Global analysis of proteins
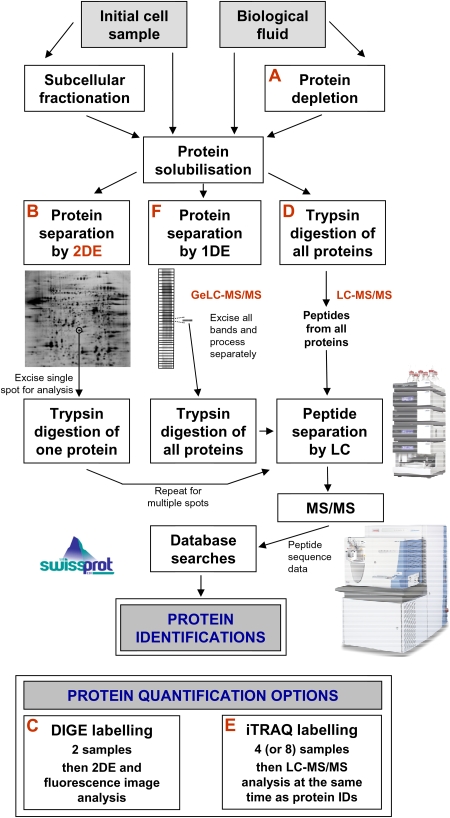
The application of proteomics (the study of protein products expressed by the genome) has become one of the leading post-genomic technologies given the increased understanding of the central role of proteins and protein–protein interactions in all aspects of cellular function [2]. Systematic global identification and quantification of proteins can, not only inform improved biomedical understanding of a particular system in healthy or diseased individuals, but also be used for protein biomarker discovery. The most popular of the many proteomic strategies available are summarized in Figure 1 and, whilst two-dimensional electrophoresis (2DE) gel-based approach remains popular, liquid chromatography–tandem mass spectrometry (LC-MS/MS) and gel electrophoresis then liquid chromatography mass spectrometry (geLC-MS) approaches are now preferred as state-of-the-art.
Fig. 1.
Strategies for the global identification and quantification of proteins. Proteins may be derived from biological fluids or solubilized from entire cells or their subcellular fractions. One option with biological fluids is to use proprietary immunodepletion methods (A) to remove up to 20 of the most abundant proteins in order to more readily analyse the lower abundance proteins. Following solubilization, proteins may be separated by 2DE (B). In this workflow, an individual separated protein is removed as a gel plug, trypsin digested and the resulting peptides are typically separated on the basis of relative hydrophobicity by nanoscale LC. The amino acid sequence of these peptides is then determined by MS/MS [19,20] and this data is used to search existing protein databases to achieve a match and, therefore, a protein identification (ID). One can process multiple gel plugs to identify many of the 2DE-separated proteins. The method of choice for 2DE protein quantification is DIGE (C) whereby samples are labelled with different fluorescence dyes prior to mixing together (multiplexing) and 2DE [16]. Such gel-based proteomics remains popular, but for global analysis, it is now more commonplace to trypsin digest the entire solubilized protein mixture to produce a peptide ‘soup’ of all the proteins in the sample (gel-free LC-MS/MS proteomics) (D). Peptides are then separated by LC on the basis of relative hydrophobicity and often also charge as a multidimensional separation. Then, extensive MS/MS and database searches are performed to identify many (ideally all) of the proteins in the original sample. One of the advantages of this workflow is that it is possible to achieve simultaneous quantitative data by introducing an iTRAQ labelling step (E) (or other labelling step) following trypsin digestion without the need for additional LC-MS/MS [17,18]. A further method for global protein identification is to first separate proteins by one-dimensional electrophoresis (1DE) before subjecting individual protein bands to digestion and LC-MS/MS (the geLC-MS workflow) (F). Whilst this is an excellent workflow for high numbers of protein IDs, it is not particularly amenable to protein quantification methodologies. Finally, there are many other options. For example, one may enrich for peptides or proteins of a particular type to study a particular group of proteins and this is most commonly performed for phosphorylation analysis. Peptide isoelectric focussing (IEF) as an additional step within the usual LC-MS/MS workflow is a valid option for increased numbers of IDs and protein LC can also be performed prior to trypsin digestion. Finally, SELDI-TOF MS ProteinChip technology may be potentially used for biomarker discovery, although the approach has certain limitations [21].
