Abstract
Recent studies have identified histone modifications and suggested a role for epigenetic gene regulation in Trypanosoma brucei. The histone modification H4K10ac and histone variants H2AZ and H2BV localize to probable sites of transcription initiation. Although all T. brucei histones have very evolutionarily divergent N-terminal tails, histone H3 shows conservation with other eukaryotic organisms in 6 of 8 amino acids encompassing lysine 4. Tri-methylation of H3K4 is generally associated with transcription. We therefore generated a specific antibody to T. brucei H3K4me3 and performed chromosome immunoprecipitation and high-throughput sequencing. We show that H3K4me3 is enriched at the start of polycistronic transcription units at divergent strand-switch regions and at other sites of RNA Polymerase II transcription reinitiation. H3K4me3 largely co-localizes with H4K10ac, but with a skew towards the upstream side of the H4K10ac peak, suggesting that it is a component of specific nucleosomes that play a role in Pol II transcription initiation.
Keywords: Trypanosoma brucei, Histone modification, Transcription regulation, Chromatin immunoprecipitation, High-throughput DNA sequencing
Trypanosoma brucei branched early in evolution and shows some conservation to higher eukaryotes, but also striking divergences, making it an interesting model for a “simpler” eukaryotic organism. One of the unique differences relates to the initiation of gene transcription. T. brucei genes do not have traditional promoter elements. Instead, RNA polymerase II transcribes polycistronic units that become mature monocistronic mRNAs after polyadenylation and trans-RNA splicing [1]. Polycistronic transcription units can be convergent or divergent, and divergent strand-switch regions (SSR) have been hypothesized to be transcription start sites (TSS) [2].
In addition to the lack of promoter elements, there appear to be no traditional primary sequences directing RNA pol II to TSS [2, 3]. This suggests that epigenetic phenomena may be key players in T. brucei gene regulation. Indeed, several histone modifications have recently been identified and have been implicated in various cellular processes, including life-cycle regulation and antigenic variation [4–11]. Direct evidence that epigenetics plays a role in gene transcription was recently provided by ChIP-sequencing (ChIP-seq), which showed that H4K10 is likely 100% acetylated in all nucleosomes at TSS of T. brucei [3]. H4K10ac represents only 10% of the total H4 and its associated acetyltransferase (HAT) is essential, supporting its role as a key player in a regulated cellular process [8]. Histone variants have also been shown to be important players in gene regulation and are often associated with sites of active transcription and more open chromatin conformation [12, 13]. T. brucei has four histone variants, two of which, H2AZ and H2BV, form a complex in vivo [14, 15] and co-localize with H4K10ac at TSS [3].
As with gene transcription, the T. brucei histone code is highly divergent. An exception is the region surrounding lysine 4 on the H3 N-terminal tail, which shows conservation to higher eukaryotes (Fig.1 Inset (A)). Although several modifications were identified in the H3 N-terminal tail by mass spectrometry, they could not be assigned to a specific amino acid: a blocked N-terminus prevented Edman sequencing and the abundance of lysines and lack of other useful cleavage sites confounded mass spectrometry assignment [4]. Nevertheless, because of sequence conservation and the key role of H3K4 tri-methylation in other organisms, antibodies were made against T. brucei H3 N-terminal nonapeptides that were either un-methylated or trimethylated at K4 [16]. H3K4me3 antibody bound to T. brucei H3 and co-immunoprecipitated with H2AZ and H2BV [4, 16]. Tri-methylated H3K4 is traditionally associated with sites of active transcription [17], suggesting that this modification could form part of the active nucleosome complex at SSR. Indeed, there is modest enrichment of H3K4me3 at divergent SSR in Trypanosoma cruzi, as demonstrated by ChIP-CHIP [18].
