Abstract
The evolutionary origin of human Hepatitis Delta Virus (HDV) replication by RNA-directed transcription is unclear. Here we identify two species of 5′ capped, ∼18-25 nucleotide small RNAs. One was of antigenomic polarity corresponding to the 5′ end of Hepatitis Delta Antigen (HDAg) mRNA and interacted with HDAg and RNA POLYMERASE II (POL II), while the other mapped to a structurally analogous region on the genomic RNA hairpin. An HDAg-interaction screen uncovered MOV10, the human homologue of the A. thaliana RNA amplification factor SDE3 and D. melanogaster RISC-maturation factor Armitage. MOV10 knockdown inhibited HDV replication, but not HDAg mRNA translation supporting a role for MOV10 in RNA-directed transcription. Together, our studies define RNA hairpins as critical elements for the initiation of HDV-related RNA-directed transcription. The identification of capped small RNAs and the involvement of MOV10 in HDV replication further suggest a conserved mechanism related to RNA-directed transcription in lower eukaryotes.
Introduction
RNA-directed transcription is the generation of an RNA transcript from an RNA template and is carried out by RNA-dependent RNA polymerases (RdRPs). RNA-directed transcription is known from RNAi-related amplification of gene silencing in non-vertebrates such as C. elegans, fungi, and plants1. It is also part of the replication cycle of vertebrate RNA viruses. For the majority of viruses, these processes are carried out by virally encoded RdRPs. There are, however, two known exceptions: plant viroids and HDV (for a review of HDV replication, ref. 2).
HDV is the smallest known animal virus and encodes only one protein, the Hepatitis Delta Antigen (HDAg). Since HDAg does not harbor polymerase activity, HDV relies on host RNA polymerases to amplify its RNA genome. This is remarkable since non-viral, cellular RNA-directed transcription is otherwise thought to be absent in vertebrates. This raises the question whether HDV replication uses an independently evolved RNA-directed transcription process, or a more general and ancient capacity for RNA-directed transcription3. In this context, the presence of MOV10/Armitage in the human and fly genomes is notable. MOV10/Armitage are the homologues of A. thaliana SDE3, a putative RNA helicase that had originally been isolated as the first non-RdRP factor required for the amplification step of RNAi in plants4. SDE3 is required for single-strand RNA-induced RNAi, but dispensable for hairpin RNA-induced RNAi. Drosophila Armitage has been shown to act as a maturation factor during RISC assembly5; human MOV10 is associated with Argonaute proteins, but its role in RNA silencing is unclear6.
Infectious HDV particles are packaged with Hepatitis B surface antigen (HBsAg), restricting infection and spread to people with HBV. Once inside a mammalian cell, HDV replication requires only HDV RNA and a source of HDAg2. Following infection, the circular genomic HDV RNA reaches the nucleus, where it becomes the template for rolling-circle replication thus generating multimers of antigenomic HDV RNAs. The multimers are cleaved into monomers by a ribozyme activity in the antigenomic RNA which then circularize by end-ligation. Antigenomic RNA in turn becomes the template for analogous rolling-circle replication thereby yielding more genomic HDV RNAs. Due to >70% intramolecular Watson-Crick base-pair complementarity, both genomic and antigenomic HDV RNAs assume a compact unbranched, rod-like structure. HDAg mRNA is capped and polyadenylated and therefore most likely generated by Pol II. Partly due to the circular nature of full-length antigenomic and genomic HDV RNA, the HDAg mRNA 5′ end is also the only defined 5′ end of an HDV RNA indicative of transcription initiation7-9, while the initiation sites for full-length antigenomic and particularly genomic HDV RNA, have not been determined. Although it had been speculated that Pol I may mediate full-length antigenomic HDV RNA synthesis10, recent nuclear run-on11 and HDV RNA immunoprecipitation12 studies favor a model in which the core Pol II enzyme mediates all HDV RNA synthesis.
