
Introduction
The emergence of resistance to antiretroviral drugs and the transmission of drug-resistant HIV strains to newly infected persons are now major public health concerns.1 Resistant variants comprising as little as 1% of the viral population in an HIV-infected person are clinically important because they can rapidly reemerge under drug selection pressure and hasten the occurrence of antiretroviral treatment failure.2,3 Population genotyping (PG) of HIV is routinely used to help guide treatment of HIV-infected patients.4,5 PG can reliably detect resistant variants only if they make up at least 20% of the circulating viral population, and, thus, it may be inadequate in the detection of low abundance HIV variants containing resistance mutations.6 These variants may possess a slight replication advantage under drug-specific selection pressure, permitting a low level of replication from which those variants may emerge as the dominant viral population. Additional mutations may also be selected during this period of low-level replication, thereby further impacting the patient's future therapeutic options.
To test this hypothesis, the mutational profiles of HIV assessed by PG and clonal analysis (CA) were compared for patients meeting virologic failure (VF) criteria in a 48-week, open-label clinical trial that evaluated two once-daily (qd) ritonavir-boosted protease inhibitor (PI) regimens, in combination with tenofovir/emtricitabine (TDF/FTC), in the treatment of antiretroviral therapy (ART)-naive HIV-infected patients.7
Materials and Methods
Study design
ALERT (COL103952) was a 48-week open-label, efficacy/safety study conducted at 16 U.S. centers in which antiretroviral-naive patients 18–65 years of age with documented HIV infection were randomized to all-qd regimens of either atazanavir (ATV) or fosamprenavir (FPV) boosted with ritonavir (r) 100 mg (ATV/r and FPV/r, respectively), in combination with TDF/FTC.7 Enrollment was stratified at screening by plasma HIV-1 RNA to one of two strata (<100,000 and ≥100,000 copies/ml). All sites received institutional review board (IRB) approval to participate in the study and all patients provided their written informed consent agreeing to participate in the study and to the testing procedures.
Determination of plasma viremia
Plasma samples were collected and assayed for all enrolled patients for HIV-1 RNA using Roche Amplicor HIV-1 Monitor Ultrasensitive assay 1.0 (lower limit of detection: 50 copies/ml) and by Standard assay (lower limit of detection: 400 copies/ml) at baseline (day 1) and at weeks 2, 4, 8, 16, 24, 32, 40, and 48. All patients with protocol-defined virologic nonresponse (VF; plasma HIV-1 RNA ≥400 copies/ml beginning with the week 24 assessment visit) had a second confirmatory measurement performed 2–4 weeks after the first measurement of HIV-1 RNA ≥400 copies/ml.
Genotypic and phenotypic analysis
For those patients meeting VF criteria, HIV population genotype and phenotype analysis was performed by Monogram (South San Francisco, CA). Phenotypic resistance was assessed for select PIs, nucleo(t)side reverse transcriptase inhibitors (NRTIs), and nonnucleoside reverse transcriptase inhibitors (NNRTIs) using the PhenoSense GT assay, and was expressed as the fold-change (FC) in IC50 values (i.e., clinical isolate IC50 divided by the IC50 of a fully drug-sensitive “wild-type” virus). Reduced susceptibility (RS) for each drug was defined by the FC in IC50 values above the cut-off values for each drug established by Monogram. Only values above the Monogram FC cut-offs are presented. HIV clonal analysis was performed at baseline and at select time points by Research Think Tank, Inc. (RTT, Alpharetta, GA) for baseline and on-therapy samples from patients with VF. RTT was blinded as the study arm in which the patients were enrolled during this analysis. RTT also analyzed HIV population genotype at baseline from each patient using their Genetanker assay in conjunction with the OpenGene System (Siemens Healthcare Diagnostics Inc., Deerfield, IL) as per the manufacturer's instructions. Genotypic resistance mutations are reported as codons associated with the resistance to the study drugs as defined by the International AIDS Society–USA (IAS-USA),8 as well as reversion mutations of the thymidine analogue mutation (TAM) T215.9 Phylogenetic analysis was performed by aligning the nucleotide sequence of the clonal species from each patient's samples at the baseline and on-therapy time points, followed by tree construction based on evolutionary distances using MEGA version 4.10,11
Results
Patient characteristics and disposition
A total of 106 patients were enrolled, with 53 randomized to the FPV/r regimen and 53 to the ATV/r regimen. The populations of both study arms were similar in most baseline characteristics (Table 1). As the patients did not have HIV genotyping performed at screen, HIV genotype was not a factor in the randomization process. Baseline HIV population genotype revealed that the FPV/r arm had a disproportionately higher number of patients (10/53; 19%) who entered the study with IAS-USA-defined resistance mutations compared with the ATV/r arm (3/53; 6%). In particular, in the FPV/r randomization arm, Subjects 1, 4, 5, and 7 and Patient 1 from the ATV/r arm had major IAS-USA-defined protease mutations at baseline with the potential to decrease protease susceptibility. The baseline HIV mutational profiles for those patients with HIV with IAS-USA-defined resistance mutations are shown in Table 2, and some of the mutations detected are known to impact response to the study drugs under investigation. Detection of any resistance-associated mutation at baseline in an ART-naive patient is concerning, since it may indicate the possibility of additional underrepresented archived virus-containing resistance mutations that are at a replication disadvantage in the absence of specific ART.