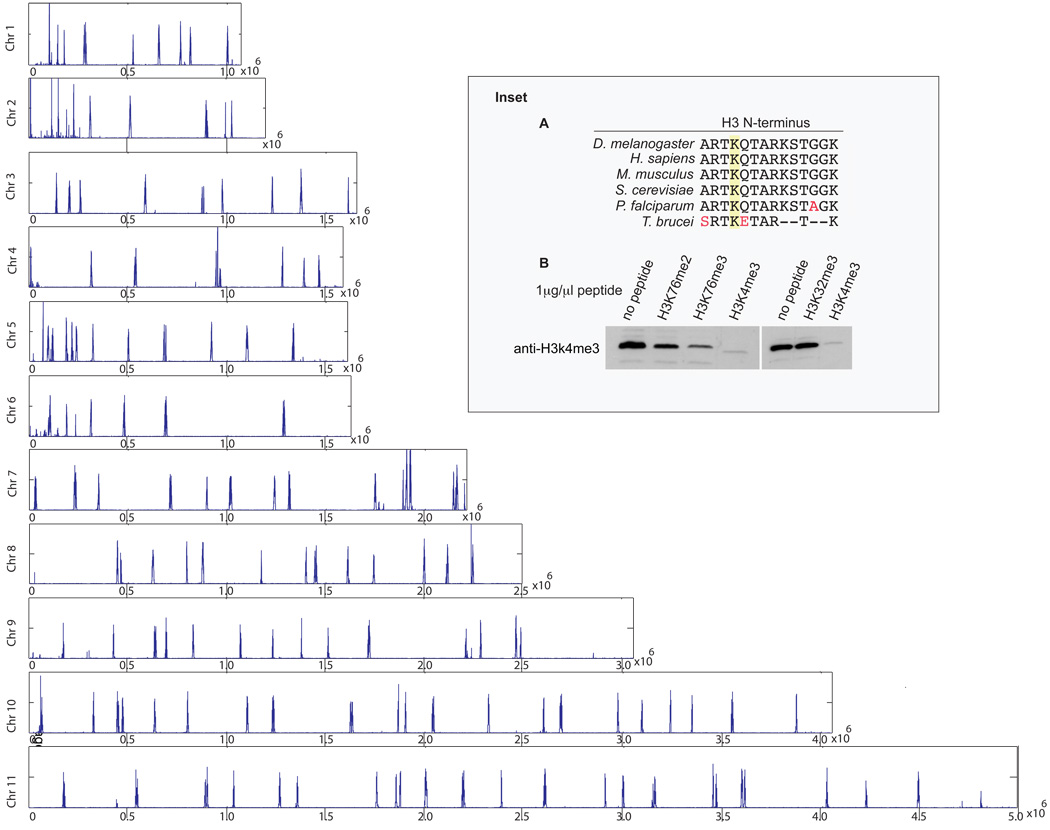
Fig. 1.
Genome-wide distribution of H3K4me3. Nucleotide hits from 36-bp sequences were averaged over a 100-bp window. Inset (A) The N-terminal sequence of T. brucei H3 is evolutionarily conserved. Divergent residues are shown in red and the conserved K4 lysine is highlighted. (B) Specificity of H3K4me3 ChIP-grade antibody. The total extract from 2×106 Dot1B null cells was electrophoresed and blotted with purified H3K4me3 antibody that was preincubated for 1 h with 1 µg/µl of the indicated peptides. Peptide sequences are as follows: H3K4me3 RTKETARTC; H3K73me3 VSGAQKEGLRFC; H3K76me2 VSGAQKEGLRFC (provided by Christian Janzen); H3K32me3 ASGVKTAQRC.
To investigate the distribution of H3K4me3 in T. brucei, ChIP-grade antibody was generated by affinity purification with positive selection for an H3K4me3 peptide and negative selection against an unmodified H3K4 peptide. As assayed by peptide competition, the resulting antibody showed strong specificity for the H3K4me3 peptide and no reactivity to unmodified H3K4 (data not shown). However, the antibody cross-reacted with tri-methylated H3K76, a known trypanosome modification [4]. When purified on an H3K76me3 column, the resulting antibody concentration was too low for ChIP analysis and no further selectivity was achieved. H3K76 is tri-methylated by the Dot1B enzyme [6] and this modification is not essential. We thus pursued all further experiments using a Dot1B null strain. The purified antibody showed no significant cross reactivity to either H3K76me2 or to tri-methylated H3K32 (Fig. 1 Inset (B)).
