Abstract
Background and Aims
Black cherry (Prunus serotina) is a North American tree that is rapidly invading European forests. This species was introduced first as an ornamental plant then it was massively planted by foresters in many countries but its origins and the process of invasion remain poorly documented. Based on a genetic survey of both native and invasive ranges, the invasion history of black cherry was investigated by identifying putative source populations and then assessing the importance of multiple introductions on the maintenance of gene diversity.
Methods
Genetic variability and structure of 23 populations from the invasive range and 22 populations from the native range were analysed using eight nuclear microsatellite loci and five chloroplast DNA regions.
Key Results
Chloroplast DNA diversity suggests there were multiple introductions from a single geographic region (the north-eastern United States). A low reduction of genetic diversity was observed in the invasive range for both nuclear and plastid genomes. High propagule pressure including both the size and number of introductions shaped the genetic structure in Europe and boosted genetic diversity. Populations from Denmark, The Netherlands, Belgium and Germany showed high genetic diversity and low differentiation among populations, supporting the hypothesis that numerous introduction events, including multiple individuals and exchanges between sites, have taken place during two centuries of plantation.
Conclusions
This study postulates that the invasive black cherry has originated from east of the Appalachian Mountains (mainly the Allegheny plateau) and its invasiveness in north-western Europe is mainly due to multiple introductions containing high numbers of individuals.
Keywords: Microsatellites, putative sources, invasive tree species, phylogeography, population genetics, black cherry, Prunus serotina var. serotina
INTRODUCTION
The success of non-indigenous plants in areas distant from their native range have caused considerable economic and ecological costs worldwide (e.g. Wilcove et al., 1998; Forman, 2003; Simberloff, 2003; Pimentel et al., 2005). Different stages are usually recognized in the invasion process, ranging from the introduction of a species to its spread into the new range (With, 2002; Lockwood et al., 2005; Theoharides and Dukes, 2007). Because invasive species are introduced by human activities either intentionally or unintentionally, introduction modes can greatly vary among species. Propagule pressure constitutes a particularly important factor in the first steps of invasion. This composite term accounts for both the number of individuals – propagule size – within a single introduction event and the number of independent introductions – propagule number (Lockwood et al., 2005). Both propagule size and number play a major role in the dynamics of species' establishment. The introduction of a small number of individuals in a new environment can potentially limit the establishment of a species if there is not enough genetic variation for the adaptive evolutionary changes required by the new selection pressures to take place (Suarez and Tsutsui, 2008; Okada et al., 2009). On the other hand, multiple introductions and subsequent gene flow between populations may maintain part or all genetic variation present in native populations (Carlton, 1996; Novak and Mack, 2001; Lockwood et al., 2005; Facon et al., 2008; Prentis et al., 2008; Henry et al., 2009; Wilson et al., 2009). The influence of these multiple introductions has not yet been fully resolved and a recent review has pointed out that the influence of propagule number on the ability of a species to spread is dependent on both spatial and temporal aspects of the multiple introductions (Dlugosch and Parker, 2008). Nevertheless, multiple introductions from genetically differentiated source populations have been increasingly found in successful plant invasions (Hufbauer and Sforza, 2008; Prentis et al., 2009). The time needed for populations resulting from different introductions to become connected is then dependent on the initial size of the introduced populations and the geographical distance separating them.
Key steps in the understanding and management of biological invasions are the frequency with which a species is introduced into a specific area, the size of each introduction and the subsequent pattern of spread across the new range. Several population genetic studies have been devoted to answer these questions by identifying source populations (e.g. Carlton, 1996; Meekins et al., 2001; Besnard et al., 2007; Schlaepfer et al., 2008), comparing genetic structure of invasive populations with those in the native range (e.g. Eckert et al., 1996; Prentis et al., 2009), unravelling invasion histories (e.g. Carlton, 1996; Neuffer and Hurka, 1999; Meekins et al., 2001; Gaskin et al., 2005; Le Roux et al., 2008; Henry et al., 2009), or seeking the role played by evolutionary processes in establishment and spread (for reviews, see Prentis et al., 2008; Suarez and Tsutsui, 2008).
Combining molecular markers with different modes of inheritance can be highly useful to assess population structure in invasive species (e.g. Gaskin et al., 2005; Grapputo et al., 2005). Chloroplast DNA (cpDNA) is generally maternally inherited in angiosperms, and cpDNA markers thus record gene flow coming from seed movement only, whereas nuclear markers record gene flow of both pollen and seeds (Devos et al., 2003). In addition, polymorphisms in haploid genomes are more affected by genetic drift than those from the nuclear genome (Petit et al., 1993). Patterns of the spatial distribution of cpDNA polymorphism established by seed dispersal during range expansion should therefore be more slowly eroded by subsequent gene flow, when compared with spatial patterns in nuclear genetic markers, especially in trees (e.g. McCauley et al., 2003; Petit and Hampe, 2006).
Trees have long been transferred across countries and continents without being considered as potential threats (Petit et al., 2004; Bucharova and van Kleunen, 2009). In most cases, invasive trees have a long history of successive introductions before being eventually classified as problematic aliens. Among these trees is the black cherry (Prunus serotina; Rosaceae), which was one of the first North American tree species to be introduced into Europe (Starfinger et al., 2003). Five varieties are usually recognized in its natural range that covers most of eastern north America (i.e. var. serotina, eximia, rufula, virens and salicifolia; McVaugh, 1951). The most common and widespread variety serotina produces valued furniture wood, but only in a limited portion of its range on the Allegheny plateau of Pennsylvania, New York and West Virginia (Marquis, 1990). The first record of the species in Europe dates back to 1623 when the tree was planted for ornamental purposes near Paris (Starfinger, 1997). Until the late 18th century, P. serotina was sparsely planted in parks and gardens in several European countries. Recommended in 1810 for sylvicultural practices, it was then more intensively planted for growing high quality timber trees and increasing forest production mainly on poor sandy soils (Muys et al., 1992). These experimental plantations rarely produced good quality wood. Between 1900 and 1930, black cherry was planted for multiple uses such as wind and firebreaks, improving the sandy soils under coniferous plantations, or providing shelter (Starfinger, 1997; Starfinger et al., 2003; Pairon et al., 2006a; Verheyen et al., 2007; Vanhellemont et al., 2010). Massive underplanting and filling of conifer stands occurred until the late fifties in Belgium, Germany and The Netherlands (Muys et al., 1992). Black cherry has been planted for forestry purposes since the 1980s in Poland and Romania and local introductions were performed in Italy and the UK (Muys et al., 1992). Presently, the species grows spontaneously in many European countries from France to Poland and Romania as well as from Denmark to Italy. Prunus serotina is spreading throughout temperate forests in north-western Europe, especially on well-drained poor soils. This tree is now considered as one of the 100 most invasive alien species in Europe (DAISIE, 2009). It competes for resources with native plant species, especially during forest regeneration and under high herbivore pressure (e.g. Verheyen et al., 2007; Chabrerie et al., 2008; Vanhellemont et al., 2009). Invaded stands have higher levels of phosphorus, a shallower litter layer, and lower pH values than non-invaded stands (Chabrerie et al., 2008). Shading out light-demanding species, especially seedlings of other tree species, it can impede natural regeneration of native tree species and induces a decrease in species richness, mainly in disturbed stands. Fruits are produced in high quantities and are well dispersed, mainly by birds. More than 50 % of the seeds are dispersed at higher distances than 50 m; the average dispersal distance being estimated to 257 m (Pairon, 2007; Chabrerie et al., 2008). Despite important investment, the control of its spread remains ineffective (Vanhellemont et al., 2009).