Protein biomarker discovery has been a major driving force for the field and abundant scientific evidence from proteomics and other disciplines strongly suggests that most, if not all, diseases will result in changes in certain proteins in plasma [3]. This primary clinical specimen is usually chosen for analysis but other biological fluids will also likely contain protein profiles indicative of homeostasis or diseases in that locale or organ system. For example, urine has been the focus of a number of studies in relation to kidney and bladder diseases [4,5] and cerebrospinal fluid has also received a good deal of attention for neurological conditions [6]. There have been very few proteomic investigations of clinical effusion materials (pleural, pericardial and peritoneal) [7] with preliminary reports on peritoneal fluid limited to studying endometriosis [8] and ovarian cancer [7,9].
Proteomics to study peritoneal dialysis
One of the reasons why there have been so few studies on peritoneal effusions is that the material is not straightforward to obtain. Peritoneal dialysis (PD) is a highly effective and convenient mode of renal replacement therapy and offers the unique opportunity for access to and analysis of peritoneal dialysis effluents (PDE) from all types of patients. Whilst not the same as peritoneal effusions, this fluid is nonetheless a clinically relevant sample that can be analysed to provide critical information about PD specifically as well as the peritoneal environment more generally. Several groups have recently taken on this challenge and produced preliminary proteomics studies on PDE [1,10–14].
The paper in this edition by Wang et al. [1] is the first study to show proteomic differences between diabetic peritoneal PDE and normal peritoneal fluid (five patients and two controls). Following 2DE analysis, a number of these proteins were confirmed by western blotting and included vitamin D-binding protein, haptoglobin and α-2 macroglobulin at raised levels and complement component C4A and immunoglobulin κ at lower levels. The authors propose that the loss of vitamin D-binding protein, haptoglobin and α-2 macroglobulin may be due to a change in the permeability of the peritoneal membrane to medium-sized proteins or leakage due to peritoneal inflammation. They also suggest that lower levels of C4A in the dialysate may shed light on the mechanisms responsible for the initiation of peritoneal membrane scleroses. Only limited studies on the role of immunoglobulin κ in the peritoneum have been reported and lower levels in the dialysate may provide a novel aspect for peritoneal change during PD therapy. Whilst preliminary in nature, the identification of protein differences between normal fluid and diabetic PDE provides useful information for more detailed follow-up studies.
Altered levels of C4A and immunoglobulin κ were also revealed in the first published proteomic analysis on PDE using a cohort of 20 chronic PD patients with varying transport rates [12]. Following 2DE, five proteins were identified with significantly different levels among the transport groups. In particular, increased levels of C4A and immunoglobulin κ in higher transport versus lower transport patients were confirmed in a further validation set of 24 patients by ELISA. This further emphasizes the value in further exploring these two proteins and demonstrates that proteomics can be used to compare differences between different patient sub-groups.
The only other significant PD proteomic study used a more sophisticated methodology (geLC-MS) with nine paediatric PD patients to identify a total of 189 PDE proteins with 88 shared by all patients [10]. Unlike the previous two studies, no comparisons were made between different sample types but analysis of the protein list revealed that the majority of proteins identified were derived from the extracellular matrix (84 compared with 11% plasma proteins) reflecting the clear retention of PD fluid within the extracellular space. Hence, proteomics can also potentially provide clinically relevant information about localization of and changes in proteins in the peritoneal membrane. There is clear relevance here to pathological alterations in the peritoneal membrane described in the biopsy registry study [15]. This proteome also revealed a number of new proteins such as gelsolin and intelectin that had not been previously reported in PDE. One of the exciting aspects of proteomics technology is that it allows for the discovery of previously unknown proteins in a particular sample which can then lead to the generation of new hypotheses. In this case, gelsolin has been proposed to be a marker for sepsis but in PD might play a protective role in mesothelial cell damage and against infection. Intelectin (also called omentin) is an adipocytokine with a possible role in defence against intestinal bacterial permeation and parasites and may be of relevance to host defence in the peritoneal cavity.