Table 1.
HIV Population Genotyping Profiles for All Patients with Baseline IAS-USA-Defined Major Resistance Mutations or TAMS Reversion Mutationsa
| FPV/r patients (n = 53) | ATV/r patients (n = 53) | Baseline RT mutations | Baseline protease mutations |
|---|---|---|---|
| 1b | L210W, T215C | K20I, M46I, I62V, L63P, A71V, L76V, V77I, I93L | |
| 2 | T215N | L63P | |
| 3 | A62V, V75I, Y188L | M36I/M, I64V | |
| 4 | V108I/V, Y115F, L210W, T215D | L10F, M36I, L63P, I64I/L, A71V, N88D, L90M, I93M | |
| 5 | M41L, V75I, F77L, K103N/S, F116Y, Q151M | M46I, L90M | |
| 6 | F77F/L | WT | |
| 7 | WT | I47I/V, L63P, V77I, I93L | |
| 8 | Y181C | M36I, I62V, I64M, A71V, I93L | |
| 9 | WT | I13V, D60D/E, I62I/V, L63P, I64I/L, N88N/S | |
| 10 | M41L, T215C/S | M36I, I62V, L63P, I93L | |
| 1 | WT | D60E, L63P, A71V, L90M, I93L | |
| 2 | M41L, K103N, T215S | L63L/P, V77I | |
| 3 | K219E/Q | L10V, I13V, G16E, I64L |
All 106 enrolled patients had HIV population genotypic analysis performed. Of those, 13 patients (10 randomized to the FPV/r arm; 3 randomized to the ATV/r arm) had IAS-USA-defined major resistance mutations or TAMS reversion mutations at baseline. Their genotypes are shown. Major IAS-USA resistance-associated mutations are underlined. Mutations that could have impacted the specific study regimen, e.g., TAMs and IAS-USA-defined mutations associated with ATV resistance or FPV resistance, are shown in bold.
This patient (Patient FPV/r 1 in Table 2) later experienced VF while on FPV/r-based therapy.
Table 2.
Comparison of Population Genotypic Results to Genotypic Results Obtained by Clonal Analysis at Baseline and the Failure Time Points for the Seven Patients Meeting VF Criteriaa
| |
Baseline |
On therapy/virologic failure |
||||
|---|---|---|---|---|---|---|
| Patient | Phenotype | Population genotype | Clonal analysis | Phenotype | Population genotype | Clonal analysis |
| FPV/r-1b | FPV FC = 3.8 | RT: L210W + T215C; PR: M46I + L76V |
RT:L210W, T215C; PR: M46I, L76V (50/53) |
FPV FC = 3.0 | RT: M184V,L210W + T215C | RT: M184V,L210W, T215C; PR: M46I, I54M or L,L76V (48/55) |
| TDF FC = 1.0 | TDF FC = 0.6 | |||||
| FTC FC = 1.5 | RT: M41L,L210W, T215C; PR: M46I, L76V (1/53) RT: L210W, T215C or Y; PR: M46I, L76V (2/53) |
FTC FC = 78 | PR: M46I, I54I/L/M,L76V | RT: M184V,L210W, T215C; PR: M46I, I47V (1/55) RT: M184V,L210W, T215C; PR: M46I, L76V(2/55) WT (1/55) RT: M184V, L210W, T215C; PR:M46I,I47V,L76V (3/55) |
||
| FPV/r-2 | FPV FC = 1.4 | WT | WT (53/58) | FPV FC = 1.3 | RT: M41L, M184V,L210W, T215S/Y; PR: WT | RT: M41L, M184V,T215Y; PR: WT (1/51) |
| TDF FC = 0.8 | RT:Y181I + 215P + 236L; PR:WT (1/58) |
TDF FC = 1.8 | RT: M41L, M184V,L210W, T215Y; PR: WT(39/51) | |||
| FTC FC = 1.1 | RT: L210W, T215Y; PR:WT (1/58) RT: M41L; PR: WT (3/58) |
3TC FC > MAX | RT: M184V, PR: WT (8/51) RT: K103N, M184V, P225H; PR: WT (2/51) RT: Y181C, M184V; PR: WT (1/51) |
|||