The evolutionary origin and molecular details of HDV-related RNA-directed transcription, including its initiation and associated non-POL II host factors, remain largely unknown. We hypothesized that HDV might compensate for its limited coding capacity by utilizing non-coding small RNAs similar to the ones implicated in RNAi-related processes13. Such an RNA may either function in modulating viral or host transcript levels by RNA silencing and/or function in the initiation of RNA-directed transcription, possibly similar to the RdRP-dependent triphosphorylated small RNAs in C. elegans RNA silencing14.
Results
A capped HDV small RNA related to the HDAg mRNA 5′ end
The pre-microRNA-like appearance of the folded genomic and antigenomic HDV RNA hairpin ends (‘pode’ and ‘antipode’) prompted us to probe for small RNAs corresponding to either the top or bottom strands of these hairpins (Fig. 1a,b). In this initial Northern blot screen, we detected an HDV-derived and replication-dependent small RNA with an oligonucleotide probe targeting the bottom strand of the antigenomic pode (Fig. 1b, boxed). This ∼24-25nt RNA was observed in both HDV-replicating Huh-7 (Fig. 1c, lanes 1-3) and 293 (lanes 4-7) cells, regardless of whether HDV replication had been induced by plasmid or RNA transfection (Fig. 1c, lanes 5 and 7). Transfection with a plasmid containing an early nonsense mutation in HDAg and that consequently does not support HDV replication, did not result in such small RNAs (Fig. 1c, lanes 2 and 6). We therefore consider this RNA to be a bona fide HDV small RNA.
Figure 1.
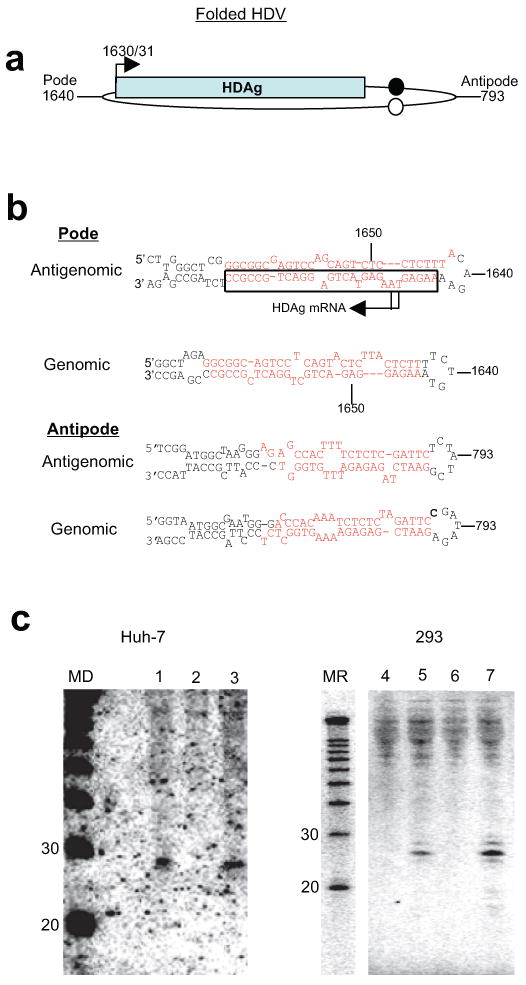
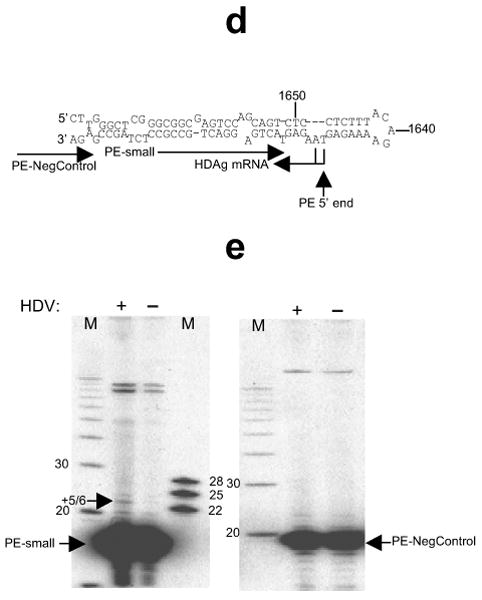
Discovery and mapping of an HDV-derived small RNA. (a-c) Discovery of an HDV small RNA. (a) Schematic of HDV secondary structure (applies to both genomic and antigenomic HDV RNA). HDAg: HDAg mRNA; arrow: HDAg mRNA start site. Circles: antigenomic (closed)/genomic (open) ribozymes; nucleotide numbering according to Kuo et al.29. (b) Predicted secondary structures of both genomic and antigenomic RNA hairpin ends. Top and bottom strands of all four hairpins were screened for corresponding small RNAs. Target sequences for the Northern blot oligonucleotide probes in this screen marked in red, the region from which the small RNA was detected by a black box; reported7-9 HDAg mRNA start sites indicated by arrows. (c) An HDV-derived small RNA from the antigenomic pode (Northern Blot). 