Finally, two other studies have used proteomic methods on PDE. In one study, 2DE and surface-enhanced laser desorption/ionization mass spectrometry (SELDI-TOF MS) were performed on PDE from 16 patients with peritonitis and β2-microglobulin was further proposed as a biomarker for PD peritonitis [11]. In the other study, 2DE and SELDI-TOF MS were again used and a total 21 proteins were identified from PDE but no significant conclusions were drawn [14].
Time now for more extended studies
The proteomic studies to date on PDE have only been very preliminary and further prospective studies with greater numbers of patients might enable sub-group analyses to yield additional information about changes in peritoneal dialysate proteins that are associated with specific phenotypes, for example, association with membrane function, residual renal function, nutritional status, the risk of peritoneal infection and fibrosis or the onset of encapsulating peritoneal sclerosis [12]. From this, conclusions might be directly drawn or this information could be used to inform further more focused investigation.
The number of proteins identified in PDE studies to date is small with only one managing over 30 proteins [10] and many much less than this. With so many more proteins remaining to be discovered or confirmed, the challenge now is to adopt the range of workflow choices (see Figure 1) in order to produce more comprehensive protein lists. Each workflow will reveal certain unique proteins not found with the other methodologies; so, for maximum proteomic coverage, a combination of several approaches is recommended. Whilst expensive and technically challenging, such efforts will enable novel proteins to be identified to enable new hypotheses to be generated or novel potential biomarkers or biomarker signatures to be more carefully investigated.
In addition better approaches are required to quantify differences between samples and the PD field need to adopt these. The proteomics community has largely moved on to 2DE-based difference gel electrophoresis (DIGE) [16] or LC-based MS labelling workflows, such as isobaric tags for relative and absolute quantification (iTRAQ) [17,18], as gold standards for relative protein quantification (see Figure 1). These are more statistically robust than 2DE gel-to-gel comparison approaches used to date on PDE and necessary for the quantification of more subtle changes in proteins.
The Achilles’ heel of proteomics
As a note of caution, the analysis of biological fluids, including PDE, is not straightforward. The huge potential for biomedical and biomarker discovery is limited by the considerable challenge of identifying the lowest abundant proteins which may often be the most biologically relevant or the likely source of biomarkers. In biological fluids, there are typically well over 10 orders of magnitude differences in abundance between the most and the least abundant proteins [3]. As the analytical technologies available generally have 2–4 orders of magnitude dynamic range for protein detection, there is clearly a shortfall and not all proteins can currently be identified using these methods alone.
All is not lost, however, as these technologies still do identify proteins that have not been previously identified in given systems and hence produce highly valuable information. Immunodepletion methods can be used to remove the 20 or so most abundant proteins (∼97% of the total protein content for plasma), but this is not sufficient to solve the problem and additionally interesting proteins might be lost that bind to these common proteins. There are also certain other sample fractionation approaches that may be employed to improve specificity, but paradigm changes in proteomics technology will be necessary for all proteins to be identified.
Looking to the future
Whilst preliminary in nature, the PDE proteomics studies performed to date do reveal how proteomics can be used to potentially further understand PD. Whilst PD replaces the function of the kidney, pathological damage of the peritoneum is a frequent occurrence and cannot be easily identified without invasive techniques. Proteins identified from PDE may provide insights and facilitate the non-invasive discovery of potential biomarkers for measuring peritoneal damage and changes in transport. Additionally, the effect of different dialysis fluids or the impact of infection and/or prolonged PD duration on protein profiles might also be investigated to provide improved understanding of the pathological processes that remain the barrier to wider acceptance and utilization of the therapy.
Further detailed proteomics studies are now warranted in this field and will undoubtedly provide improved biomedical understanding and may also lead to novel biomarker discovery for the diagnosis, prognosis and therapeutic monitoring of pathological events related to PD. Clinical proteomics is still in its infancy but does have the potential for ‘bedside’ applications. In time, there is every reason to believe that this technology platform will also make key contributions to both the understanding and utilization of PD.
Conflict of interest statement. None declared.