In this study, the invasion history of black cherry in Europe was inferred from genetic patterns. Both cpDNA markers and nuclear microsatellite loci were used to genotype native and invasive populations. This study was undertaken to (a) find the putative source(s) of European populations and (b) test whether invasive populations have lower genetic diversity than native populations.
MATERIALS AND METHODS
Sampling scheme
Prunus serotina Ehrh. var. serotina plant material was collected from 23 populations in the invasive range and from 22 populations in the native range (Table 1) for a total number of 442 and 321 trees, respectively. The collection covered nearly the entire geographical distribution of the species (Fig. 1A, B). Samples were collected in countries where the species is known to be abundant and/or of commercial value in northern America and where it is classified as invasive or naturalized in Europe. In each population, two or three leaves were sampled from 10–20 randomly chosen trees. The leaves were either dried for shipping or frozen in liquid nitrogen. DNA was extracted from 1 cm2 of leaf tissue using a modified CTAB protocol (Pairon and Jacquemart, 2005).
Table 1.
Geographical location of the Prunus serotina populations sampled in this study together with their basic genetic information content at eight microsatellite loci
| Pop ID | Location | Country (state) | Latitude | Longitude | N | NA | NPA† | A | HO | H | FIS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Native range | |||||||||||
| CA | Guénette | Canada (Quebec) | 46·50 °N | −75·25 °W | 15 | 6·63 | 4 (0) | 5·92 | 0·75 | 0·74 | −0·02 |
| IN | Bloomington | USA (Indiana) | 39·19 °N | −86·51 °W | 15 | 8·00 | 4 (1) | 6·75 | 0·62 | 0·70 | 0·12 |
| IA1 | Ames | USA (Iowa) | 42·04 °N | −93·68 °W | 15 | 8·25 | 4 (0) | 7·17 | 0·83 | 0·81 | −0·02 |
| IA2 | New Sharon | USA (Iowa) | 41·58 °N | −92·50 °W | 15 | 6·88 | 6 (1) | 5·92 | 0·60 | 0·70 | 0·14*** |
| MO | Springfield | USA (Missouri) | 37·17 °N | −93·33 °W | 15 | 8·00 | 13 (2) | 6·93 | 0·69 | 0·75 | 0·08 |
| NC | Otto | USA (North Carolina) | 35·03 °N | −83·48 °W | 10 | 6·50 | 4 (0) | 6·31 | 0·67 | 0·76 | 0·12 |
| NE1 | Lincoln | USA (Nebraska) | 40·81 °N | −96·64 °W | 14 | 7·50 | 16 (2) | 6·64 | 0·66 | 0·73 | 0·09 |
| NE2 | Omaha | USA (Nebraska) | 41·26 °N | −96·01 °W | 15 | 8·25 | 14 (1) | 7·15 | 0·75 | 0·78 | 0·03 |
| OH1 | Ashtabula | USA (Ohio) | 41·89 °N | −80·79 °W | 15 | 7·50 | 4 (0) | 6·58 | 0·72 | 0·74 | 0·03 |
| OH2 | Delaware | USA (Ohio) | 40·37 °N | −83·05 °W | 14 | 8·50 | 14 (1) | 7·27 | 0·73 | 0·79 | 0·07 |
| OH3 | Ashville | USA (Ohio) | 39·67 °N | −82·93 °W | 14 | 7·75 | 8 (1) | 6·72 | 0·74 | 0·73 | −0·01 |
| OK | Broken Bow | USA (Oklahoma) | 33·97 °N | −94·54 °W | 13 | 6·13 | 13 (3) | 5·61 | 0·64 | 0·63 | −0·02 |
| PE1 | Behrend | USA (Pennsylvania) | 42·11 °N | −79·98 °W | 15 | 7·38 | 2 (0) | 6·38 | 0·74 | 0·76 | 0·02 |
| PE7 | Denton Hill | USA (Pennsylvania) | 41·77 °N | −77·82 °W | 15 | 8·50 | 12 (0) | 7·45 | 0·82 | 0·81 | 0 |
| PE2 | Buckaloons | USA (Pennsylvania) | 41·80 °N | −79·25 °W | 15 | 9·38 | 11 (2) | 7·83 | 0·74 | 0·76 | 0·03 |
| PE3 | Chapman Dam | USA (Pennsylvania) | 41·75 °N | −79·17 °W | 15 | 7·75 | 10 (0) | 6·66 | 0·71 | 0·79 | 0·1 |
| PE4 | Tionesta | USA (Pennsylvania) | 41·71 °N | −78·94 °W | 15 | 8·25 | 8 (0) | 7·27 | 0·84 | 0·80 | −0·05 |
| NY | Pawling | USA (Pennsylvania) | 41·58 °N | −73·61 °W | 15 | 7·88 | 3 (0) | 6·74 | 0·77 | 0·78 | 0 |
| SC | Charleston | USA (South Carolina) | 32·75 °N | −79·91 °W | 15 | 7·13 | 12 (1) | 6·32 | 0·68 | 0·78 | 0·13* |
| WI1 | Saukville | USA (Wisconsin) | 43·39 °N | −88·02 °W | 15 | 8·13 | 6 (1) | 6·83 | 0·71 | 0·74 | 0·05 |
| WI2 | Madison | USA (Wisconsin) | 43·04 °N | −89·44 °W | 15 | 8·63 | 14 (2) | 7·31 | 0·77 | 0·77 | 0·01 |
| WV | Spruce Knob | USA (West Virginia) | 38·71 °N | −79·54 °W | 14 | 7·38 | 5 (0) | 6·70 | 0·82 | 0·79 | −0·04 |
| Mean | 7·74 | – | 6·75 | 0·72 | 0·76 | 0·03 | |||||
| Invasive range | |||||||||||
| BE | Lagland | Belgium | 49·40 °N | 05·46 °E | 20 | 7·13 | 0 (1) | 5·69 | 0·72 | 0·72 | 0·007 |
| DA1 | Copenhagen | Denmark | 55·95 °N | 12·27 °E | 20 | 6·88 | 0 | 5·83 | 0·74 | 0·75 | 0·006 |
| DA2 | Tisvilde | Denmark | 56·05 °N | 12·08 °E | 14 | 7·00 | 0 | 6·28 | 0·79 | 0·74 | −0·069 |
| DA3 | Aarhus | Denmark | 56·18 °N | 9·65 °E | 14 | 6·38 | 0 | 5·82 | 0·71 | 0·73 | 0·04 |
| FR1 | Fontainebleau | France | 48·38 °N | 2·75 °E | 20 | 4·63 | 0 (1) | 4·14 | 0·57 | 0·59 | 0·035 |
| FR2 | Alsace | France | 48·72 °N | 7·68 °E | 16 | 6·50 | 0 | 5·65 | 0·66 | 0·72 | 0·09 |
| FR3 | Compiègne | France | 49·37 °N | 2·88 °E | 20 | 7·25 | 0 (1) | 6·08 | 0·74 | 0·74 | −0·004 |
| FR4 | Bordeaux | France | 44·13 °N | 0·90 °W | 20 | 6·38 | 0 (2) | 5·29 | 0·71 | 0·70 | −0·023 |
| GE1 | Potsdam S | Germany | 52·73 °N | 12·30 °E | 20 | 7·5 | 0 (1) | 5·46 | 0·76 | 0·75 | 0·008 |
| GE2 | Potsdam T | Germany | 52·47 °N | 13·22 °E | 20 | 6·38 | 0 | 6·16 | 0·71 | 0·72 | −0·003 |
| GE3 | Neuwig | Germany | 50·43 °N | 7·63 °E | 20 | 7·13 | 0 | 5·94 | 0·72 | 0·72 | 0·008 |
| GE4 | Mannheim N | Germany | 49·65 °N | 8·53 °E | 20 | 7·25 | 0 | 5·95 | 0·69 | 0·74 | 0·068 |
