Abstract
The extent of human genomic structural variation suggests that there must be portions of the genome yet to be discovered, annotated and characterized at the sequence level. We present a resource and analysis of 2,363 novel insertion sequences corresponding to 720 genomic loci. We show that a substantial fraction of these sequences are either missing, fragmented or mis-assigned when compared to recent de novo sequence assemblies from short-read next-generation sequence data. We determine that 18–37% of these novel insertions are copy-number polymorphic, including loci that show extensive population stratification among Europeans, Asians and Africans. Complete sequencing of 156 of these insertions identifies novel exons and conserved non-coding sequences not yet represented in the reference genome. We develop a method to accurately genotype these novel insertions by mapping next-generation sequencing datasets to the breakpoint thereby providing a means to characterize copy-number status for regions previously inaccessible to SNP microarrays.
Introduction
The human genome reference assembly is a mosaic of distinct haplotypes sampled from multiple individuals1. As a result of both gaps in the assembled sequence and the structural differences that exist among different humans, individual genome projects are expected to uncover human sequences present in some (or all) individuals that are not represented in the assembly. Consistent with this prediction, the first sequences of individual genomes2, 3 revealed 23–29 Mb of sequence that do not map against the reference assembly. The short-read, high-throughput approaches currently being employed are also expected to uncover unrepresented insertions4–7. However, these sequences often assemble only as short (median length of 220 to 314 bp 7) contiguous sequences (contigs) that are difficult to anchor and incorporate into existing genome assemblies. Thus, while thousands of novel sequences may be discovered over the next few years, their annotation and complete integration into the human genome will remain a significant bottleneck 8. Since genotyping and expression microarrays are fundamentally dependent upon the reference genome for array probe design, a small fraction of the human genome effectively can not be assayed.
We recently reported efforts to systematically map and sequence human genome structural variation using a fosmid end-sequence pair mapping approach9–11. We fragmented genomic DNA from nine human individuals and subcloned 40-kb segments. Using standard capillary sequencing, reads were generated from both ends of each fragment (end-sequence pairs) and clones were mapped to the human reference genome. Structural differences (inversions, deletions, insertions and translocations) between the reference genome assembly and library source were identified based on the mapped location of the end-sequence pairs. Since the individual fosmid clones were retained, the procedure allowed simultaneous discovery and complete sequence characterization of a subset of structural variant loci including novel insertion sequences common to most individuals but not represented in the human reference genome. Here, we present a detailed sequence and copy-number analysis of these segments missing from the human reference genome.
Results
Discovery
We systematically searched 9.7 million end-sequence pairs, corresponding to 92-fold physical coverage of the human genome, for sequences that failed to map to the reference sequence (NCBI build35). The end-sequence dataset was derived from nine individual genomes (4 Yoruba individuals from Ibadan Nigeria (YRI), 2 individuals with European ancestry (CEU), 2 individuals with Han Chinese or Japanese ancestry (CHB+JPT), and 1 individual of unknown ethnicity). We distinguished clones that only mapped onto the assembly with one end (one-end anchored or OEA, clones) and orphan clones where neither end mapped. After eliminating low-quality sequence and obvious viral and bacterial contaminants, we identified 44,415 high-quality fosmid end sequences that do not map onto the genome reference sequence (NCBI build35)11. This set includes individual sequences from 26,001 OEA clones and 9,207 orphan clones. Using phrap (http://www.phrap.org), we initially assembled these individual sequences into 3,963 sequence contigs (total size = 4.47 Mb, N50 = 1,148 bp) (Table 1) but after applying additional experimental and computational filters, this was reduced to 2,363 distinct sequence contigs (Supplementary Note).
Table 1. Assembling novel sequence contigs.
The number of novel sequence contigs, their size, and the number of corresponding loci with contributions from each sample is shown. Results are given for the initial set of 3,963 assembled contigs as well as for the 2,363 contigs that pass all filters. The sample origin of 222 sequenced clones (corresponding to 192 distinct loci) is also shown.
| Assembled Sequences | Typed by ArrayCGH | |||||||
|---|---|---|---|---|---|---|---|---|
| Sample | Population | Contigs | Contig Size (Mb) |
Loci | Contigs | Contig Size (Mb) |
Loci | Sequenced Clones |
| NA15510 | -- | 768 | 0.904 | 345 | 387 | 0.512 | 177 | 9 |
| NA18517 | Yoruba | 726 | 0.925 | 307 | 529 | 0.700 | 229 | 15 |
| NA18507 | Yoruba | 1,386 | 1.752 | 534 | 904 | 1.208 | 363 | 22 |
| NA18956 | Japan | 885 | 1.140 | 342 | 597 | 0.815 | 243 | 65 |
| NA19240 | Yoruba | 1,034 | 1.295 | 400 | 682 | 0.910 | 295 | 44 |
| NA18555 | China | 953 | 1.187 | 380 | 615 | 0.825 | 269 | 20 |
| NA12878 | CEPH | 977 | 1.232 | 386 | 653 | 0.879 | 279 | 26 |
| NA19129 | Yoruba | 990 | 1.277 | 359 | 678 | 0.932 | 266 | 13 |
| NA12156 | CEPH | 996 | 1.278 | 377 | 667 | 0.914 | 266 | 8 |
| Total (non- redundant) |
3,963 | 4.465 | 1,182 | 2,363 | 2.834 | 720 | 192 | |
40% (1,019/2,363) of the contigs contain sequence contributed by at least one orphan clone, suggesting that these contigs represent segments longer than 40 kb (Supplementary Table 1). Using OEA anchoring information and the mate-pair relationships from the orphan clones, we identified 720 contigs (400 of which have a mapped genomic position) corresponding to ~2.8 Mbp of sequence with a median contig size of 1 kb (Supplementary Note). Interestingly, 80 of the 400 anchored loci (20%) map within 5 Mb of the ends of a chromosome (a significant 2.9-fold subtelomeric enrichment, p=1.0e-18, binomial test) (Supplementary Fig. 1, Supplementary Table 2). In addition to these 720 loci, we identified 19,038 singleton OEA sequences (average length 790 bp) as well as 5,654 orphan clones that did not contribute to any contigs. By convention, we refer to these sequences as “novel insertions” based on the fact that they are not present within the public reference genome assembly.