1-3 (Huh-7, day 11): 1, DNA induction, wt HDAg; 2, DNA induction, mutant HDAg; 3, DNA induction with construct engineered to express only the small HDAg (note: only the small, but not large isoform of HDAg is required for HDV replication); 4-7 (293, day 6): 4, untransfected; 5, DNA induction, wt HDAg; 6, DNA induction, mutant HDAg; 7: RNA induction. MD: DNA size marker; MR: RNA size marker. (d-e) HDV small RNA 5′ end mapping by primer extension (PE). (d) Primer Extension strategy. Arrows towards the left indicate the HDAg mRNA initiation site(s), the vertical arrow the major 5′ end mapped by PE. The relative positions of the extension primers ‘PE-small’ and ‘PE-NegControl’ are indicated. (e) A largely 5-6nt extension maps the HDV small RNA to position 1630/1. The absence of an HDV-dependent extension with ‘PE-NegControl’ confirms the specificity of the assay with regards to the HDV small RNA. Numbers indicate nucleotide sizes and ‘+(number)’ the extent of the extension; HDV+: DNA induction, wt HDAg; HDV-: DNA induction, mutant HDAg; M: DNA Marker.
To more precisely map the small RNA, we used a primer extension assay specific for detecting the 5′ end of the small RNA, but not that of the co-linear, approximately 800nt HDAg mRNA (Fig. 1d,e). The HDAg mRNA 5′ end is the only established 5′ end of an HDV RNA and had been mapped to positions 1630 and 1631 (refs. 7-9). We therefore used RNA preparations depleted of molecules larger than 200nt (Methods). As a negative control, we chose a primer that should anneal downstream of the predicted HDV small RNA, but that would still anneal to HDAg mRNA. We initially chose an HDV small RNA-directed primer of which the 3′ end was at position 1627, and obtained a 3-4 nucleotide extension that was only seen in the HDV-containing samples (data not shown). Confirming the specificity of these results, a primer shortened by 2 nucleotides at the 3′ end yielded a ∼5-6 nucleotide extension (Fig. 1e, left panel), mapping the 5′ end of the small RNA to around position 1630/31 (Fig. 1d). No HDV-dependent product was seen with the negative control primer, supporting the efficient depletion of HDAg mRNA in the samples (Fig. 1e, right panel). We conclude that the 5′ end of the HDV small RNA coincides with that of HDAg mRNA and is within the evolutionarily conserved RNA secondary structure thought to harbor promoter activity15.