| GE5 | Karlsruhe | Germany | 48·97 °N | 8·37 °E | 20 | 7·13 | 0 | 5·87 | 0·73 | 0·70 | −0·042 |
| GE6 | Saarbrucken | Germany | 49·30 °N | 7·23 °E | 19 | 6·75 | 0 | 5·78 | 0·68 | 0·74 | 0·08 |
| GE7 | Cuxhaven | Germany | 53·82 °N | 8·63 °E | 20 | 6·63 | 0 | 5·67 | 0·69 | 0·74 | 0·07 |
| HO1 | Arnhem | The Netherlands | 52·00 °N | 5·83 °E | 20 | 7·25 | 0 | 5·82 | 0·73 | 0·71 | −0·014 |
| HO2 | Amsterdam | The Netherlands | 52·35 °N | 4·55 °E | 19 | 6·63 | 0 | 5·54 | 0·63 | 0·71 | 0·123 |
| IT1 | Ticino | Italy | 45·52 °N | 8·72 °E | 20 | 4·50 | 0 | 4·08 | 0·66 | 0·64 | −0·018 |
| IT2 | Udine | Italy | 46·12 °N | 13·17 °E | 20 | 3·13 | 0 | 2·98 | 0·60 | 0·55 | −0·088 |
| PO1 | Poznan | Poland | 52·17 °N | 17·15 °E | 20 | 5·38 | 0 | 4·76 | 0·67 | 0·68 | 0·015 |
| PO2 | Wroclaw | Poland | 51·37 °N | 16·55 °E | 20 | 5·00 | 0 | 4·49 | 0·69 | 0·61 | −0·119 |
| UK1 | Shakelford | UK | 51·18 °N | 0·65 °W | 20 | 4·63 | 0 | 3·91 | 0·59 | 0·59 | 0·006 |
| UK2 | Bagshot | UK | 51·38 °N | 0·73 °W | 20 | 5·50 | 0 (2) | 5·03 | 0·76 | 0·75 | −0·011 |
| Mean | 6·36 | – | 5·31 | 0·69 | 0·70 | 0·01 | |||||
N is the number of individuals sampled per population, NA is the mean number of alleles per locus, A is the mean allelic richness per locus (based on n = 10) and NPA is the number of private alleles. Estimates of observed heterozygosity (HO), gene diversity (H) and inbreeding coefficient (FIS) are also given.
* 0·01 < P < 0·05, *** P < 0·001
† For the private alleles, the first number is the number of alleles private to a given range (invasive or native) in the considered population, while the number in parenthesis is the number of alleles that are private to the considered population in its range.
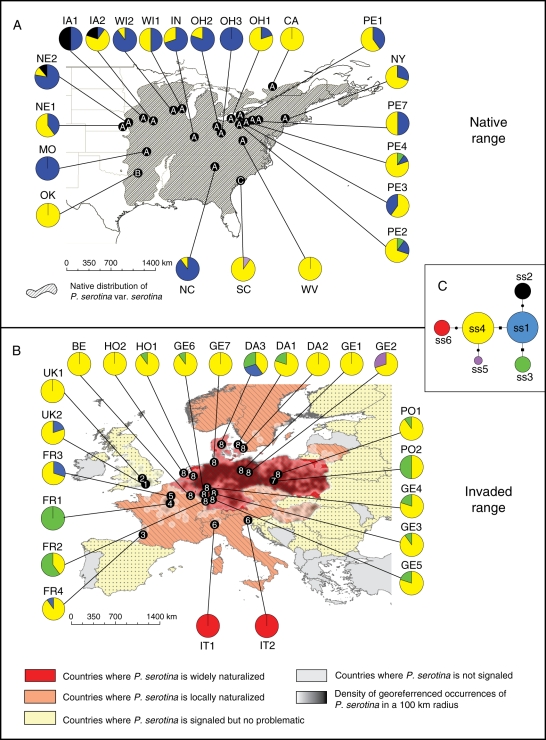
Fig. 1.
Geographical distribution of nuclear and chloroplast genetic variation in Prunus serotina. (A) Native range with sampling locations. The hatched area indicates the natural occurrence of P. serotina var. serotina in the wild. Dots on the map represent the three genetic clusters defined by Baps based on nuclear microsatellite data. These dots are labelled from A to C according to the cluster they belong to. Pie charts represent the relative proportions of cpDNA haplotypes in a population, as indicated in (C). (B) Invasive range with sampling locations. Dots on the map represent the eight genetic clusters defined by Baps based on the nuclear microsatellite data. The dots are numbered from one to eight according to the cluster they belong to. (C) Median-joining network of cpDNA haplotypes. The size of the circles represents the relative frequency of the haplotypes. The biggest size corresponds to haplotypes with a frequency >10 %, the intermediate size corresponds to haplotypes with a frequency between 1 and 10 % and the smallest size corresponds to haplotypes with a frequency <1 %. Steps between haplotypes are either insertion/deletion (i.e. length variation – dots) or nucleotide substitutions without length variation (squares).
Microsatellite and cpDNA typing
Eight nuclear microsatellite loci previously described for P. serotina (Table 2; Pairon et al., 2008) were amplified in all sampled individuals. Polymerase chain reactions (PCR) were performed in a total volume of 15 µL using the methods previously described (Pairon et al., 2006b). PCR products were separated on an ABI3100 genetic analyser (Applied Biosystems) and individuals genotyped using GeneMapper 3·5 (Applied Biosystems).
Table 2.
Genetic diversity detected at eight microsatellite loci in invasive and native Prunus serotina
| Allele size range |
Total no. of alleles |
HS |
FIS |
FST |
F′ST |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Invasive | Native | Invasive (n = 442) | Native (n = 321) | Invasive | Native | Invasive | Native | Invasive | Native | Whole dataset | Invasive | Native | Whole dataset |
| UDP96-005 | 82–88 | 82–96 | 4 | 7 | 0·57 | 0·60 | 0·025ns | 0·09ns | 0·06* | 0·09* | 0·09* | 0·16* | 0·24* | 0·24* |
| UDP96-025 | 109–137 | 108–137 | 13 | 18 | 0·78 | 0·84 | −0·008ns | 0·07* | 0·09* | 0·03* | 0·08* | 0·41* | 0·20* | 0·42* |
| UDP96-405 | 109–123 | 109–123 | 6 | 8 | 0·51 | 0·63 | 0·119ns | 0·01ns | 0·15* | 0·09* | 0·14* | 0·32* | 0·26* | 0·32* |
| UCD-CH14 | 130–168 | 124–182 | 19 | 29 | 0·81 | 0·90 | −0·002ns | 0·02ns | 0·10* | 0·04* | 0·09* | 0·56* | 0·47* | 0·60* |
| PceGA34 | 135–175 | 131–175 | 19 | 24 | 0·80 | 0·89 | −0·030ns | 0·01ns | 0·08* | 0·05* | 0·08* | 0·38* | 0·45* | 0·53* |
| Pchgms2 | 127–159 | 125–163 | 11 | 18 | 0·69 | 0·55 | 0·007ns | −0·02ns | 0·09* | 0·13* | 0·12* | 0·30* | 0·29* | 0·32* |
| Pchpgms3-1 | 168–198 | 166–198 | 11 | 12 | 0·65 | 0·79 | −0·012ns | 0·05ns | 0·10* | 0·05* | 0·10* | 0·30* | 0·23* | 0·33* |
| Pchpgms3-2 | 200–238 | 200–238 | 15 | 18 | 0·80 | 0·86 | 0·011ns | 0·08ns | 0·07* | 0·05* | 0·06* | 0·35* | 0·32* | 0·37* |
| Mean | 0·70 | 0·76 | 0·01ns | 0·03ns | 0·09* | 0·06* | 0·09* | 0·35* | 0·31* | 0·39* | ||||
The Nei's gene diversity per locus (HS) and Wright's inbreeding coefficient (FIS) were averaged over the native and invasive populations. The Wright's fixation index (FST) and an analogue unconstrained by gene diversity (F′ST) were averaged over the native and invasive populations or the whole dataset.