| FPV/r-3 | FPV FC = 2.2 | WT | WT (48/50) | FPV FC = 38 | RT: M184V | RT: M184V; PR:M46I + I50V (54/58) |
| TDF FC = 0.9 | RT: K65R, K70R; PR: WT (1/50) |
TDF FC = 0.5 | PR: M46I, I50V | RT: M184V + Y188H; PR:M46I + I50V (1/58) |
||
| FTC FC = 0.8 | RT: Y188C; PR: WT (1/50) |
FTC FC = 76 | RT: D67N + M184V; PR:M46I + I50V (1/58) RT: M184V; PR:L33F + M46I + I50V (1/58) RT: M184V; P225H; PR:M46I + I50V (1/58) |
|||
| FPV/r-4 | FPV FC = 0.8 | WT | WT (51/53) | FPV FC = 19 | RT: K65R, D67N | RT: K65R; PR:V32I, M46I,I47V (35/55) |
| TDF FC = 0.7 | RT: WT, PR:N88S (1/53) | TDF FC = 1.8 | PR: V32I, M46I, | RT: K65R; PR:V32I,I47V (1/55) | ||
| FTC FC = 0.7 | RT: WT, PR: I47V (1/53) | FTC FC = 8.5 | I47V | RT: K65R, M184I or V; PR:V32I, M46I,I47V (5/55) RT: K65R, D67N; PR:V32I, M46I,I47V (14/55) |
||
| ATV/r-1 | ATV FC = 1.2 | WT | WT (51/52) | ATV FC = 1.0 | RT: M184V, | WT (34/60) |
| TDF FC = 1.0 | RT: WT; PR: N88S(1/52) | TDF FC = 0.6 | PR: WT | RT: D67N; PR: WT (1/60) | ||
| FTC FC = 1.1 | FTC FC = 18 | RT: WT; PR: V82A; (1/60) RT: M184V; PR: WT (24/60) |
||||
| ATV/r-2 | ATV FC = 1.2 | WT | WT (49/55) | ATV FC = 3.9 | WT | WT (52/52) |
| TDF FC = 0.9 | RT: K65R; PR: WT (1/55) | TDF FC = 0.8 | ||||
| FTC FC = 1.2 | RT: WT; PR:V82S (1/55) RT: T215P, P236L; PR:WT (1/55) RT: WT; PR: I84V (2/55) RT: Y181C; PR: WT (1/55) |
FTC FC = 0.9 | ||||
| ATV/r-3 | ATV FC = 0.9 | WT | WT (48/50) | ATV FC = 2.1 | WT | WT (56/59) |
| TDF FC = 1.1 | RT: T215P, P236L; PR:WT (5/50) |
TDF FC = 1.1 | RT: T215R, P236L; PR: WT (2/59) | |||
| FTC FC = 1.0 | RT: P236L; PR WT (1/50) | FTC FC = 1.1 | RT: V106A; PR: WT (1/59) | |||
Major IAS-USA-defined resistance-associated mutations or TAMs reversion mutations are shown and IAS-USA-defined mutations associated with resistance to the specific study regimen are in bold. Minor IAS-USA-defined protease mutations are not shown. Mutations detected at baseline by either method that were also present at the failure time points are underlined. Population phenotypic susceptibility fold-changes (FC) for the study drug regimen are also presented; phenotypic results that exceed the Monogram lower limit cut-offs for reduced susceptibility (TDF: 1.4 FC; FTC: 3.5 FC; ATV: 2.2 FC; and FPV: 2.0 FC) are in bold and underlined. A minimum of 50 clones was analyzed at each time point; the number of clones with that genotype for that sample is shown in parentheses after each genotype.
Patient FPV/r-1 is Patient 1 as described in Table 1. Week 12 on-therapy results compared (insufficient week 24 sample was available for clonal analysis); all other CG and PG obtained at VF.
At week 48, by ITT-observed analysis, 93% (42/45) of the patients on the FPV/r arm vs. 96% (46/48) on the ATV/r arm achieved VL <400 copies/ml and 89% (40/45) on the FPV/r arm vs. 92% (44/48) on the ATV/r arm achieved VL <50 copies/ml.5 A total of seven patients met VF criteria through 48 weeks, including four in the FPV/r arm and three in the ATV/r arm.