We next determined the nature of the small RNA 5′ and 3′ ends. The 3′ end was studied with the chemical β-elimination assay16. The increased gel mobility following β-elimination is consistent with a 2′,3′-hydroxyl 3′ end similar to miR-15a (Fig. 2a). The 5′ end was investigated by enzymatic means (Fig. 2b,c). As for the 5′-monophosphorylated miR-15a, T4 PNK which adds a phosphate to 5′-hydroxylated RNAs did not affect the mobility of the HDV small RNA, excluding a 5′-OH terminus (Fig. 2b, lane 3). Tobacco Acid Pyrophosphatase (TAP) cleaves various pyrophosphate bonds including those in triphosphorylated and methylguanosine-capped RNAs, leaving a 5′ monophosphate group. A change from triphosphate to monophosphate has a negligible effect on small RNA mobility under comparable conditions14, the loss of a cap, however, should increase its mobility because of the additional loss of the guanosine moiety. Consistent with a cap, the mobility of the HDV small RNA was increased by TAP (Fig. 2b, lane 4), while miR-15a was unchanged. A triphosphorylated end was further excluded by treatment with Antarctic Phosphatase which had no effect on HDV small RNA gel mobility (Fig. 2c, lane 3) and neither did subsequent treatment with PNK (lane 4), while the internal control miR-15a was slightly retarded by Antarctic Phosphatase and restored to its original mobility by PNK. The disappearance of miR-15a and HDV small RNA with T4 RNA Ligase can be explained by ligation of the 3′ hydroxyl ends of the HDV small RNA to a heterogeneous population of 5′ monophosphorylated RNAs in the sample14. Finally, the HDV small RNA proved resistant to Terminator Exonuclease, an enzyme that selectively removes 5′ monophosphorylated RNAs such as miR-15a (Fig. 2b, lane 6). While these data indicated that the HDV small RNA was 5′ capped, we wished to more precisely determine its identity by performing RNA immunoprecipitation with an antibody directed against trimethylguanosine (TMG) and that is known to weakly recognize 7-methylguanosine caps17. Consistent with an mRNA-like monomethylguanosine cap, the TAP-sensitive HDV small RNA precipitation efficiency was intermediate to that of the 5′-phosporylated microRNAs (not recognized) and TMG-capped U5 snRNA (efficiently recognized; Fig. 2d). In conclusion, the 2′-3′ hydroxylation and 5′ cap of the HDV small RNA, in addition to its particular position within the HDV sequence, place the HDV small RNA in a position where it could be involved in HDV transcription initiation (Fig. 2e).
Figure 2.
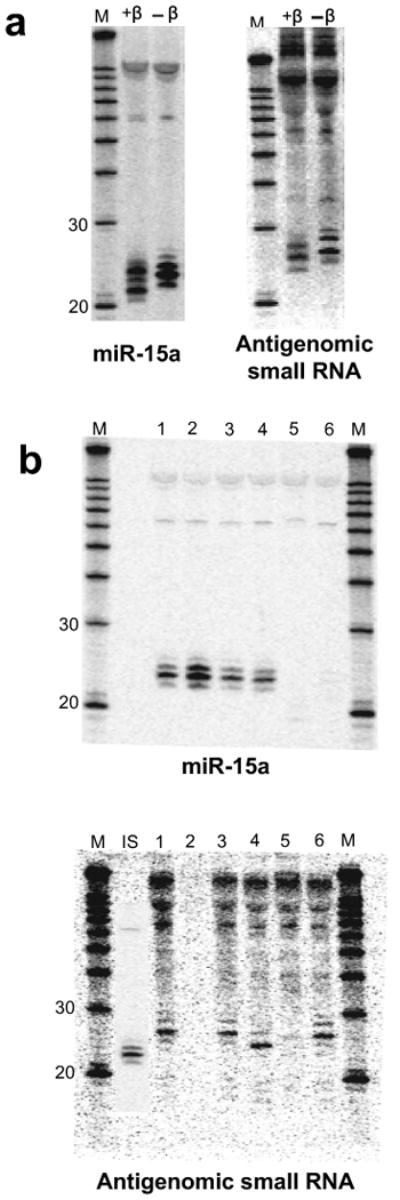