ns, not significant; *, P < 0·001.
Five cpDNA loci previously described for P. serotina (Petitpierre et al., 2009) were used on a subsample of ten randomly chosen individuals per population (i.e. 230 individuals in the invasive range and 220 in the native range). These markers are derived from non-coding plastid regions and correspond either to length variation in polyT/A stretches (ccmp5, trnT-trnL-poly-T and trnT-trnL-poly-A) or to restriction fragment length polymorphisms due to a single nucleotidic substitution (trnD-trnT-TasI and trnS-trnG-TasI).
Data analyses
Analysis of nuclear microsatellite data
For each microsatellite locus, the total number of alleles, Nei's unbiased estimate of gene diversity (HS; Nei, 1987), and the inbreeding coefficient (FIS) were separately estimated for invasive and native populations using Fstat 2·9 (Goudet, 1995). To characterize further the genetic structure within loci, Wright's fixation index (FST) and its analogue based on allele size (RST; Michalakis and Excoffier, 1996) were computed using Spagedi 1·2 (Hardy and Vekemans, 2002). To detect whether FST and RST were significantly different from 0, Spagedi was also used to perform a 10 000-permutations test on alleles. FST values cannot be higher than the homozygosity level, 1 – HS (Hedrick, 2005). F′ST, an estimator unconstrained by genetic diversity, was therefore calculated as FST/1 – HS. 10 000 randomizations tests were performed to assess the differences of genetic diversity and differentiation between the native and invaded range. The partitioning of the gene diversity at different geographic scales (i.e. within population, among populations and between invasive and native ranges) was also measured using an AMOVA with Arlequin 3·1·1 (Excoffier et al., 2005).
To characterize the genetic diversity at the population level over the eight loci, the observed heterozygosity (HO) was calculated with Genalex 6·0 (Peakall and Smouse, 2006), as well as the allelic richness (A), Nei's unbiased estimate of gene diversity (HS; Nei, 1987) and the inbreeding coefficient (FIS) with Fstat. Significance of the inbreeding coefficient was assessed with Fstat using permutations. Hardy–Weinberg equilibrium was tested using Genepop 3·4 (Rousset, 2008). The number of private alleles was computed for each population in the native range.
Introduced populations are often subject to founder effects that result in reduced genetic diversity (i.e. a bottleneck). To assess the decrease in diversity from native to invasive populations, three different methods were used. First, the values of allelic richness and gene diversity averaged over the whole invasive and native ranges were compared, postulating that a genetic bottleneck should decrease the number of alleles present in the invasive range. Differences in A and HS were assessed using a paired t-test in the SAS software version 8·2 (SAS Institute, 1999). Secondly, Bottleneck 1·2 (Cornuet and Luikart, 1996) and the Wilcoxon test for heterozygote excess were used under the two-phase model and the stepwise mutation model. The Wilcoxon test is preferable when few loci are used (Cornuet and Luikart, 1996; Cornuet et al., 1999). Thirdly, the allele frequencies were plotted for each invasive population based on the method described in Luikart et al. (1998). This method postulates that recent bottlenecks should cause a decrease in rare alleles, creating a characteristic mode-shift distortion in the distribution of allele frequencies.
The population genetic structure at the geographic level is shaped by different forces such as genetic drift and the possibility of inter-population gene exchanges. The characterization of genetic relationships between populations can be informative about their histories. The relatedness among populations was first assessed by computing F-statistics (Weir and Cockerham, 1984) and Nei's standard genetic distance (D; Nei, 1972) between pairs of invasive populations. A significance test of 10 000 permutations of the estimates of FST and D obtained was performed using Genetix 4·052 (Belkhir et al., 2000). The genetic structure in the invasive range was further described, together with that of the native range, by using the Bayesian clustering method implemented in Baps 4·14 (Corander and Marttinen, 2006). Baps uses an analytical integration strategy combined with stochastic search methods to infer the number of clusters K and was run using both genotype and sample group information (group mode). The maximum number of populations (prior information) was set to 40.
The genetic clusters defined by Baps in the native range were then used as reference populations in Geneclass2 (Piry et al., 2004) to assign individuals from invasive populations. The frequency-based method of Paetkau et al. (1995) and Monte-Carlo resampling was used to compute assignment probabilities with the simulation algorithm of Piry et al. (2004). To gain insight into the source of the clusters defined by Baps in the invasive range, the proportion of individuals assigned to the different clusters of the native range was calculated within each cluster of the invasive range.
Analysis of cpDNA data
To characterize the genetic diversity of the chloroplastic genome, the allelic richness (A), cpDNA haplotype diversity within populations (HS), total cpDNA haplotype diversity (HT) and the level of genetic differentiation (FST) within both ranges were calculated using Fstat. Ten thousand randomizations tests were performed to assess the differences of genetic diversity and differentiation between the native and invaded range. Relationships among cpDNA haplotypes were visualized by constructing a statistical median-joining network implemented in Network 4·112 (Bandelt et al., 1999).
Haplotypes were then plotted on maps to illustrate their geographical distribution and the putative introduction sources. Their percentage distribution was also estimated within both ranges.
RESULTS
Genetic diversity
Within microsatellite loci
All eight nuclear microsatellite loci were polymorphic in each population, and a total number of 134 alleles was observed on the whole sample (Table 2). Ninety-eight alleles were observed in invasive populations, while all of them were detected in native populations (Table 2). Thirty-six alleles (27 %) were private to the native range. Overall gene diversity (HS) was also higher in the native range at the majority of loci (Table 2). FIS ranged from −0·02 to 0·09 and a significant deviation from Hardy–Weinberg equilibrium was found at locus UDP96-025 but only in the native range. FST showed an overall differentiation of 0·06 in the native range and 0·09 in the invasive range. RST values were not significantly different from FST and were thus not shown. F′ST values also showed that the overall differentiation in the native range (0·31) was lower than in the invasive range (0·35; Table 2) but this difference was not significant. The AMOVA analysis indicated that genetic diversity was mainly partitioned within populations (89 ± 2·9 %), then among populations (8 ± 0·3 %), and then between the native and invasive ranges (3 ± 0·1 %).