HIV-1 PG and phenotypic resistance in patients with VF
HIV-1 PG and phenotypic and viral load response profiles for the four patients with VF on the FPV/r arm are shown in Fig. 1, and response profiles for the three patients with VF on the ATV/r arm are shown in Fig. 2. Six of these seven patients were infected with clade B HIV-1; one was infected with clade G HIV-1. All patients experienced a >1.5 log drop in viremia postbaseline prior to rebound.
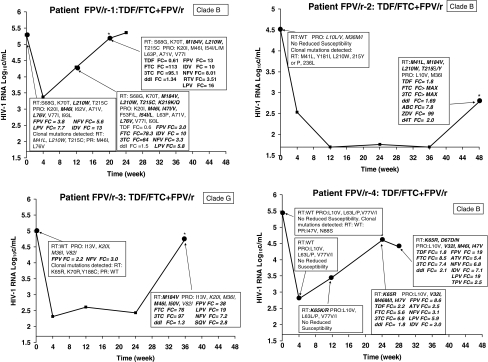
FIG. 1.
Longitudinal population genotypic and phenotypic changes for patients with virologic failure treated with FPV/r + TDF/FTC. Major IAS-USA-defined resistance mutations detected by population sequencing are shown in bold italics. The fold changes in IC50 are shown for any time point at which it was above the drug cut-off, indicating reduced susceptibility to that drug; although evaluated, fold changes below the cut-offs are not presented. All IAS-USA-defined major resistance mutations detected at baseline by clonal analysis are also presented.
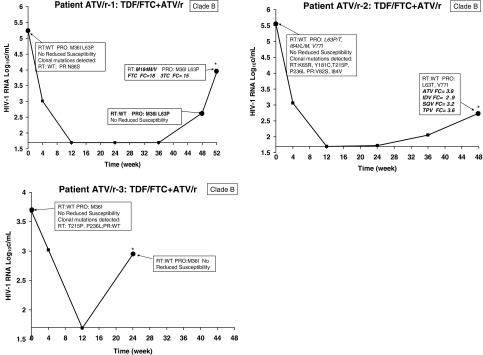
FIG. 2.
Longitudinal population genotypic and phenotypic changes for patients with virologic failure treated with ATV/r + TDF/FTC. Major IAS-USA-defined resistance mutations detected by population sequencing are shown in bold italics. The fold changes in IC50 are shown for any time point at which it was above the drug cut-off, indicating reduced susceptibility to that drug; although evaluated, fold changes below the cut-offs are not presented. All IAS-USA-defined major resistance mutations detected at baseline by clonal analysis are also presented.
Population resistance in the FPV/r arm
As seen in Fig. 1, HIV from patient FPV/r-1 had evidence by population sequencing of mutations prior to antiviral initiation that could impact study response. These included the TAM L210W, the TAM reversion mutation T215C, and the major protease mutation M46I plus additional minor protease mutations. RS to FPV and to other PIs, including nelfinavir (NFV), indinavir (IDV), and lopinavir (LPV), was also observed. Additional RT and protease mutations were selected at weeks 12 and 28, by which time RS to FTC and FPV was detected.
HIV-1 from patient FPV/r-2 had no PG-detectable major resistance-associated mutations at baseline, and was susceptible to all study drugs. This patient experienced a rebound in viremia at week 48, at which time PG detected the TAMs M41L, M184V, L210W, and T215S/Y, plus the M184V mutation; the virus had RS to all marketed NRTIs.
Patient FPV/r-3 was infected with clade G HIV-1 and at baseline had RS to both FPV and NFV but had no PG-detectable major resistance-associated mutations. At week 36, virus from this patient had selected for the M184V mutation and the protease mutations M46I and I50V, and had RS to the study drug FTC in addition to newly observed cross-resistance to several other NRTIs and PIs.
HIV-1 from patient FPV/r-4 had no PG-detectable major resistance-associated mutations at baseline and at week 4, and was susceptible to all study drugs. At week 12, a K65K/R mutation mixture was observed by PG, although this mixture did not result in RS for either FTC or TDF. K65R was detected at week 24, resulting in RS to both NRTI study drugs, and the protease mutations I32V, M46M/I, and I47V were detected, resulting in RS to FPV, in addition to newly observed cross-resistance to several other NRTIs and PIs. Postfailure, selection for the TAM D67D/N mutation was also detected, although the RS pattern remained similar to that observed at VF.