FISH Analysis
Our analysis distinguished two different types of novel human sequences: 400 loci that were anchored within euchromatin based on OEA assignments and 320 unassigned loci where a clear anchor position could not be identified. We explored the genomic distribution and assessed the accuracy of our assigned locations using individual fosmid clones as FISH probes. Although limited to larger regions, this analysis provided us valuable high-level mapping information with respect to the distribution of insertions in heterochromatin and euchromatin. We selected 33 contigs derived only from orphan clones (assigned to seven distinct unmapped loci) and mapped these loci to metaphase chromosomes by FISH. Three loci mapped separately to telomeric regions on chromosomes 10q, 7p, and Xp; one locus mapped to 6q1; and three loci mapped to the p-arms of the acrocentric chromosomes (Supplementary Table 1).
As a complement to these studies we also tested an additional 68 large orphan contigs, which were constructed based on a detailed fingerprint analysis of all orphan clones from a single individual human genome library (NA15510) (Supplementary Note). After excluding 31 contigs assigned to genome assembly gaps12, we found that 15 of the contigs mapped interstitially, with the remainder (22/37) mapping to telomeric, pericentromeric or acrocentric positions (Supplementary Table 3, Supplementary Fig. 2).
Finally, we considered sequence contigs that had been anchored by OEA clones, but also had contributions from at least one orphan clone, to positions in human euchromatin by using 37 fosmid clones (20 OEA and 17 orphan clones) as FISH probes. We found that 78% (29/37) of the clones support the predicted position while 11% (4/37) map to a different interstitial location and 11% (4/37) map to the p-arms of the acrocentric chromosomes. We additionally tested a limited number (n = 3) of smaller insertions (<30 kb) that had been completely sequenced and confirmed all three by metaphase oligo-FISH, finding that 2/3 were copy-number polymorphic among the four individuals tested (Supplementary Note). Our FISH results indicate that mega-bases of uncharacterized sequence remain within the heterochromatin and euchromatin-heterochromatin transition regions of the human genome but also confirm the presence of missing euchromatic sequences that are copy-number polymorphic.
Assembly Comparisons
We searched for evidence of the identified 2,363 sequence contigs in other human and non-human primate genome assemblies. 600 contigs (71 loci)have a match against the newest human reference genome assembly, GRCh37 and 1,467 contigs (54 loci) have a match against the HuRef assembly 2 (Supplementary Note). We find partial support for 1,700–2,000 of the contigs in sequence data from the JDW, YH, and NA18507 genomes3–5 (Supplementary Note). One of the genomes in our study, NA18507, was sequenced to high coverage using the Illumina platform4 and subjected to a SOAP de novo sequence assembly 7. Surprisingly, we found that the 94% of our smallest insertions identified from single unmapped reads (~790 bp) were not identified as part of the de novo assembly (Supplementary Note). 32% of our larger contigs had no representation and only 25% had complete sequence coverage (defined as more than 95% bp representation). When we restricted our analysis to insertions from sequenced NA18507 clones, we found that 52% (11/21 sequenced fragments) were either not present (n = 4) or mapped to different scaffolds (n = 7) in the de novo assembly. We find that this fragmentation often corresponds to the presence of large common repeat sequences that disrupt the contiguity and complicate map assignment. Regions largely devoid of common repeats or segmental duplications showed the greatest correspondence in length and coverage.
In order to determine the ancestral state of each of these sequences, we also searched the 2,363 contigs against available whole-genome sequence data from chimpanzee and orangutan13. 74% (1,745/2,363) of the contigs had a match against one of these datasets with 68% (1,599/2,363) of the contigs identified within chimpanzee. We were concerned that these sequences may have characteristics leading to their underrepresentation in genome-sequencing datasets, so we performed an arrayCGH experiment using DNA from a single chimpanzee and tested whether the DNA in fact hybridized. This experiment indicated that 84% (1,985/2,363) of the contigs were present in the single chimpanzee analyzed (Supplementary Note). This includes 624 contigs that do not have a match to the chimpanzee genome sequence data. In total, we find experimental or computational support for 94% (2,223/2,363) of the contigs in the chimpanzee and 96% (2,266/2,363) in either chimpanzee or orangutan. The absence of these new insertions in the current reference genome represents either genome assembly errors or deletions that have emerged within the human lineage and are now copy-number polymorphic in our species.