Chloroplastic DNA haplotypes
Considering allelic combinations at the five cpDNA loci, six haplotypes were detected in the whole dataset. Both native and invasive ranges shared four haplotypes (Table 3). Haplotype ss2 was private to the native range and only found in the western part of the United States (Fig. 1A). On the other hand, haplotype ss6 was private to the invasive range and exclusively found in northern Italy (Fig. 1B). Haplotypes ss1 and ss4 were by far the most abundant (Table 3), and displayed a central position in the median-joining network (Fig. 1C). This network also suggests that ss2 and ss3 have derived from ss1 while ss5 and ss6 have derived from ss4. The level of genetic differentiation measured by FST averaged 0·40 and 0·52 in native and invasive populations (Table 4), respectively, but these values were not significantly different (P = 0·49).
Table 3.
Compared occurrences of the six cpDNA haplotypes (with their frequency given in parenthesis) in both native and invasive ranges of Prunus serotina
| Haplotype no. | Native range | Invasive range |
|---|---|---|
| ss1 | 96 (43·8 %) | 9 (3·9 %) |
| ss2 | 8 (3·7 %) | – |
| ss3 | 2 (0·9 %) | 34 (14·8 %) |
| ss4 | 112 (51·1 %) | 164 (71·3 %) |
| ss5 | 1 (0·5 %) | 3 (1·3 %) |
| ss6 | – | 20 (8·7 %) |
Table 4.
Chloroplast DNA diversity in native and invasive Prunus serotina populations
| Range | N | NA | A | HS | HT | FST |
|---|---|---|---|---|---|---|
| Native | 219 | 5 | 1·953 | 0·333 | 0·547 | 0·402* |
| Invasive | 230 | 5 | 1·651 | 0·225 | 0·461 | 0·519* |
Haplotypes were defined by the combination of alleles at five loci. Number of individuals (N), number of haplotypes (NA), mean allelic richness per population (A), haplotype diversity within populations (HS), total haplotype diversity (HT) and the level of genetic differentiation (FST) are given for each range.
* P < 0·001.
Sources of invasive populations
Nuclear microsatellite data
Baps in group mode found three clusters in the native range (Fig. 1A) and eight clusters in the invasive range (Fig. 1B). The three clusters found in the native range only showed that one cluster (A) was formed by all populations except OK and SC, which accounted for the two remaining groups (B and C, respectively). The genetic structure was more pronounced in the invasive range and the output of the program suggested that the two populations from the United Kingdom (UK1-UK2), populations FR1, FR3 and FR4 (France), and population PO2 (Poland) were all distinct genetic clusters. The two populations from Italy represented another independent cluster, and the last cluster was formed by the remaining populations (i.e. populations from Denmark, Germany, The Netherlands and Belgium and populations FR2 and PO1). When the individual genotypes from the invasive range were used to assign each individual to one of the three clusters of the native range, very few trees were assigned to clusters B and C, and the vast majority was assigned to the main cluster (A) in the native range. When the clusters found in the invasive range were reconstructed and the proportion of individuals within each cluster that were assigned to the three main clusters of the native range were looked at, it was noted that 83–95 % of the individuals within clusters 2, 3, 5, 6, 7 and 8 were assigned to the main cluster (A) in the native range (Table 5). Clusters 1 and 4, comprising populations UK2 and FR1, respectively, showed a relatively low proportion of individuals assigned to the main cluster in the native range (65 % and 0 %, respectively), and even to any of the two other native clusters (0–15 % and 0–50 %, respectively).
Table 5.
Proportion of individuals belonging to the eight clusters from the invasive range (Fig. 1B) assigned to each of the three clusters defined in the native range (Fig. 1A)
| Clusters in native range |
|||
|---|---|---|---|
| Clusters in invasive range | A (293) | B (13) | C (15) |
| 1 (20) | 0·65 | 0·00 | 0·15 |
| 2 (20) | 0·95 | 0·00 | 0·05 |
| 3 (20) | 0·95 | 0·00 | 0·05 |
| 4 (20) | 0·00 | 0·50 | 0·00 |
| 5 (20) | 0·95 | 0·00 | 0·15 |
| 6 (40) | 0·83 | 0·00 | 0·25 |
| 7 (20) | 0·95 | 0·05 | 0·30 |
| 8 (282) | 0·90 | 0·00 | 0·20 |
The number of individuals within each cluster (for both native and invasive ranges) is given in parenthesis. Note that individuals can be assigned to more than one cluster.
Chloroplastic DNA data
cpDNA haplotype distribution gave more information as to which regions in the native range may have acted as source(s) for invasive populations (Table 3). This distribution was indeed quite different among the two ranges, and in the invasive area, a much lower proportion of the ss1 haplotype was detected, whereas ss3 and ss4 were overrepresented. The vast majority of populations located to the east of the Appalachian Mountains showed a predominant proportion of ss4, as observed in the invasive range, whereas most populations located to the west to the Appalachian Mountains showed a predominant proportion of ss1. Interestingly, ss3 was found in half of the European populations and only detected in two populations of the Allegheny plateau in the USA (PE2 and PE4; Fig. 1A). Finally, ss2 was not observed in the invasive range and was only found in the western part of the native range.
Partition of genetic diversity within native and invasive ranges
Within the native range
The 36 alleles that were private to the native range were relatively well distributed among the native populations (Table 1). Populations PE1 and NY (Pennsylvania) displayed, respectively, only two and three alleles that were not found in the invasive range whereas a maximum of 16 private alleles were found within population NE1 (Nebraska). The number of alleles per locus (NA) was 7·74 in average, ranging from 6·13 to 9·38 per population. The mean allelic richness (A) ranged from 5·61 to 7·83 with a mean of 6·75. Gene diversity (HS) ranged from 0·63 to 0·81 with a mean of 0·76 whereas mean observed heterozygosity (HO) ranged from 0·60 to 0·84 among populations with a mean of 0·72. The inbreeding coefficient (FIS) varied widely among populations, ranging from −0·05 to 0·14 with a mean of 0·03. Significant deviations from Hardy–Weinberg genotypic proportions associated with positive FIS values were found for populations IA2 and SC.
Within the invasive range
The mean allelic richness (A) was significantly lower in the invasive range (5·31) than in the native range (6·75) (t = 6·79, P < 0·0001) as expected by the higher number of alleles found in the native range (Table 1). This also resulted in a significantly lower gene diversity in the invasive range (t = 3·75, P = 0·0005; Table 1). The number of alleles per population from the invasive range varied more widely than in the native range. Populations FR1 (France), IT1, IT2 (Italy), PO1, PO2 (Poland), UK1 (United Kingdom) displayed the lowest amount of genetic diversity. Gene diversity (HS) varied from 0·55 to 0·75 whereas mean observed heterozygosity (HO) ranged from 0·57 to 0·79 among populations. None of the populations showed significant departure from Hardy–Weinberg proportions and inbreeding coefficients (FIS) ranged from −0·12 to 0·07, with a mean of 0·01.
Even though some populations were identified as having a low number of alleles and a low gene diversity, only five populations at the margins of the invasive range (IT1, IT2, PO2, DA1 and UK2) were significantly detected as bottlenecked based on the Wilcoxon test under the two-phase model, and only two populations (IT2 and UK2) under the stepwise mutation model. In addition, the graphical method allowed two recently bottlenecked populations (PO1 and PO2) to be detected, with a marked distortion in the distribution of allele frequencies.
A maximum of two cpDNA haplotypes was found per population in the invasive range, whereas some populations of the native range exhibit three haplotypes (Fig. 1A and B). Allelic richness was significantly lower in the invasive range (A = 1·65) than in the native range (A = 1·95) (permutation test, P = 0·04; Table 4). The haplotype diversity within populations (HS) and the total haplotype diversity (HT) were lower in the invasive than in the native range (Table 4), and this difference was significant for HT (permutation tests, P = 0·02) but not for HS (permutation tests, P = 0·054).