Population resistance in the ATV/r arm
HIV-1 from patient ATV/r-1 had no PG-detectable major resistance-associated mutations at baseline, and was susceptible to all study drugs (Fig. 2). This patient experienced a rebound in viremia at week 48, at which time PG detected M184M/V mixture and RS to study drugs FTC and 3TC.
HIV-1 from patient ATV/r-2 had no PG-detectable major resistance-associated mutations at baseline, and was susceptible to all study drugs. This patient experienced a rebound in viremia at week 48, at which time by population genotype no major resistance-associated mutations were detected, although RS to ATV plus indinavir (IDV), saquinavir (SQV), and tipranavir (TPV) was observed.
HIV-1 from patient ATV/r-3 had no PG-detectable major resistance-associated mutations at baseline or at VF (week 24), and was susceptible to all study drugs.
HIV-1 CA and PG results comparison for patients with VF
HIV-1 CA was performed using samples obtained at baseline and at a virologic failure time point by Research Think Tank and those results were compared with the paired PG results. A minimum of 50 clones was examined per time point. For the baseline study visit, any major IAS-USA defined or T215 reversion mutations detected by clonal analysis are presented in the response profiles shown in Figs. 1 and 2.
As shown in Table 2, Patient FPV-1's HIV PG at baseline included TAMs L210W and T215C and protease mutations M46I and L76V, while additional TAMs (M41L and T215Y) were detected by CA. These mutations were present at week 12, and both PG and CA detected the same major resistance-associated mutations. By PG, HIV from patient FPV/r-2 at baseline was WT (wild type). However, CA detected variants containing one or more of the following TAMs (M41L, L210W, T215Y) and the NNRTI mutation Y181I. These same major NRTI resistance-associated mutations were detected at VF, with CA also detecting additional NNRTI mutations and both methods detecting the M184V mutation. HIV-1 from patient FPV/r-3 was also WT by PG at baseline (although this clade G virus had baseline FPV RS). By CA, clones with NRTI (K65R and K70R) and an NNRTI mutation (Y188C) were detected at baseline. However, these mutations were not detected at VF and study drug resistance-associated mutations emerged that were detected by both methods. For patient FPV/r-4 whereas by PG HIV was WT at baseline, CA detected protease mutations variants with N88S or I47V. At VF, this I47V mutation was also detected by both PG and CA. Additional major resistance-associated mutations were observed at VF by both methods, with CA detecting one additional mutation (M184I).
In the ATV/r arm, baseline PG showed that patient ATV/r-1 had WT HIV-1, but CA detected protease mutations variants with N88S or I47V. At VF, PG and CA detected M184V but not the protease mutations variants. Both methods detected M184V, but only the CA detected the PR V82A and RT D67N. Patient ATV/r-2 also had WT HIV-1 at baseline, whereas CA detected clones containing NRTI mutations K65R or T215P and protease mutations V82S and I84V. At VF, however, no resistance mutations were detected in this patient by either method. Patient ATV/r-3 similarly had WT HIV-1 at baseline; at VF CA alone detected clones containing NRTI mutations T215P and NNRTI mutation P236L, but no study drug-specific resistance mutations were detected by either method; low-abundance variants containing the P236L mutation were also present at VF.
The presence of baseline low abundance mutation-containing viral quasispecies detectable only by CG was more closely associated with poor outcomes than what could be predicted utilizing only PG. Of the seven subjects who met VF criteria, only one patient had a baseline HIV population sequence that detected mutations with the potential to impact the study drug regimen; five of seven additional patients with VF had baseline low abundance mutation-containing viral quasispecies at prevalences of <10% with the potential to impact the specific study drug regimen used.