Copy-number Polymorphism
We designed two customized oligonucleotide microarrays in order to provide an assessment of copy-number polymorphism among these novel insertion sequences. In the first, we designed a microarray targeting the 19,038 single OEA sequences that did not assemble into sequence contigs and tested them against the sample genomes used for discovery. After filtering additional contaminants, we found that 38% (7,240/19,038) of these unassembled sequences were represented by at least three probes with signal intensities sufficiently above the background level. Based on a comparison of the intensity values for the eight analyzed samples, we estimate that 31% (2,228/7,240) of the assayable single OEA sequences are copy-number polymorphic (Supplementary Table 4).
In the second design, we investigated copy-number polymorphism for the 2,363 sequence contigs that had been assigned to 720 distinct loci and tested a larger collection of 28 unrelated HapMap individuals (9 CEU, 11 YRI, 8 JPT+CHB). These experiments clearly identified sets of sequences that are copy-number polymorphic or apparently fixed among the analyzed individuals (Fig. 1). Polymorphic contigs were identified using two alternative calling schemes: a noise-multiplier approach that compares the median probe log-ratios for each contig with the results of a control self-self hybridization (using reference sample NA15510, Supplementary Table 5) and a clustering approach that assigns contigs to log-ratio clusters14 that are then fitted to distinct, small integer copynumber states (Fig. 2, Supplementary Table 6). The noise-multiplier approach identifies 37% of the contigs as being copy-number polymorphic. 518 contigs could be fitted to a copy-number state, of which 461 contigs are fitted to two or more distinct copy-number states. 443 contigs (18.7%) were identified as polymorphic by both approaches, an indication of the challenges in assigning discrete copy numbers to all copy-number variable loci.
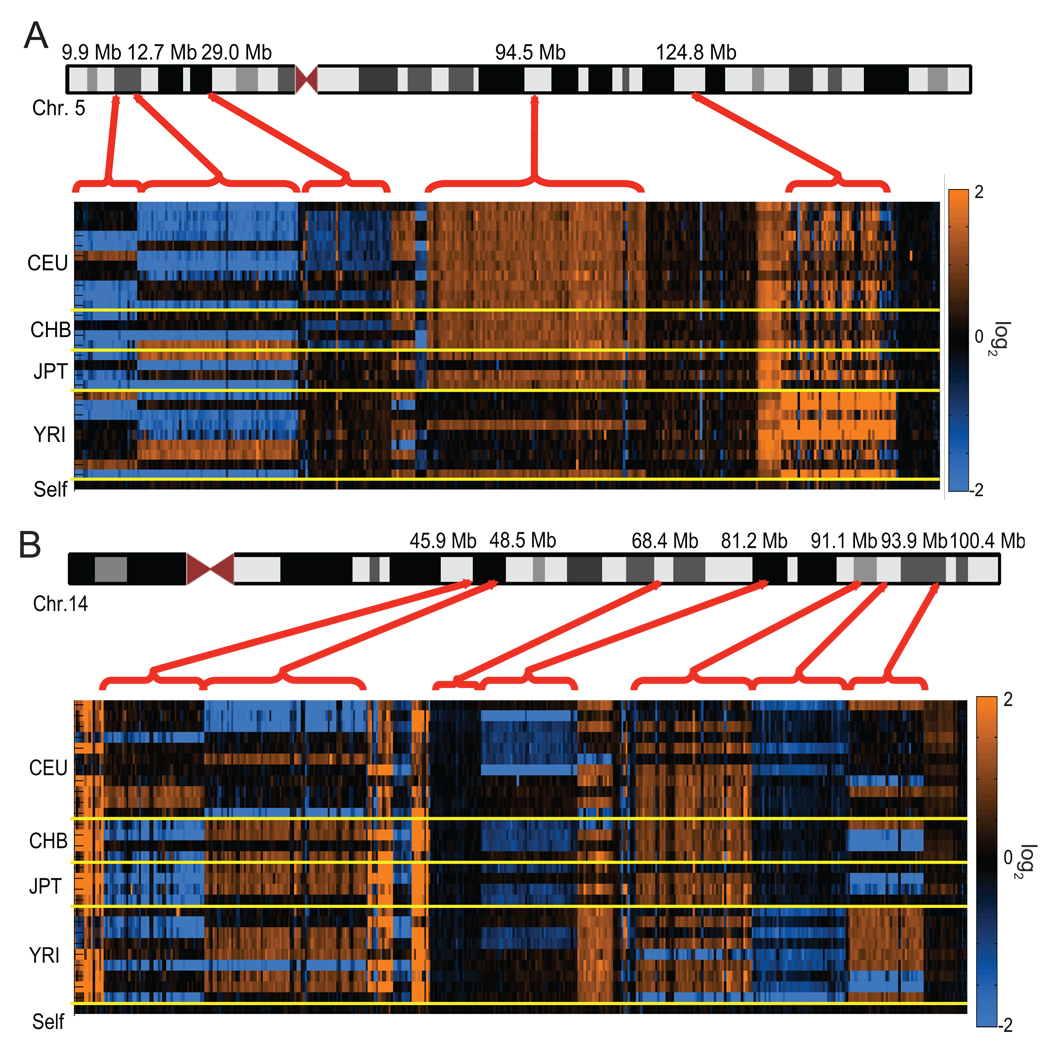
Figure 1. Copy-number polymorphism of novel insertions.