DISCUSSION
The combined use of cpDNA and nuclear microsatellite loci aimed at comparing the genetic diversity of P. serotina var. serotina populations between native and invasive ranges and detecting the putative sources of invasive populations. Previous studies have demonstrated the importance of extensive sampling in both ranges (e.g. Novak and Mack, 1993; Eckert et al., 1996; Meekins et al., 2001; Okada et al., 2009) and the usefulness of combining markers with two different modes of inheritance (Williams et al., 2005) to achieve these goals, but as far as is known, the present study is one of the first attempts to answer these questions for a long-lived species.
Sources of invasive P. serotina
The presence of five cpDNA haplotypes in Europe suggests there were multiple introductions of individuals. The distribution of haplotypes in the native range can be used to identify putative sources of invasive populations (e.g. Besnard et al., 2007; Hufbauer and Sforza, 2008). First, frequencies of haplotypes ss1 and ss4 in both ranges and the distribution of haplotype ss2 all suggest that invasive P. serotina trees mainly originated from the eastern United States. Haplotype ss4 is very frequent in the invasive range (Table 3) and mainly distributed to the east of the Appalachian Mountains whereas ss1 is almost absent in Europe and very frequent in the western part of the native range (Fig. 1A). Moreover, ss2 is absent in Europe and only found in the north-western states of Indiana and Nebraska, further suggesting that such populations should not be considered as putative source regions. Secondly, ss3 is overrepresented in many invasive populations in Europe suggesting that the Allegheny plateau was an important source of introduction as this haplotype is only found in native populations PE2 and PE4 (where ss4 and ss1 are also present). Prunus serotina is harvested for timber only in the Allegheny plateau (Hough, 1965; Marquis, 1990) and it is not surprising that this area was an important source for introductions to Europe.
No strong geographic differentiation based on nuclear loci in the native range was detected. Only two populations in the southern native range (SC and OK) were distinguished from other populations (Fig. 1A). Their geographic isolation in the native range probably explains their high genetic differentiation from the main native cluster A. Human movement of the species for forestry purposes and high seed dispersal in the native range may in part be responsible for the low level of genetic differentiation within the large cluster A. Assignment tests of invasive individuals were not very informative as they only supported the assertion that cluster A was probably involved in most introductions to Europe. Nevertheless, the low number of native-range private alleles in some populations from the Allegheny plateau (e.g. PE1 and NY) again suggests, as cpDNA haplotypes, that this area within cluster A was the main source of introductions to Europe.
Genetic structure and low reduction of gene diversity in the invaded range
The overall genetic structure displayed by microsatellite loci in the present study was more pronounced in the invasive range than in the native range. Populations from Denmark, Germany, The Netherlands and Belgium had higher genetic diversity and lower differentiation among populations, while some populations from the United Kingdom, France and Italy had low allelic richness, low gene diversity and high genetic differentiation among populations. These two different patterns were supported by the output of Baps as populations from Denmark, Germany, The Netherlands and Belgium were clustered together (cluster 8) whereas the other populations belonged to different separated clusters.
A reduction in the overall number of alleles, allelic richness and heterozygosity was found at microsatellite loci in the invasive range. Most introductions result in a temporary high reduction in the effective size of populations, and a reduction in diversity within invasive populations should therefore often be observed (Dlugosch and Parker, 2008). This reduction was particularly evident at microsatellite loci as these markers contain many rare alleles, and are therefore more sensitive to founder effects (Spencer et al., 2000). Despite the observed reduction in allelic diversity, strong evidence of population bottlenecks was only observed in some populations at the margins of the invaded range that probably correspond to recent introductions (PO2, DA1, IT1, IT2 and UK2). Indeed, the tests performed look at recent bottleneck events by searching for the allele depletion that should be generated by a sudden decrease of reproducing individuals in a newly formed population. Because allele depletion is a complex function that depends on several parameters, including the time since a population bottleneck (Cornuet and Luikart, 1996; Prentis et al., 2008; Bucharova and van Kleunen, 2009), populations that had gone through a bottleneck several generations ago may not display allelic depletion any more (Dlugosch and Parker, 2008). This points to the importance of the temporal nature of introductions in genetic diversity studies as the particular time of study might affect the ability to detect past bottlenecks (Dlugosch and Parker, 2008).
Despite the reduction in genetic diversity in some marginally distributed populations, the invasive range in general possessed high levels of diversity (73 % of allelic richness), with an overall high gene diversity with microsatellite markers and the same number of cpDNA haplotypes in both ranges. The low reduction in genetic diversity can be attributed to multiple introductions with large numbers of individuals and the relatively long time since introduction (Bucharova and van Kleunen, 2009).
The initial propagule size and the occurrence of gene flow between established populations seem in the present case to be important factors explaining the genetic patterns observed in the invasive range. All species are likely to experience periods when founding populations are isolated from other conspecific populations, but this relative isolation cannot always be recorded, especially when natural gene flow has been rapidly restored among expanding populations, multiple introductions are geographically close to one another and/or human-mediated seed exchanges occur among introduced populations (Suarez and Tsutsui, 2008). Interestingly, genetically less diverse populations from France, Italy, Poland and England (FR1, IT1–IT2, PO1–PO2, UK1–UK2) are all situated on the margins of the species range in Europe and in countries where no records of massive plantations for forestry purposes are known (Starfinger, 1997). Populations from The Netherlands, Belgium, Germany and Denmark were more diverse and historical records support that propagule number was more important, with several massive introductions of the species for different purposes during the 19th century (Muys et al., 1992; Starfinger, 1997). The relatively high genetic diversity in populations from north-western Europe probably contributed to the adaptive success and the invasive behaviour of P. serotina.
ACKNOWLEDGEMENTS
This study was supported by the Fonds Spéciaux de Recherche (FSR) of the Université catholique de Louvain (UCL), the Belgian Scientific Policy (BelSPo–InPlanBel) and the National Centre of Competence in Research (NCCR) Plant Survival, research programme of the Swiss National Science Foundation. Collection of some samples was partly funded by grants to K. O. Reinhart from Highlands Biological Station and from the AW Mellon Foundation. We are grateful for the kind collaboration of many foresters and scientists who helped us to collect leaves of Prunus serotina. Samples from France were provided by P. Geldreich, J. M. Ansolabehere, R. Rodriguez and J. Jaminon (Office National des Forêts), and G. Decocq (University of Picardie Jules Verne); from Germany by T. Heinken (University of Potsdam), U. Starfinger (Technische University of Berlin) and J. Meyer (Forestrevier Altenwalde); from The Netherlands by W. van der Putten (NIOO-KNAW) and A. Ehrenburg (Amsterdam Water Supply); from Italy by F. Caronni and L. Hildebrand (Parco Lombardo della Valle del Ticino) and S. Del Fabbro; from Denmark by E. Kjaer (Royal Veterinary and Agricultural University), N. P. Revsbech (University of Aarhus) and O. Raspé (Jardin botanique national de Belgique); from Poland by B. Suszka (Polish Academy of Sciences) and A. Halarewicz (University of Agriculture of Wroclaw); from the USA by G. Meyer (University of Wisconsin–Milwaukee Field Station), A. K. Buthod (Robert Bebb Herbarium, The University of Oklahoma), D. A. Lewis (Ada Hayden Herbarium, Iowa State University), L. Iverson (Northern Research Station, USDA Forest Service), R. B. Kaul and D. M. Sutherland (Bessey Herbarium University of Nebraska), T. S. Cochrane (University of Wisconsin–Madison Herbarium), K. O. Reinhart (United States Department of Agriculture, Agricultural Research Service), R. A. Klips (Ohio State University) and L. M. Bowe (Ozarks Regional Herbarium, Missouri State University). We thank L. Dhondt, C. Noël and D. Savova-Bianchi for technical help in the laboratory. We would like to thank O. Hardy, O. Raspé, U. Starfinger and two anonymous referees for helpful comments and revision of previous versions of the manuscript.