Using the purified H3K4me3 antibody, we performed ChIP-seq using previously described methodology [3]. Briefly, chromatin was extracted from 1×108 Dot1B-null cells, cross-Page linked with formaldehyde, sonicated (Bioruptor, Wolf Laboratories Ltd.: 15 cycles of 30 sec on /30 sec off, on the highest setting) to generate DNA fragments < 300 bp, then immunoprecipitated with 10 µg of purified H3K4me3 antibody [19]. The resulting DNA was prepared for sequencing by amplification with standard adapters and oligonucleotidess (Illumina), and sequenced with a Solexa sequencer (Illumina). DNA sequences were aligned to the trypanosome genome using the BLAT algorithm [20] and enrichment was determined by graphing the number of hits per 100 base pair windows for each chromosome (using MATLAB, Mathworks). As the BLAT analysis assigns repetitive regions to the first chromosome hit, these sequences can result in artifactual enrichment peaks. To eliminate this, we excluded repetitive regions from the analysis.
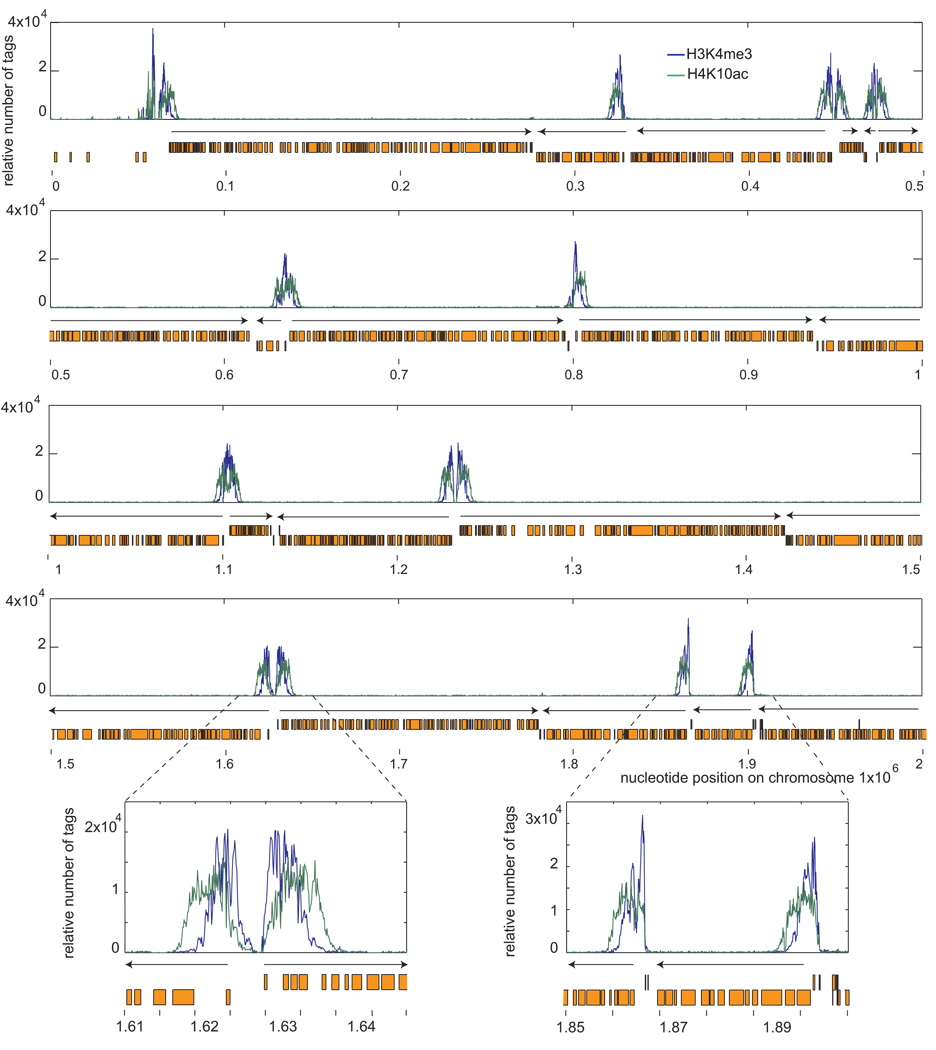
ChIP-seq with the H3K4me3 antibody resulted in a series of peaks of strong enrichment across the genome (Fig. 1). When compared with peaks generated from an H4K10ac antibody [3], there was extensive overlap of each modification, as illustrated in a higher resolution rendering of a portion of chromosome 10 (Fig. 2). The overlap included all divergent SSR, as well as other peaks of H4K10ac enrichment, such as the regions flanked by tRNA genes, which are transcribed by Pol III (Fig. 2). Closer inspection reveals that H3K4me3 has a narrower peak of enrichment than H4K10ac, which extends slightly in the direction of transcription (Fig. 2).