ArrayCGH intensity data is displayed for novel sequences ordered along (a) chromosome 5 and (b) chromosome 14 based on anchored map locations (build35 coordinates, UCSC). Copy-number gains (orange) and losses (blue) are shown relative to the reference sample (NA15510). Each column in the heat map represents a probe on the array, and each row represents a sample ordered and separated (yellow lines) by corresponding HapMap population (CEU, CHB, JPT and YRI). The bottom row depicts a reference self-self hybridization as control. The red brackets group multiple contigs into loci that generally show a consistent hybridization pattern by arrayCGH.
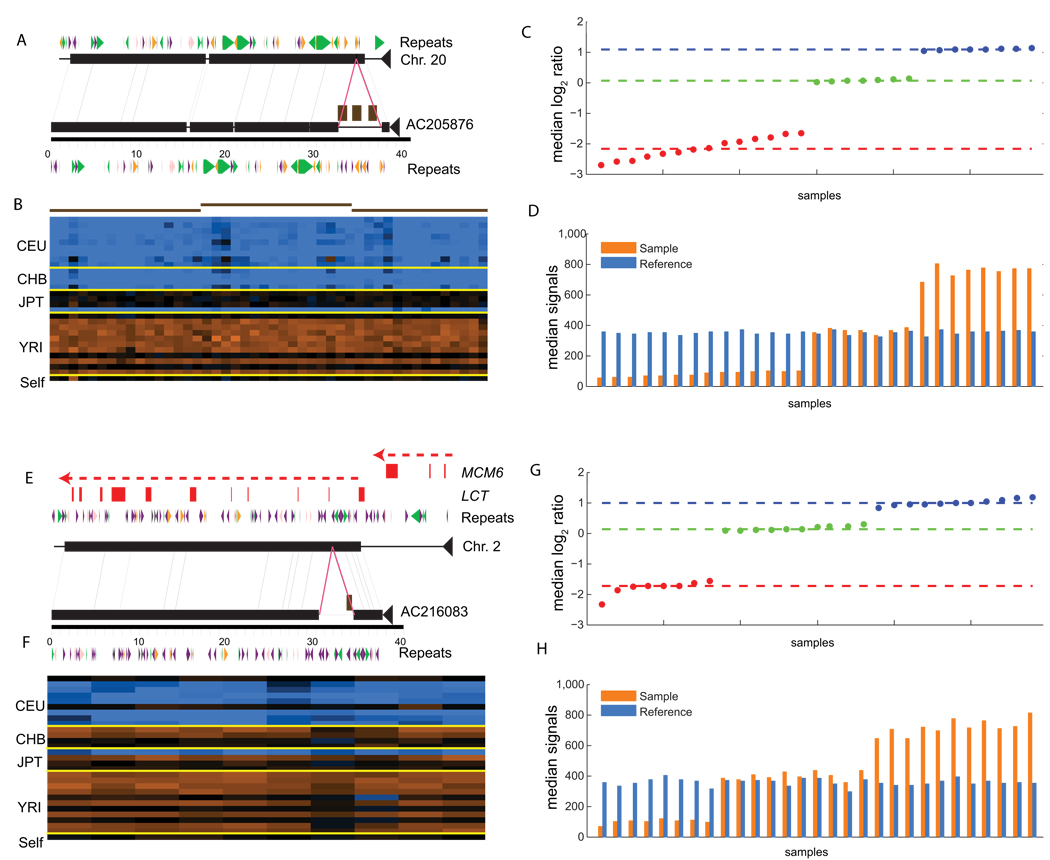
Figure 2. Sequencing and genotyping insertions.
(a) The complete sequence of a clone (AC205876) carrying a 4.8-kbp novel insertion sequence is compared to the corresponding segment from chromosome 20 using miropeats (black lines connect segments of matching sequence; colored arrows correspond to common repeats; green: LINEs; purple: SINEs; orange: LTR elements; pink: DNA elements). The magenta lines denote the insertion breakpoints. The brown boxes correspond to the mapped position of three assembled novel sequence contigs. (b) ArrayCGH hybridization results represented as a heat map suggest that the deletion is fixed in CEU and CHB populations. The brown-red lines correspond to the three sequence contigs depicted in part (a) and are represented by 16, 15, and 18 arrayCGH probes respectively. The median log2 ratios (c) and single channel intensities (d) are shown for all probes matching AC205876. Note that the reference (blue bars) channel shows similar intensity across hybridizations. For this example the reference sample is inferred to have a copy number of 1. The signals form three distinct clusters that are assigned integer copy-number states of 0, 1, and 2. The dotted red, green, and blue lines correspond to the median intensities of each defined cluster. Using these genotypes an FST of 0.70 is calculated for this insertion. (e–h) A second example as described above depicting a 3.9-kb insertion (AC216083) within the first intron of the LCT (lactase) gene (red boxes represent exons as indicated).