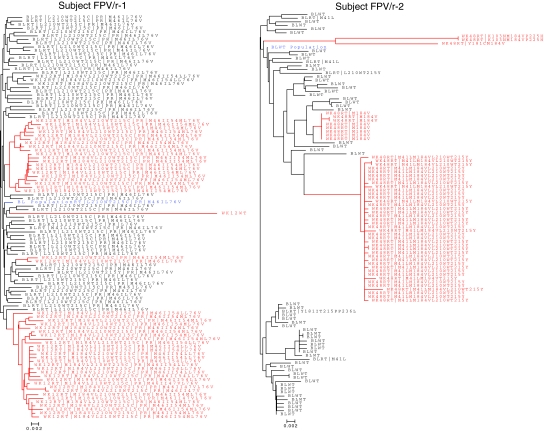
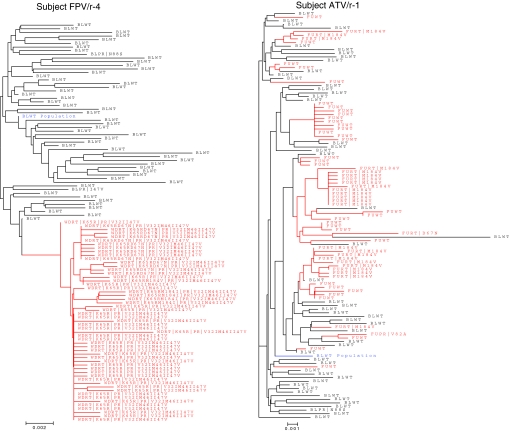
Phylogenetic analysis of clonal HIV from patients with VF
Phylogenetic analysis was performed for HIV from all VFs. Four representative phylogeny trees are shown in Fig. 3. Phylogenetic distance analysis for FPV/r-treated patients 1, 2, and 4 demonstrated that clones from VFs in this arm exhibited minimal diversity with just a few clusters that included highly related clones. In contrast, phylogenetic distance analysis for ATV/r arm, as shown for patient ATV/r-1, revealed that clones detected at VF do not cluster together and that they display a relatively high degree of genetic diversity, possibly indicating many pathways for de novo selection of M184V rather than a situation in which the VF clones arise from a single or small number of variants.
FIG. 3.
Phylogenetic analysis of representative patients FPV/r-1, FPV/r-2, FPV/r-4, and ATV/r-1. Blue: Baseline population sequence (labeled as BL population). Black: Baseline time point clones (labeled BL followed by clone identification number). Red: Virologic failure time point clones [labeled either as week 12, week 48, withdrawal (WD). or follow-up (FU) followed by the clone identification number]. Clonal analysis of samples from all FPV/r-treated patients exhibits a high degree of clustering of failure sequences, indicating expansion from relatively few parental clones, whereas clones from the phylogenetic tree of the ATV/r-treated patient (ATV/r-1) exhibit a high degree of genetic diversity.
Discussion
Transmission of drug-resistant HIV has been increasing in the United States.12 Current DHHS and European treatment guidelines now recommend that genotyping be performed in ART-naive patients before ART is initiated.13,14 These revisions occurred after ALERT was started, so genotyping was not performed at the time of study screening and was not a randomization criteria.
Retrospective baseline analysis of HIV from all patients demonstrated that patients randomized to the FPV/r + TDF/FTC arm were >3-fold more likely to have enrolled with RT or major protease mutations than patients randomized to the ATV/r + TDF/FTC arm (19%; 10/53 patients versus 6%; 3/53 patients, respectively). Despite this imbalance in the percentage of patients entering the study with preexisting drug resistance mutations that favored patients randomized to the ATV/r arm, similar numbers of patients on either arm experienced VF (four patients in the FPV/r arm and three in the ATV/r arm), and similar virologic suppression was observed with both qd treatment regimens through 48 weeks in the ITT-observed analysis. This finding was notable, because although there have been many studies comparing the efficacy of PIs boosted with 100 mg of ritonavir twice daily, there are fewer studies examining the efficacy using 100 mg ritonavir once-daily boosting in one or more study arms for ATV/r or FPV/r.15–18
Population resistance profiles at virologic failure from three of the four ALERT study patients with VF in the FPV/r + TDF/FTC arm strongly suggested that HIV isolated from these patients contained drug resistance mutations that impacted response to one or more study drugs prior to the start of their antiviral therapy. However, only one patient (patient FPV/r-1) had HIV with major RT and protease drug resistance-associated mutations that were detectable by PG. CA revealed that HIV from an additional two patients (FPV/r-2 and FPV/r-4) also had low abundance species with resistance mutations to one or more study drugs prior to initiating treatment with FPV/r + TDF/FTC. These mutations allowed the virus some escape from drug selection pressure that impacted virologic response and resulted in VF, and were detectable by population sequencing at VF. This contrasts with what was observed for the patients with VF on the ATV/r arm, where no outgrowth from baseline of low abundance resistance mutation-containing variants was observed at VF.