Fig. 2.
H3K4me3 overlaps with H4K10ac peaks of enrichment. H4K10ac and H3K4me3 hits per 100 base pair windows were normalized to the total number of hits per chromosome. Orange boxes represent ORFs and arrows indicate the direction of transcription. H3K4me3 co-enriches with H4K10 at all peaks, including SSR (bottom left) and tRNA gene regions (bottom right). H3K4me3 shows a narrower range of enrichment, with H4K10ac broadening in the direction of transcription (bottom panels)
These results strongly suggest that the H3K4me3 histone associates with H4K10ac, H2AZ, and H2BV. The presence of another histone modification at divergent SSR, especially one that is known to correlate with active transcription in other organisms [17], provides further support for the role of several histone modifications in transcriptional activation in T. brucei. Each of these modifications has been linked to a more open chromatin structure. As there is significant gene regulation at the post-transcriptional level in T. brucei, it is possible that a more open chromatin conformation is sufficient to direct transcription. Although no traditional promoter elements have been identified in SSR, sequence analysis determined that a stretch of 9 to 15 guanines is enriched in the sense strand upstream of H4K10ac, suggesting a motif for Pol II recruitment that may also impart directionality [3].
Questions remain as to what directs nucleosome modification or exchange at these probable TSS, and whether this is a regulated process or if TSS are “always on”. There are some clues to suggest an order of events. H4K10 is acetylated by HAT2, a MYST-family protein that contains a chromodomain [21], which suggests that H3K4 may be tri-methylated first and recruit HAT2.
There are two possibilities for the order of addition of the histone variants. The first is that H3K4me3 and H4K10ac are modified first. In this scenario, BDF3, which is also associated with the H4K10ac modification at SSR [3], could recruit a remodeling complex that would exchange H2A and H2B for an H2AZ-H2Bv dimer. BDF proteins are effector molecules containing bromodomain motifs that associate with acetylated lysines and BDF1 is responsible for H2AZ incorporation through the Swr1 remodeling complex in yeast [22]. Alternatively, incorporation of H2AZ-H2Bv could precede H3K4 tri-methylation. In yeast, ubiquitination of H2B is a pre-requisite for H3K4 tri-methylation [23]. There is no known histone ubiquitination in T. brucei. The replacement of H2B with H2Bv, however, could provide an alternative signal for H3K4 modification [16]. The enzyme responsible for H3K4 methylation remains to be identified in trypanosomes, and could provide the answer to some of these questions.
Effector proteins are the readers of the histone code, playing multiple downstream roles, including transcription-factor recruitment [24]. In addition to BDF3, whose association with nucleosomes at probable TSS has been described, several other proteins with bromodomain motifs have been identified in T. brucei [10]. The PHD finger is a motif that is known to associate with H3K4me3, and has been shown to recruit a transcription factor in yeast [25]. Several proteins with putative PHD motifs are present in T. brucei, but their ability to bind this modification has yet to be confirmed (unpublished data). What effector proteins act at TSS, and how they contribute to gene regulation, is another interesting question for further investigation.