The antigenomic HDV small RNA is 2′-3′ hydroxylated and has an mRNA-like 5′ cap (Northern Blot, 293 cells, RNA induction). (a) 3′ end by β-elimination. The mobility of the HDV small RNA is increased following β-elimination. miR-15a: 2′-3′ hydroxylated positive control; +β: +β-elimination; -β: untreated RNA. (b) 5′ end by enzymatic analysis. 1: mock-treated (+HDV); 2: mock-treated (no HDV); 3: PNK (+HDV); 4: Decapping enzyme (TAP; +HDV); 5: T4 RNA Ligase (+HDV); 6: Terminator Exonuclease (+HDV). The size of the HDV small RNA was estimated to be ∼24nt based on the largely 22nt, 5′ phosphorylated miR15-a shown in the inset (IS). (c) Confirmation that the 5′ end of the HDV small RNA is capped, not triphosphorylated (enlarged image to emphasize changes in gel mobility for miR-15a, but not HDV small RNA). 1: mock-treated (+HDV); 2: mock-treated (no HDV); 3: Antarctic Phosphatase (+HDV); 4: Antarctic Phosphatase followed by T4 PNK (+HDV). (d) RNA immunoprecipitation with anti-2,2,7-trimethylguanosine antibody K121. The immunoprecipitation efficiency of the HDV small RNA, U5 snRNA (positive control) and microRNAs miR-15a and let-7a (negative controls) was analysed by Northern blot. ‘S’: supernatant; ‘I’: IP fraction. (e) Predicted structure of the HDV small RNA. The various RNAs in a-d were detected after stripping and rehybridisation to the same blot. M: RNA marker.
Sequencing uncovers a corresponding genomic small RNA
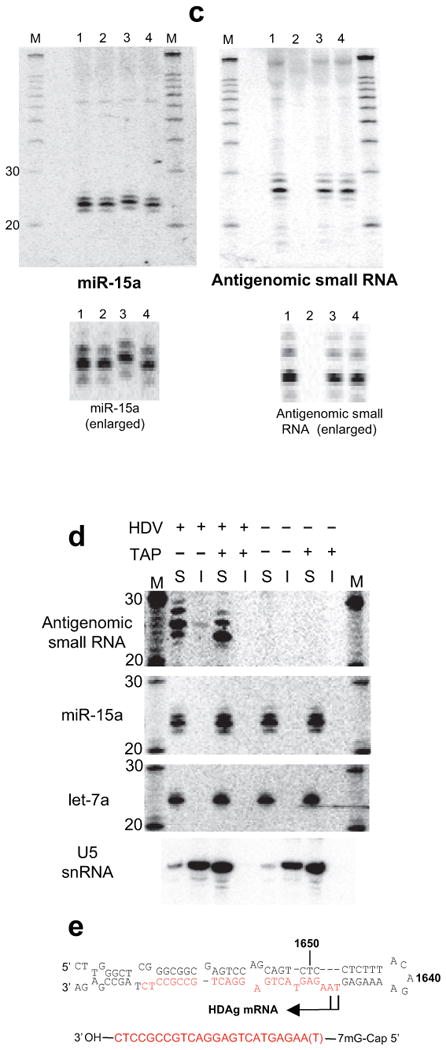
As part of efforts to uncover structurally related endogenous small RNAs (unpublished data), we sequenced small RNAs, including those from HDV replicating cells (Supplementary Table 1 online). Importantly, five of the seven antigenomic HDV RNAs recovered following a protocol designed to enrich for capped small RNAs and which consequently was largely depleted of microRNAs (Methods) corresponded to the antigenomic small RNA (Fig. 3a; Supplementary Table 1 online). The sequence further supports that the small RNA starts at around position 1630 with a non-templated terminal nucleotide addition accounting for the extra nucleotide in the primer extension7. Interestingly, a second distinct HDV small RNA was identified, in this case of genomic polarity (Supplementary Table 1 online), and was subsequently confirmed by Northern Blot (Fig. 3b). Strikingly, this small RNA falls into the same pode hairpin as the antigenomic small RNA (Fig. 3a). Although the enzymatic 5′ modification analysis indicated that the genomic small RNA was predominantly capped (Fig. 3c), a 20-21 nucleotide species was cloned without prior selection for capped RNAs (Supplementary Table 1 online), while the 18-19 nucleotide species was detected in the 5′ cap-enriched fraction. A minor 5′ phosphorylated subpopulation would also be consistent with the partially persistent signal from the most slowly migrating genomic small RNA species following decapping (Fig. 3c, lane 6). Additional dispersed HDV small RNAs of both genomic and antigenomic polarity were also cloned. These were obtained particularly following the 5′ ligation-independent cloning method14, which would also pick up 5′-hydroxylated RNAs, and likely represent RNA turnover intermediates.