Data from both the PG and CA suggest that baseline low-level drug resistance to one or more study drugs impacted the initial response in the patients with VF on the FPV/r arm and contributed to their VF, whereas the reasons for VF for those patients on the ATV/r arm remain unclear, and could have been impacted by a number of other factors, e.g., suboptimal adherence, reduced plasma drug levels, and potency or therapy interruption. In a recent editorial, Steve Deeks has suggested that a sizable proportion of treatment-naive HIV-infected individuals harbor minority drug-resistant HIV species, and in some patients these infrequent variants may reflect natural variability and are unlikely to compromise treatment, whereas in others these variants likely reflect transmission of an HIV population that has been exposed to suboptimal drug pressures, and can compromise future responses in some but not all individuals.19 Phylogenetic analysis of the clonal data from patients on the FPV/r + TDF/FTC arm at VF demonstrated a much lower degree of genetic diversity relative to baseline clones, suggesting that replication occurred by only a few mutational pathways, with genetic bottlenecking probably due to incomplete suppression of replication in the presence of drugs for those viruses already carrying study drug resistance-associated mutations. It should be noted that to perform the phylogenetic analysis and establish the mutational linkage between the protease and RT regions in each clone, higher throughput methods to evaluate low abundance variants by the examination of individual mutations or analysis of short stretches of nucleic acid sections were eschewed and traditional methods of CA were employed. Because of this, a limitation of this study is that any low abundance mutations present at either baseline or at VF occurring at an incidence of <1/50 clones may not have been detected.
As recently presented, ultradeep sequencing was used by Lataillade et al. to examine the prevalence of protease and RT mutations pre-therapy in ART-naive patients randomized to receive TDF/FTC plus either ATV/r or lopinavir/r.20 Samples from all patients who later met VF criteria (n = 53) and a subset of samples from patients who successfully suppressed were analyzed (n = 141), and the data were censored such that only mutations occurring at a prevalence of ≥1% were included. A high incidence of drug resistance mutations was observed, with 30.5% of patients having detectable drug resistance and 15.6% of the samples having mutations detectable at <20% of the viral population. NRTI resistance was the most commonly detected of the three drug classes, and was detected in 24 vs. 25% of the two groups. The presence of TAMs + M184V at baseline was highly correlated with VF in this TDF/FTC-treated cohort, and all patients (3/3) whose virus contained this genotype experienced VF, whereas 6/9 (66%) of the patients with detectable M184V/I experienced VF and 7/11 (44%) of subjects with multiple TAMs detected in their baseline viral genotype experienced VF. In contrast, only 1/14 (10%) of patients whose baseline viral genotype had protease mutations went on to experience VF. Although the overall incidence of detectable drug resistance at baseline in our study is much lower, the conclusions are in agreement with what was observed in the ALERT study. The majority of patients who experienced VF (5/7) in ALERT had low abundance viral quasispecies containing TAMs, TAMs reversion mutations, or K65R detected at baseline. Only 2/7 patients who experienced VF had low abundance viral quasispecies containing major protease mutations at baseline in the absence of TAMs, TAMs reversion mutations, or K65R.
In conclusion, the data from this analysis suggest that the majority of patients treated with either FPV/r or ATV/r-based regimens achieved and maintained virologic suppression, whereas in a minority of patients who experienced virologic rebound, low-abundance HIV variants containing mutations that rendered them less sensitive to the study drugs were present in baseline samples. Some of these archived viruses that reemerged under selection pressure acquired additional mutations, thus increasing their impact on drug resistance. These findings underscore the utility of assays for low abundance variants, and the need for the development of rapid and inexpensive methods for their detection to provide better guidance of therapy selection and thereby prevent outgrowth of low-abundance HIV species and their subsequent evolution.
Acknowledgments
We are grateful to all the patients, investigators, and staff at the following U.S. research sites: Cynthia Brinson, Austin, TX; Cal Cohen, Boston, MA; Orlando Immunology Center (DeJesus site), Orlando, FL; University of Miami School of Medicine (Fischl site), Miami, FL; Joseph Horvath, Columbia, SC; Ricky Hsu, New York, NY; Lewis McCurdy, Charlotte, NC; Cheryl McDonald, Ft. Worth, TX; Bruce Rashbaum, Washington, DC; Robert Scott, Oakland, CA; Rush-Presbyterian-St. Luke's Medical Center (Smith site), Chicago, IL; Ford Kinder, Miami, FL; Kaiser Permanente (Weinberg site), Atlanta, GA; and Ben Young, Denver, CO, to Robert Lloyd and Rodney Mathis at Research Think Tank and to Brian Wine, Linda Yau, Qiming Liao, and Jay Li of GlaxoSmithKline for their help with data analysis.
Author Disclosure Statement
This study was funded by a grant from GlaxoSmithKline.