We assessed the extent of population differentiation for these sequences using both the FST and VST statistics15, 16. For 189 loci with a simple autosomal insertion-deletion variant, we found 20 loci having an FST greater than 0.35 (Fig. 3, Supplementary Table 7, Supplementary Table 8, and Supplementary Fig. 3). Among these, we identified a 3.9-kb insertion sequence within the first intron of the lactase gene (LCT)(Fig. 2). Interestingly, this 3.9-kb insertion is prevalent among the YRI samples tested (allele frequency=0.86) but is largely absent among the CEPH Europeans (allele frequency = 0.11) where it is in complete linkage disequilibrium (D’ = 1) with the functional SNP that has been associated with lactase persistence17. We repeated the analysis using the VST statistic, for all 720 loci (Supplementary Fig. 4). We identified 27 loci that have a VST value greater than 0.35 with ten having a value greater than 0.5 (Supplementary Table 9). Fosmid clones corresponding to several of the most stratified loci have been completely sequenced, including a 4.8-kb insertion on chr20 (AC205876, Fig. 2, VST = 0.73, FST = 0.70) and an 11.4-kb insertion on chr1 near the ATP6V1G3 gene (AC212752, VST = 0.48, FST = 0.37). These sites represent structures that show a high level of differentiation among human populations but are absent from the genome reference.
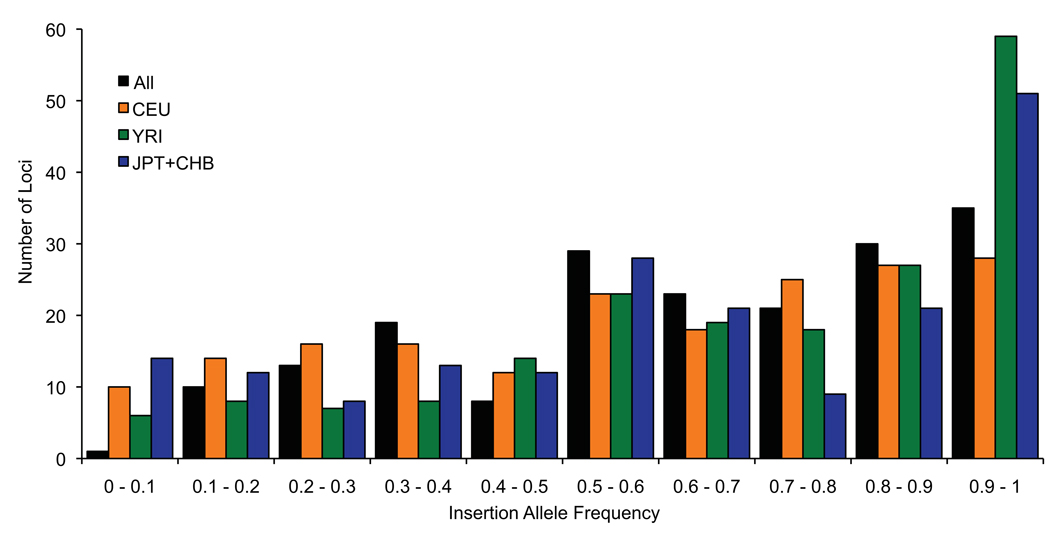
Figure 3. Insertion allele frequency distribution.
The frequency of the insertion allele is shown for 189 loci that are fitted to distinct copy numbers and are consistent with a simple autosomal insertion-deletion variant. Values are shown for all 28 individuals (black bars) and separately for each HapMap population as indicated.
Sequencing and Genotyping Novel Insertions
The complete sequence of insertions smaller than 40 kb can be directly obtained by sequencing an appropriate fosmid clone, while an iterative strategy is required to capture the sequence of larger insertions. 222 fosmid clones (53 OEA clones and 169 spanned insertions) were sequenced using a traditional capillary sequencing and assembly approach (Supplementary Table 10, Supplementary Fig. 5). The 222 clones correspond to 192 distinct genomic loci and contain a total of 1.67 Mb of inserted sequence (Supplementary Fig. 6) subsuming 475 of our original 2,363 contigs. Four of the completely sequenced insertions, ranging in size from 41–65 kb, were larger than a single clone insert (Supplementary Note). The sequenced insertions are similar in composition to segments sampled from the reference genome assembly, with a slight enrichment for common repeats, particularly LINEs (Supplementary Table 11). Only five of the 192 loci (Supplementary Table 12) have been updated in GRCh37, thus the majority (97%) of these insertions await integration into the next version of the human genome.