LITERATURE CITED
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Goudet J, Chikhi L, Bonhomme F. Genetix 4·03, logiciel sous WindowsTM pour la génétique des populations. 2000 Laboratoire Génome, Populations, Interactions CNRS UMR 5000, Université de Montpellier II, Montpellier, France. [Google Scholar]
- Besnard G, Henry P, Wille L, Cooke D, Chapuis E. On the origin of the invasive olives (Olea europaea L., Oleaceae) Heredity. 2007;99:608–619. doi: 10.1038/sj.hdy.6801037. [DOI] [PubMed] [Google Scholar]
- Bucharova A, van Kleunen M. Introduction history and species characteristics partly explain naturalization success of North American woody species in Europe. Journal of Ecology. 2009;97:230–238. [Google Scholar]
- Carlton JT. Pattern, process and prediction in marine invasion ecology. Biological Conservation. 1996;78:97–106. [Google Scholar]
- Chabrerie O, Verheyen K, Saguez R, Decocq G. Disentangling relationships between habitat conditions, disturbance history, plant diversity, and American black cherry (Prunus serotina Ehrh.) invasion in a European temperate forest. Diversity and Distributions. 2008;14:204–212. [Google Scholar]
- Corander J, Marttinen P. Bayesian identification of admixture events using multi-locus molecular markers. Molecular Ecology. 2006;15:2833–2843. doi: 10.1111/j.1365-294X.2006.02994.x. [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuet JM, Piry S, Luikart G, Estoup A, Solignac M. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics. 1999;153:1989–2000. doi: 10.1093/genetics/153.4.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAISIE. Handbook of alien species in Europe. Knoxville, TN: Springer; 2009. [Google Scholar]
- Devos N, Tyteca D, Raspé O, Wesselingh RA, Jacquemart AL. Pattern of chloroplast diversity among western European Dactylorhiza species (Orchidaceae) Plant Systematics and Evolution. 2003;243:85–97. [Google Scholar]
- Dlugosch KM, Parker IM. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Molecular Ecology. 2008;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- Eckert CG, Manicacci D, Barrett SCH. Genetic drift and founder effect in native versus introduced populations of an invading plant, Lythrum salicaria (Lythraceae) Evolution. 1996;50:1512–1519. doi: 10.1111/j.1558-5646.1996.tb03924.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3·0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Facon B, Pointier JP, Jarne P, Sarda V, David P. High genetic variance in life-history strategies within invasive populations by way of multiple introductions. Current Biology. 2008;18:363–367. doi: 10.1016/j.cub.2008.01.063. [DOI] [PubMed] [Google Scholar]
- Forman J. The introduction of American plant species into Europe: issues and consequences. In: Child L, Brock J, Brundu G, et al., editors. Plant invasions: ecological threats and management solutions. Leiden: Backhuys; 2003. pp. 17–33. [Google Scholar]
- Gaskin JF, Zhang DY, Bon MC. Invasion of Lepidium draba (Brassicaceae) in the western United States: distributions and origins of chloroplast DNA haplotypes. Molecular Ecology. 2005;14:2331–2341. doi: 10.1111/j.1365-294X.2005.02589.x. [DOI] [PubMed] [Google Scholar]
- Goudet J. Fstat (Version 1·2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Grapputo A, Boman S, Lindstrom L, Lyytinen A, Mappes J. The voyage of an invasive species across continents: genetic diversity of North American and European Colorado potato beetle populations. Molecular Ecology. 2005;14:4207–4219. doi: 10.1111/j.1365-294X.2005.02740.x. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. Spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2:618–620. [Google Scholar]
- Hedrick PW. A standardized genetic differentiation measure. Evolution. 2005;59:1633–1638. [PubMed] [Google Scholar]
- Henry P, Le Lay G, Goudet J, Guisan A, Jahodova S, Besnard G. Reduced genetic diversity, increased isolation and multiple introductions of invasive Giant Hogweed in the western Swiss Alps. Molecular Ecology. 2009;18:2819–2831. doi: 10.1111/j.1365-294X.2009.04237.x. [DOI] [PubMed] [Google Scholar]
- Hough AF. Black cherry (Prunus serotina Ehrh.) In: Fowells HA, editor. Silvics of forest trees of the United States. Washington DC: US Department of Agriculture; 1965. pp. 539–545. Agriculture Handbook No. 271. [Google Scholar]
- Hufbauer RA, Sforza R. Multiple introductions of two invasive Centaurea taxa inferred from cpDNA haplotypes. Diversity and Distribution. 2008;14:252–261. [Google Scholar]
- Le Roux JJ, Wieczorek AM, Meyer JY. Genetic diversity and structure of the invasive tree Miconia calvescens in Pacific islands. Diversity and Distributions. 2008;14:935–948. [Google Scholar]
- Lockwood JL, Cassey P, Blackburn T. The role of propagule pressure in explaining species invasions. Trends in Ecology & Evolution. 2005;20:223–228. doi: 10.1016/j.tree.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Luikart G, Allendorf FW, Cornuet JM, Sherwin WB. Distortion of allele frequency distributions provides a test for recent population bottlenecks. Journal of Heredity. 1998;89:238–247. doi: 10.1093/jhered/89.3.238. [DOI] [PubMed] [Google Scholar]
- McCauley DE, Smith RA, Lisenby JD, Hsieh C. The hierarchical spatial distribution of chloroplast DNA polymorphism across the introduced range of Silene vulgaris. Molecular Ecology. 2003;12:3227–3235. doi: 10.1046/j.1365-294x.2003.01992.x. [DOI] [PubMed] [Google Scholar]
- McVaugh R. A revision of the north American black cherries (Prunus serotina Ehrh., and relatives) Brittonia. 1951;7:279–315. [Google Scholar]
- Marquis D. Prunus serotina Ehrh. black cherry. In: Burns RM, Honkala BH, editors. Silvics of North America. Vol. 2. Washington, DC: US Department of Agriculture, Forest Service; 1990. pp. 594–604. Hardwoods. [Google Scholar]
- Meekins JF, Ballard HE, McCarthy BC. Genetic variation and molecular biogeography of a North American invasive plant species (Alliaria petiolata, Brassicaceae) International Journal of Plant Sciences. 2001;162:161–169. [Google Scholar]
- Michalakis Y, Excoffier L. A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics. 1996;142:1061–1064. doi: 10.1093/genetics/142.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muys B, Maddelein D, Lust N. Ecology, practice and policy of black cherry (Prunus serotina Ehrh.) management in Belgium. Silva Gandavensis. 1992;27:28–45. [Google Scholar]
- Nei M. Genetic distance between populations. American Naturalist. 1972;106:283–292. [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York, NY: Columbia University Press; 1987. [Google Scholar]
- Neuffer B, Hurka H. Colonization history and introduction dynamics of Capsella bursa-pastoris (Brassicaceae) in North America: isozymes and quantitative traits. Molecular Ecology. 1999;8:1667–1681. doi: 10.1046/j.1365-294x.1999.00752.x. [DOI] [PubMed] [Google Scholar]