Acknowledgements
This work was supported by fellowships from Boehringer Ingelheim Fonds and the David Rockefeller Graduate Program [TNS] and by Grant No. R01AI021729 from the National Institute of Allergy and Infectious Diseases (NIAID) of the U.S. National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the NIH.
Abbreviations
- BDF
bromodomain-containing proteins
- ChIP
chromatin immunoprecipitation
- ChIP-seq
ChIP-sequencing
- SSR
strand-switch regions
- TSS
transcription start sites
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liang XH, Haritan A, Uliel S, Michaeli S. Trans and cis splicing in Trypanosomatids: mechanism, factors, and regulation. Eukaryot Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clayton CE. Life without transcriptional control? From fly to man and back again. Embo J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, et al. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandava V, Fernandez JP, Deng H, Janzen CJ, Hake SB, Cross GAM. Histone modifications in Trypanosoma brucei. Mol Biochem Parasitol. 2007;156:41–50. doi: 10.1016/j.molbiopara.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janzen CJ, Fernandez JP, Deng H, Diaz R, Hake SB, Cross GAM. Unusual histone modifications in Trypanosoma brucei. FEBS Lett. 2006;580:2306–2310. doi: 10.1016/j.febslet.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 6.Janzen CJ, Hake SB, Lowell JE, Cross GAM. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol Cell. 2006;23:497–507. doi: 10.1016/j.molcel.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 7.Siegel TN, Kawahara T, Degrasse JA, Janzen CJ, Horn D, Cross GAM. Acetylation of histone H4K4 is cell cycle regulated and mediated by HAT3 in Trypanosoma brucei. Mol Microbiol. 2008;67:762–771. doi: 10.1111/j.1365-2958.2007.06079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawahara T, Siegel TN, Ingram AK, Alsford S, Cross GAM, Horn D. Two essential MYST-family proteins display distinct roles in histone H4K10 acetylation and telomeric silencing in trypanosomes. Mol Microbiol. 2008;69:1054–1068. doi: 10.1111/j.1365-2958.2008.06346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figueiredo LM, Janzen CJ, Cross GAM. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 2008;6:e161. doi: 10.1371/journal.pbio.0060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueiredo LM, Cross GA, Janzen CJ. Epigenetic regulation in African trypanosomes: a new kid on the block. Nat Rev Microbiol. 2009;7:504–513. doi: 10.1038/nrmicro2149. [DOI] [PubMed] [Google Scholar]
- 11.Figueiredo LM, Cross GAM. Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot Cell. 9:148–154. doi: 10.1128/EC.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein E, Hake SB. The nucleosome: a little variation goes a long way. Biochem Cell Biol. 2006;84:505–517. doi: 10.1139/o06-085. [DOI] [PubMed] [Google Scholar]
- 13.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowell JE, Cross GAM. A variant histone H3 is enriched at telomeres in Trypanosoma brucei. J Cell Sci. 2004;117:5937–5947. doi: 10.1242/jcs.01515. [DOI] [PubMed] [Google Scholar]
- 15.Lowell JE, Kaiser F, Janzen CJ. Cross GAM. Histone H2AZ dimerizes with a novel variant H2B and is enriched at repetitive DNA in Trypanosoma brucei. J Cell Sci. 2005;118:5721–5730. doi: 10.1242/jcs.02688. [DOI] [PubMed] [Google Scholar]
- 16.Mandava V, Janzen CJ, Cross GAM. Trypanosome H2Bv replaces H2B in nucleosomes enriched for H3 K4 and K76 trimethylation. Biochem Biophys Res Commun. 2008;368:846–851. doi: 10.1016/j.bbrc.2008.01.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 18.Respuela P, Ferella M, Rada-Iglesias A, Aslund L. Histone acetylation and methylation at sites initiating divergent polycistronic transcription in Trypanosoma cruzi. J Biol Chem. 2008;283:15884–15892. doi: 10.1074/jbc.M802081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korber P, Horz W. SWRred not shaken; mixing the histones. Cell. 2004;117:5–7. doi: 10.1016/s0092-8674(04)00296-x. [DOI] [PubMed] [Google Scholar]
- 23.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 24.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]