We searched the sequenced insertions against the RefSeq gene database18 to identify previously uncharacterized exons. We found that segments from 22 genes matched 21 of the insertions (Supplementary Table 13) including support for structures not represented in the build36 assembly (eg. MINK1, FSCN2, PECAM1, and VPRBP genes (Figure 4). We further searched for expressed elements using mRNA-seq data derived from multiple human tissues that do not map onto the build36 genome assembly19. We mapped these previously unmapped reads onto the sequenced clones and found that 26 insertions contained segments supported by at least three mRNA-seq reads (Supplementary Fig. 5, Supplementary Note). We searched against an alignment of nine mammalian genomes to identify segments matching the sequenced insertions (Ensembl Compara 51) 20, 21. Using these alignments, we identified 477 constrained elements from 104 different loci (Fig. 4), a signature that identifies segments of possible functional importance22. Six of the constrained elements intersect with mapped RefSeq exons with the remainder having an unknown functional importance. Using Genomic Evolutionary Rate Profiling (GERP) scores as a metric, we note that the conserved elements found in the insertions show a similar level of constraint compared to elements identified across the rest of the alignments (Supplementary Fig. 7).
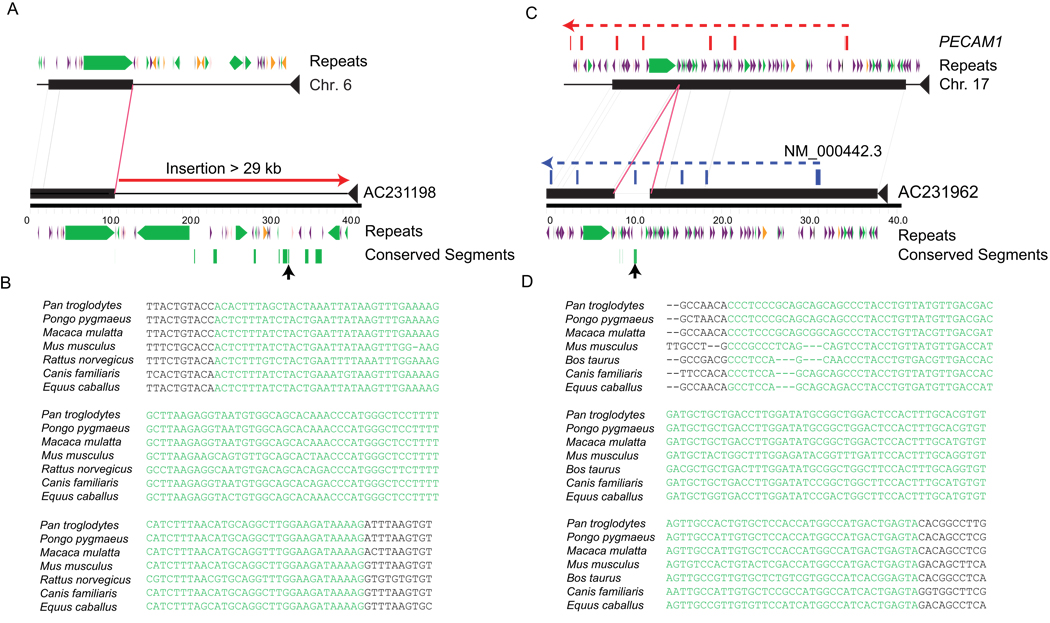
Figure 4. Annotation of conserved and functional elements.
(a) The complete sequence of an OEA clone carrying 29 kbp of novel sequence is compared by miropeats to the reference genome. We identify a 95-bp conserved element within this sequence (green rectangles) as defined by a GERP analysis of 8 species (see Online Methods). A multiple sequence alignment of one of these conserved elements (black arrow) is highlighted. (b) A novel exon is predicted within the sequence of a 4.3-kbp insertion based on comparison with the PECAM1 transcript (NM_000442.3), as shown in blue. This alternate exon is supported by RNA-seq data and corresponds to a conserved element identified by alignment comparisons.
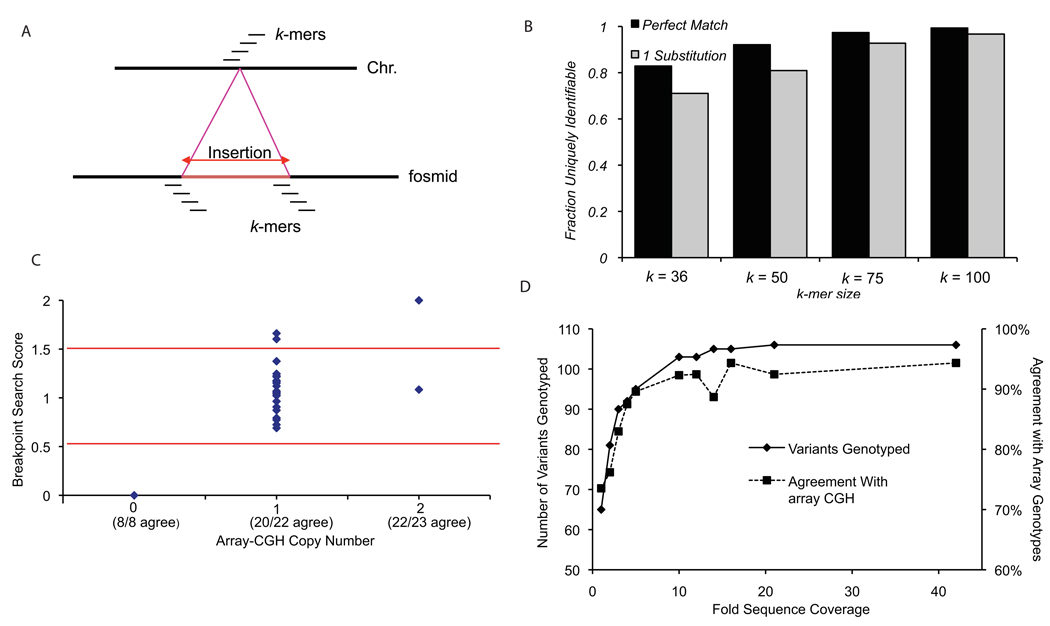
High-quality sequence across the variant breakpoints permitted a detailed assessment of exact variant boundaries and associated sequences. We used the breakpoint sequence data obtained from 152 insertions spanned by individual fosmid clones to identify a set of unique, diagnostic k-mers specific to the insertion and deletion alleles of each variant (Fig. 5). We found that 108 of the sequenced loci could be uniquely identified using a k-mer length of 36 and a search stringency of one substitution. 29% of the loci (44/152) could not be uniquely identified using this approach, although we note that this method assumes that the genome reference assembly accurately represents the structure of the deletion allele and all instances of the variant have identical breakpoints. If k-mer lengths increased to 100 bp, there would still be five loci that remained recalcitrant to analysis using this approach (Fig. 5b). We determined genotypes for 106 loci by searching Illumina sequence data from NA18507 against these diagnostic k-mers4. We observed agreement at 94.3% of the genotypes determined for this individual by arrayCGH (Fig. 5c, Supplementary Table 14). We simulated the effect of genome coverage by sampling subsets of the total sequence data from NA18507 (Fig. 5d). We found a rapid increase in the number and accuracy of the sites genotyped with increasing coverage, followed by a plateau of approximately 94% genotype agreement when sequencing coverage reached 10-fold sequence coverage. This indicates that high-quality breakpoint sequence data can be used to genotype structural variants in samples that have been analyzed by next-generation sequencing.