Figure 3.
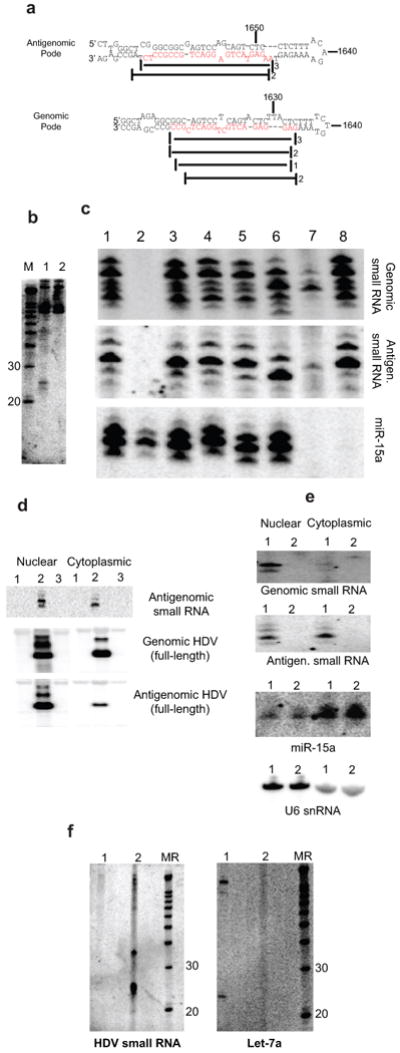
Cloning and characterization of an HDV small RNA of genomic polarity. (a) Relative location and cloning frequency of sequenced HDV small RNAs derived from the genomic and antigenomic pode hairpins (main species highlighted in red). (b) Detection of genomic HDV small RNA by Northern Blot (293 cells, day 5). 1: DNA induction, wt HDAg; 2: DNA induction, mutant HDAg. (c) Enzymatic analysis of genomic small RNA 5′ end. 1: mock-treated (+HDV); 2: mock-treated (no HDV); 3: PNK (+HDV); 4: Antarctic Phosphatase (+HDV); 5: Antarctic Phosphatase followed by T4 PNK (+HDV); 6: Decapping enzyme (TAP; +HDV); 7: T4 RNA Ligase (+HDV); 8: Terminator Exonuclease (+HDV). Note that unlike the antigenomic small RNA, a minor fraction of the genomic small RNA does not appear to be shifted following TAP treatment. (d-f) Localization of the HDV small RNAs. (d) Nuclear-cytoplasmic fractionation of antigenomic HDV small RNA (polyacrylamide gel) and full-length antigenomic and genomic HDV RNA (denaturing agarose gel). The main species in the full-length genomic/antigenomic RNA blot corresponds to the monomer, the higher molecular weight species to dimer, trimer etc. 1: DNA induction, mutant HDAg; 2: DNA induction, wt HDAg; 3: untransfected. (e) Genomic small RNA is restricted to the nucleus (nuclear-cytoplasmic fractionation). 1: DNA induction, wt HDAg; 2: DNA induction, mutant HDAg. miR-15a and U6 snRNA chosen as largely cytoplasmic and nuclear RNA controls, respectively. (f) The HDV small RNA can be found in the HDV virion. 1: RNA induction (same RNA as in Fig. 2c); 2: virion RNA isolated from tissue culture media (∼1.25×109 particles). MR: RNA Marker. The various RNAs in c-f were detected after stripping and re-hybridization to the same blot.