References
- 1.Wang HY, Tian YF, Chien CC, et al. Differential proteomic characterization between normal peritoneal fluid and diabetic peritoneal dialysate. Nephrol Dial Transplant. 2010;25:1955–1963. doi: 10.1093/ndt/gfp696. [DOI] [PubMed] [Google Scholar]
- 2.Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 3.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 4.Muller GA, Muller CA, Dihazi H. Clinical proteomics—on the long way from bench to bedside? Nephrol Dial Transplant. 2007;22:1297–1300. doi: 10.1093/ndt/gfl806. [DOI] [PubMed] [Google Scholar]
- 5.Thongboonkerd V. Current status of renal and urinary proteomics: ready for routine clinical application? Nephrol Dial Transplant. 2010;25:11–16. doi: 10.1093/ndt/gfp476. [DOI] [PubMed] [Google Scholar]
- 6.Lewczuk P, Wiltfang J. Neurochemical dementia diagnostics: state of the art and research perspectives. Proteomics. 2008;8:1292–1301. doi: 10.1002/pmic.200700703. [DOI] [PubMed] [Google Scholar]
- 7.Tyan Y, Liao P. Protoemics analysis of serous fluid and effusions: pleural, pericardial and peritoneal. Proteomics Clin Appl. 2007;1:834–844. doi: 10.1002/prca.200700036. [DOI] [PubMed] [Google Scholar]
- 8.Ferrero S, Gillott DJ, Remorgida V, et al. Proteomic analysis of peritoneal fluid in women with endometriosis. J Proteome Res. 2007;6:3402–3411. doi: 10.1021/pr060680q. [DOI] [PubMed] [Google Scholar]
- 9.Davidson B, Espina V, Steinberg SM, et al. Proteomic analysis of malignant ovarian cancer effusions as a tool for biologic and prognostic profiling. Clin Cancer Res. 2006;12:791–799. doi: 10.1158/1078-0432.CCR-05-2516. [DOI] [PubMed] [Google Scholar]
- 10.Raaijmakers R, Pluk W, Schroder CH, et al. Proteomic profiling and identification in peritoneal fluid of children treated by peritoneal dialysis. Nephrol Dial Transplant. 2008;23:2402–2405. doi: 10.1093/ndt/gfn212. [DOI] [PubMed] [Google Scholar]
- 11.Lin WT, Tsai CC, Chen CY, et al. Proteomic analysis of peritoneal dialysate fluid in patients with dialysis-related peritonitis. Ren Fail. 2008;30:772–777. doi: 10.1080/08860220802248969. [DOI] [PubMed] [Google Scholar]
- 12.Sritippayawan S, Chiangjong W, Semangoen T, et al. Proteomic analysis of peritoneal dialysate fluid in patients with different types of peritoneal membranes. J Proteome Res. 2007;6:4356–4362. doi: 10.1021/pr0702969. [DOI] [PubMed] [Google Scholar]
- 13.Thongboonkerd V. Proteomics in extracorporeal blood purification and peritoneal dialysis. J Proteomics. 2010;73:521–526. doi: 10.1016/j.jprot.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Dihazi H, Muller CA, Matttes H, et al. Proteomic analysis to improve adequacy of hemo- and peritoneal dialysis: removal of small and high molecular weight proteins with high- and low-flux filters or a peritoneal membrane. Proteomics Clin Appl. 2008;2:1167–1182. doi: 10.1002/prca.200780143. [DOI] [PubMed] [Google Scholar]
- 15.Williams JD, Craig KJ, Topley N, et al. Morphologic changes in the peritoneal membrane of patients with renal disease. J Am Soc Nephrol. 2002;13:470–479. doi: 10.1681/ASN.V132470. [DOI] [PubMed] [Google Scholar]
- 16.Lilley KS, Friedman DB. All about DIGE: quantification technology for differential-display 2D-gel proteomics. Expert Rev Proteomics. 2004;1:401–409. doi: 10.1586/14789450.1.4.401. [DOI] [PubMed] [Google Scholar]
- 17.Zieske LR. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot. 2006;57:1501–1508. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 18.Unwin RD, Evans CA, Whetton AD. Relative quantification in proteomics: new approaches for biochemistry. Trends Biochem Sci. 2006;31:473–484. doi: 10.1016/j.tibs.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 20.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 21.Poon TC. Opportunities and limitations of SELDI-TOF-MS in biomedical research: practical advices. Expert Rev Proteomics. 2007;4:51–65. doi: 10.1586/14789450.4.1.51. [DOI] [PubMed] [Google Scholar]