Figure 5. Genotyping sequenced variants through unique k-mer matches.
(a) Unique diagnostic k-mer sequences were identified for each variant using sequence-resolved breakpoints. For the deletion breakpoint, k-mers were required to have a single match to the reference genome and no matches to the fosmid sequences. For the insertion breakpoints, k-mers were required to have no matches to the genome and a single match to the fosmid. In order to be uniquely identifiable, a variant must have at least one deletion k-mer and at least one insertion k-mer that meet these criteria. (b) Effect of k-mer length and search stringency on ability to uniquely identify a variant. 71% (108/152) of the sequenced sites are uniquely identifiable with a criteria of k=36 and one substitution, while 97% (147/152) are assayable if k-mer length increased to 100 bp. (c) A comparison of genotypes determined using arrayCGH and breakpoint k-mer matching is depicted for sample NA18507. The search database consists of unique 36-mers (one substitution). Genotypes for 54 variants were successfully determined by both arrayCGH and breakpoint k-mer matching. Partitioning the breakpoint scores into distinct genotypes at 0.5 and 1.5 (red lines) results in 94.3% genotype agreement between the two methods. (d) Effect of sequence coverage on breakpoint k-mer genotyping. The number of variants genotyped (at least one matching read, solid line, left axis) and the percent agreement with arrayCGH results (dashed line, right axis) are shown at various sequence coverage levels (1–42X).
Discussion
Over the past five years the extent of structural variation among individual human genomes has become increasingly clear. Array-based approaches, for example, have systematically discovered and genotyped more than 50% of common copy-number polymorphic deletions23, 24. Sequence-based approaches have begun to more fully explore the size spectrum, cataloging an increasing number of smaller deletions and moving toward personalized duplication maps for individual genomes9, 11, 25 , 26. The characterization of other classes of structural variation, including inversions and insertions, however, has lagged due to technical biases in their discovery and difficulties associated with their validation. New insertions are limited, in particular, by the genetic community’s reliance on a single mosaic reference genome, which at some positions represents rare structural configurations and entirely omits sequences that are found in the majority of individuals. The absence of these sequences from the reference genome hinders their functional characterization leading to a less-than-complete understanding of the sequence content present in the majority of humans. We used a fosmid clone strategy to specifically focus on the characterization of human sequences that are not in the reference assembly and have therefore not been annotated for functional elements or systematically genotyped.
In this study we identified 720 distinct loci ranging in size from 1–20 kbp in length as well as several thousand additional smaller segments <1 kbp in length. We have determined that more than half map to the euchromatin with a disproportionate fraction mapping within the last 5 Mbp of human chromosomes (Supplementary Fig. 1). A remarkable feature of these sequences is their degree of copy-number polymorphism. ArrayCGH analysis indicates that 18–37% of the assembled sequence contigs vary in copy number, with 80% of the genotyped variants having a minor allele frequency >10% among the 28 individuals surveyed (Fig. 3). Experimental and computational comparisons with chimpanzee DNA suggest that at least 94% arose as a result of deletions that occurred within the human lineage.