- Novak SJ, Mack RN. Genetic variation in Bromus tectorum (Poaceae): comparison between native and introduced populations. Heredity. 1993;71:167–176. [Google Scholar]
- Novak SJ, Mack RN. Tracing plant introduction and spread: genetic evidence from Bromus tectorum (Cheatgrass) BioScience. 2001;51:114–122. [Google Scholar]
- Okada M, Lyle M, Jasieniuk M. Inferring the introduction history of the invasive apomictic grass Cortaderia jubata using microsatellite markers. Diversity and Distributions. 2009;15:148–157. [Google Scholar]
- Paetkau D, Calvert W, Stirling I, Strobeck C. Microsatellite analysis of population structure in Canadian polar bears. Molecular Ecology. 1995;4:347–354. doi: 10.1111/j.1365-294x.1995.tb00227.x. [DOI] [PubMed] [Google Scholar]
- Pairon MC. Ecology and population genetics of an invasive forest tree species: 2007 Prunus serotina Ehrh. PhD Dissertation, Université catholique de Louvain. [Google Scholar]
- Pairon MC, Jacquemart AL. Disomic segregation of microsatellites in the tetraploid Prunus serotina Ehrh. (Rosaceae) Journal of the American Society for Horticultural Science. 2005;130:729–734. [Google Scholar]
- Pairon M, Chabrerie O, Mainer Casado C, Jacquemart AL. Sexual regeneration traits linked to black cherry (Prunus serotina Ehrh.) invasiveness. Acta Oecologica. 2006a;30:238–247. [Google Scholar]
- Pairon MC, Jonard M, Jacquemart AL. Modeling seed dispersal of black cherry, an invasive forest tree: how microsatellites may help? Canadian Journal of Forest Research. 2006b;36:1385–1394. [Google Scholar]
- Pairon MC, Potter D, Jacquemart AL. Detection and characterization of genome-specific microsatellite markers in the allotetraploid Prunus serotina. Journal of the American Society for Horticultural Science. 2008;133:390–395. [Google Scholar]
- Peakall R, Smouse PE. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Hampe A. Some evolutionary consequences of being a tree. Annual Review of Ecology, Evolution, and Systematics. 2006;37:187–214. [Google Scholar]
- Petit RJ, Kremer A, Wagner DB. Finite island model for organelle and nuclear genes in plants. Heredity. 1993;71:630–641. [Google Scholar]
- Petit RJ, Bialozyt R, Garnier-Gere P, Hampe A. Ecology and genetics of tree invasions: from recent introductions to Quaternary migrations. Forest Ecology and Management. 2004;197:117–137. [Google Scholar]
- Petitpierre B, Pairon M, Broennimann O, Jacquemart AL, Guisan A, Besnard G. Plastid DNA polymorphisms in Prunus serotina var. serotina (Rosaceae), a North American tree invading Europe. European Journal of Forest Research. 2009;128:431–436. [Google Scholar]
- Pimentel D, Zuniga R, Morrison D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecological Economics. 2005;52:273–288. [Google Scholar]
- Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A. Geneclass2: a software for genetic assignment and first-generation migrant detection. Journal of Heredity. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM, Lowe AJ. Adaptive evolution in invasive species. Trends in Plant Science. 2008;13:288–294. doi: 10.1016/j.tplants.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Prentis PJ, Sigg DP, Raghu S, Dhileepan K, Pavasovic A, Lowe AJ. Understanding invasion history: genetic structure and diversity of two globally invasive plants and implications for their management. Diversity and Distributions. 2009;15:822–830. [Google Scholar]
- Rousset F. Genepop′007: A complete re-implementation of the Genepop software for Windows and Linux. Molecular Ecology Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- SAS Institute. SAS User's guide, version 8. Cary, NC: SAS Institute Inc; 1999. [Google Scholar]
- Schlaepfer DR, Edwards PJ, Widmer A, Billeter R. Phylogeography of native ploidy levels and invasive tetraploids of Solidago gigantea. Molecular Ecology. 2008;17:5245–5256. doi: 10.1111/j.1365-294X.2008.03980.x. [DOI] [PubMed] [Google Scholar]
- Simberloff D. Confronting introduced species: a form of xenophobia? Biological Invasions. 2003;5:179–192. [Google Scholar]
- Spencer CC, Neigel JE, Leberg PL. Experimental evaluation of the usefulness of microsatellite DNA for detecting demographic bottlenecks. Molecular Ecology. 2000;9:1517–1528. doi: 10.1046/j.1365-294x.2000.01031.x. [DOI] [PubMed] [Google Scholar]
- Starfinger U. Introduction and naturalization of Prunus serotina in Central Europe. In: Brock J, Wade M, Pysek P, Green D, editors. Plant invasions: studies from North America and Europe. Leiden: Backhuys Publishers; 1997. pp. 161–171. [Google Scholar]
- Starfinger U, Kowarik I, Rode M, Schepker H. From desirable ornamental plant to pest to accepted addition to the flora? The perception of an alien tree species through centuries. Biological Invasions. 2003;5:323–335. [Google Scholar]
- Suarez AV, Tsutsui ND. The evolutionary consequences of biological invasions. Molecular Ecology. 2008;17:351–360. doi: 10.1111/j.1365-294X.2007.03456.x. [DOI] [PubMed] [Google Scholar]
- Theoharides KA, Dukes JS. Plant invasion across space and time: factors affecting nonindigenous species success during four stages of invasion. New Phytologist. 2007;176:256–273. doi: 10.1111/j.1469-8137.2007.02207.x. [DOI] [PubMed] [Google Scholar]
- Vanhellemont M, Verheyen K, De Keersmaeker L, Vandekerkhove K, Hermy M. Does Prunus serotina acts as an aggressive invader in areas with a low propagule pressure? Biological Invasions. 2009;11:1451–1462. [Google Scholar]
- Vanhellemont M, Wauters L, Baeten L, et al. Prunus serotina unlashed: invader dominance after 70 years of forest development. Biological Invasions. 2010;12 doi 10.1007/s10530-009-9529-x. [Google Scholar]
- Verheyen K, Vanhellemont M, Stock T, Hermy M. Predicting patterns of invasion by black cherry (Prunus serotina Ehrh.) in Flanders (Belgium) and its impact on the forest understorey community. Diversity and Distributions. 2007;13:487–497. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wilcove DS, Rothstein D, Dubow J, Phillips A, Losos E. Quantifying threats to imperiled species in the United States. BioScience. 1998;48:607–615. [Google Scholar]
- Williams DA, Overholt WA, Cuda JP, Hughes CR. Chloroplast and microsatellite DNA diversities reveal the introduction history of Brazilian peppertree (Schinus terebinthifolius) in Florida. Molecular Ecology. 2005;14:3643–3656. doi: 10.1111/j.1365-294X.2005.02666.x. [DOI] [PubMed] [Google Scholar]
- Wilson JRU, Dormontt EE, Prentis PJ, Lowe AJ, Richardson DM. Something on the way you move: dispersal pathways affect invasion success. Trends in Ecology & Evolution. 2009;24:136–144. doi: 10.1016/j.tree.2008.10.007. [DOI] [PubMed] [Google Scholar]
- With K. The landscape ecology of invasive spread. Conservation Biology. 2002;16:1192–1203. [Google Scholar]