References
- 1.Hogg RS. Bangsberg DR. Lima VD, et al. Emergence of drug resistance is associated with an increased risk of death among patients first starting HAART. PLoS Med. 2006;3:1570–1577. doi: 10.1371/journal.pmed.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simen BB. Simons JF. Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199:693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JA. Li JF. Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008;5:1112–1121. doi: 10.1371/journal.pmed.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammer SM. Eron JJ., Jr. Reiss P, et al. Antiretroviral treatment of adult HIV infection: 2008 recommendations of the International AIDS Society-USA panel. JAMA. 2008;300:555–570. doi: 10.1001/jama.300.5.555. [DOI] [PubMed] [Google Scholar]
- 5.Hirsch MS. Gunthard HF. Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47:266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 6.Toni TA. Asahchop EL. Moisi D, et al. Detection of human immunodeficiency virus (HIV) type 1 M184V and K103N minority variants in patients with primary HIV infection. Antimicrob Agents Chemother. 2009;53:1670–1672. doi: 10.1128/AAC.01494-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith KY. Weinberg WG. DeJesus E, et al. Fosamprenavir or atazanavir once daily boosted with ritonavir 100 mg, plus tenofovir/emtricitabine, for the initial treatment of HIV infection: 48-week results of ALERT. AIDS Res Ther. 2008;5:5. doi: 10.1186/1742-6405-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson VA. Brun-Vezinet F. Clotet B, et al. Update of the drug resistance mutations in HIV-1. Top HIV Med. 2008;16:138–145. [PubMed] [Google Scholar]
- 9.Shafer RW. Rhee SY. Pillay D, et al. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. AIDS. 2007;21:215–223. doi: 10.1097/QAD.0b013e328011e691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura K. Dudley J. Nei M. Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 11.Kumar S. Nei M. Dudley J. Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross LL. Wine BC. Vavro C, et al. Changes in regional prevalence, clade and epidemiology of HIV-1 drug resistance mutations and clade among antiviral therapy-naive patients in the United States (US) from 2000 to 2007. Antiviral Ther. 2008;13(S3):A160. [Google Scholar]
- 13.Department of Health and Human Services: Guidelines for the use of antiretroviral agents in HIV-1-infected adults, adolescents. Nov 3, 2008. http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf. [Jul 25;2009 ]. pp. 1–139.http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf
- 14.European AIDS Clinical Society (EACS): Guidelines for the clinical management and treatment of HIV infected adults in Europe. Oct 1, 2008. http://www.europeanaidsclinicalsociety.org/guidelines.asp. [Jul 25;2009 ]. http://www.europeanaidsclinicalsociety.org/guidelines.asp [DOI] [PubMed]
- 15.Molina JM. Andrade-Villanueva J. Echevarria J, et al. Once-daily atazanavir/ritonavir versus twice-daily lopinavir/ritonavir, each in combination with tenofovir and emtricitabine, for management of antiretroviral-naive HIV-1-infected patients: 48 week efficacy and safety results of the CASTLE study. Lancet. 2008;372:646–655. doi: 10.1016/S0140-6736(08)61081-8. [DOI] [PubMed] [Google Scholar]
- 16.Elion R. Cohen C. Ward D, et al. Evaluation of efficacy, safety, pharmacokinetics, and adherence in HIV-1-infected, antiretroviral-naive patients treated with ritonavir-boosted atazanavir plus fixed-dose tenofovir DF/emtricitabine given once daily. HIV Clin Trials. 2008;9(4):213–224. doi: 10.1310/hct0904-213. [DOI] [PubMed] [Google Scholar]
- 17.Elion R. DeJesus E. Sension M, et al. Once-daily abacavir/lamivudine and ritonavir-boosted atazanavir for the treatment of HIV-1 infection in antiretroviral-naive patients: A 48-week pilot study. HIV Clin Trials. 2008;9(3):152–163. doi: 10.1310/hct0903-152. [DOI] [PubMed] [Google Scholar]
- 18.Hicks CB. DeJesus E. Sloan LM, et al. Comparison of once-daily fosamprenavir boosted with either 100 or 200 mg of ritonavir, in combination with abacavir/lamivudine: 96-week results from COL100758. AIDS Res Hum Retroviruses. 2009;25:395–403. doi: 10.1089/aid.2008.0231. [DOI] [PubMed] [Google Scholar]
- 19.Deeks SG. Transmitted minority drug-resistant HIV variants: A new epidemic? PLoS Med. 2008;5:e164. doi: 10.1371/journal.pmed.0050164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lataillade M. Chiarella J. Yang R, et al. Prevalence and clinical significance of transmitted drug-resistant (TDR) HIV mutations by ultra deep sequencing (UDS) in HIV-infected ARV-naïve subjects in CASTLE Study. Antiviral Ther. 2009;14(S1):A44. [Google Scholar]