Many of the common insertions show striking differences in allele frequency among populations, a pattern suggestive of either selection or genetic drift since the migration of humans out of Africa (Fig. 2, Supplementary Table 8, Supplementary Table 9). We observe that the average insertion allele frequency for the variable loci was significantly greater in African populations when compared to European or Asians (YRI versus CEU p = 0.0003 and YRI versus ASN p = 0.005, 1 sided t-test). The 3.9-kb novel insertion within the first intron of the LCT gene is illustrative. Our initial survey suggests that this insertion sequence is prevalent among the Yoruba (86%) and Asian samples (63%) but is present at a much lower frequency among CEPH Europeans (11%). These findings raise the possibility that the additional sequence within this haplotype may play a role in regulating expression of this gene. The complete sequence of this insertion sequence (AC20193) now allows this hypothesis to be directly tested.
An important question going forward is how well de novo assembly methods using next-generation sequence data compare to the clone-based approach we have described here. We had the opportunity to compare an Illumina SOAP de novo assembly 7 against the clone-based discovery on the same individual genome (Supplementary Note). We found that many of the larger novel contigs were only partially represented (50–60%) in a 30X de novo assembly, and in more than a third of studied cases novel contigs were fragmented—mapping to two or more scaffolds instead of being placed in the same region. In many cases, the fragmentation corresponded to common repeats disrupting the contiguity of the novel sequence. In regions largely devoid of retrotransposons, de novo sequence assemblies using NGS datasets perform quite well. These results highlight both the limitation of de novo sequence assembly using NGS and the value of high-quality clone-based data to resolve and integrate these sequences into the reference genome. Nevertheless, there are advantages to de novo assembly. The de novo sequence assembly identifies 2–3 times more novel sequence per genome when compared to our results from 0.3X sequence coverage per genome, suggesting that the methods are complementary. Surprisingly, only 2.9% of our singletons from NA18507 (average size ~790 bp) were identified in the de novo assembly. Since these smaller insertions require more characterization, the significance of this discrepancy is unclear.
The major benefit of our approach is the ability to directly obtain high-quality sequence for the insertion loci by complete sequencing of corresponding clone inserts at a quality commensurate with that of the human reference genome. While no complete missing genes were discovered, we did identify 477 elements that have been conserved over evolutionary time, six of which appear to correspond to exons from RefSeq genes as well as 26 loci having support from multiple mRNA-seq reads. Moreover, we demonstrate that these high-quality sequences can be utilized to accurately genotype these regions using next-generation sequence sets produced from the 1000 Genomes and other projects. The complete sequence of these and other loci will facilitate their functional characterization as they can now be incorporated into future genotyping platforms, expression microarrays, and ultimately future genome assemblies to provide a more accurate representation of the organization and genetic variation of the human genome.
Supplementary Material
Acknowledgments
We thank C. Campbell, G. Cooper, T. Marques-Bonet for thoughtful discussion, P. Sudmant for assistance with Illumina sequence data, and members of the University of Washington and Washington University Genomes Centers for assistance with data generation. J.M.K. is supported by a National Science Foundation Graduate Research Fellowship. This work was supported by grant HG004120 to E.E.E. E.E.E. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Accession codes
Gene Expression Omnibus: GSE20634
GenBank accessions for assembled contigs: GU266782 – GU269144
Competing financial interests
N.S. P.A. A.T. N.Y. P.T. and L.B. are employees of Agilent Technologies. E.E.E. is a SAB member for Pacific Biosciences.
Author contributions
J.M.K, N.S., F.A., A.T., R.K. and E.E.E. analyzed data, N.S., P.A, A.T., N.A.Y, P.T. and L.B. performed arrayCGH and copy-number analysis. F.A, M.V. and G.G. performed FISH experiments. C.A. assembled contigs, T.G., R.F., H.S.H, M.M, J.K., R.K, and R.K.W. performed clone characterization and sequencing. J.M.K, R.K., L.B. and E.E.E designed the study. J.M.K and E.E.E. wrote the paper with contributions from the other authors.
References
- 1.IHGSC Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Levy S, et al. The Diploid Genome Sequence of an Individual Human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 4.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKernan KJ, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009 doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li R, et al. Building the sequence map of the human pan-genome. Nat Biotechnol. 28:57–63. doi: 10.1038/nbt.1596. [DOI] [PubMed] [Google Scholar]
- 8.Hormozdiari F, Alkan C, Eichler EE, Sahinalp SC. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res. 2009;19:1270–1278. doi: 10.1101/gr.088633.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuzun E, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 10.Eichler EE, et al. Completing the map of human genetic variation. Nature. 2007;447:161–165. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bovee D, et al. Closing gaps in the human genome with fosmid resources generated from multiple individuals. Nat Genet. 2008;40:96–101. doi: 10.1038/ng.2007.34. [DOI] [PubMed] [Google Scholar]
- 13.Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 14.Perry GH, et al. The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weir BS. Genetic Data Analysis II. Sunderland MA: Sinauer; 1996. [Google Scholar]
- 16.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enattah NS, et al. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 18.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008;18:1814–1828. doi: 10.1101/gr.076554.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paten B, et al. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res. 2008;18:1829–1843. doi: 10.1101/gr.076521.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper GM, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 24.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2009 doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alkan C, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsons J. Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.