Co-localization of complementary small and full-length RNAs
We then investigated the cellular localization of the HDV small RNAs (Fig. 3d-f). As previously reported, and reflecting export of the genomic HDV-RNP to the cytoplasm for packaging with HDAg and HBsAg into the HDV virion during an infection cycle, nuclear-cytoplasmic fractionation recovered the full-length genomic RNA, particularly the mature monomer, in both the nuclear and cytoplasmic fractions (Fig. 3d). Full-length antigenomic RNA, however, was largely enriched in the nucleus, consistent with it largely serving as a nuclear replication intermediate and further indicates that the cytoplasmic fraction was reasonably free from nuclear RNA contaminants. Accordingly, control miR-15a and U6 snRNA were predominantly detected in the cytoplasmic and nuclear fractions, respectively (Fig. 3e). Strikingly, and in marked contrast to their full-length counterparts, it was the HDV small RNA of antigenomic polarity that was found in both compartments, while the genomic small RNA was recovered only in the nuclear fraction. The co-localization of antigenomic small RNA with genomic full-length RNA and vice versa, extended to the virions, where only antigenomic, but neither the genomic small RNA nor the ubiquitously expressed let-7 microRNA was readily detected (Fig.3f and data not shown). While the exact stoichiometry and biological importance of this localization remain to be determined, the ease of detection by Northern blot and preliminary quantitation analysis (data not shown) are consistent with the notion that a substantial fraction of HDV virions may contain the small RNA.
Mov10 interacts with HDAg and facilitates replication
Given their size and location within a microRNA-like precursor hairpin, we were interested in whether the HDV small RNAs exhibited silencing activity. Dual luciferase assays with reporters carrying target sequences complementary to the HDV small RNAs in their 3′ UTR (Supplementary Fig. 1 online and data not shown), however, did not show an HDV replication-dependent decrease in luminescence as such an activity would have predicted. This is consistent with the HDV small RNAs being largely 5′ capped, a modification predicted to interfere with small RNA silencing18. An HDAg-interaction screen (HDAg immunoprecipitation followed by mass-spectrometric identification of interacting proteins; Cao, Haussecker, Kay, unpublished data) identified the human homologue of the A. thaliana SDE3, MOV10 (ref. 4) both in the presence and absence of HDV replication (see also Fig. 4a). Supporting a function in HDV replication, MOV10 knockdown using a pool of MOV10-directed siRNAs inhibited viral replication (Fig. 4b, lane 12). The specificity of this effect was confirmed by the fact that not only did the siRNA pool suppress replication, but also three out of the four individual MOV10 siRNAs tested (lanes 13-16), with the non-inhibitory siRNA (lane 13) exhibiting the least MOV10 knockdown by qRT-PCR (Fig. 4b). It was conceivable that MOV10 knockdown might inhibit HDV by interfering specifically with the transcription or translation of HDAg. This was of particular interest, since MOV10 has been implicated in microRNA-related post-transcriptional regulation19. To address this, the MOV10 inhibition experiment was repeated in cells stably expressing HDAg and which therefore complement HDAg-mutant strains in trans (data not shown). MOV10 knockdown was still sufficient to inhibit HDV replication in this cell line (Fig. 4b, lanes 18-20) with no obvious effect on HDAg protein levels (Fig. 4c). These results therefore suggest a role for MOV10 in HDV replication. We next asked whether additional RNAi-related genes were required for HDV replication by targeting the primary microRNA processing factors DGCR8 and DROSHA, the microRNA export factor EXPORTIN-5, the pre-microRNA processing enzyme DICER and co-factor TRBP, plus the small RNA effector proteins, ARGONAUTE (AGO) 1-4. In agreement with the observed lack of silencing activity of the HDV small RNAs, all of these knockdown treatments, except for one, had no appreciable effect on HDV replication, although we formally cannot rule out insufficient knockdown of these factors. The exception was AGO4 where all 4 individual siRNAs and the siRNA pool directed against AGO4 inhibited HDV replication (Fig. 4d). To our knowledge, this is the first process for which a genetic requirement of AGO4 has been identified. Since both Argonaute proteins and MOV10 have been placed at the downstream stages of microRNA silencing, it is possible that these factors function together in HDV replication.
Figure 4.
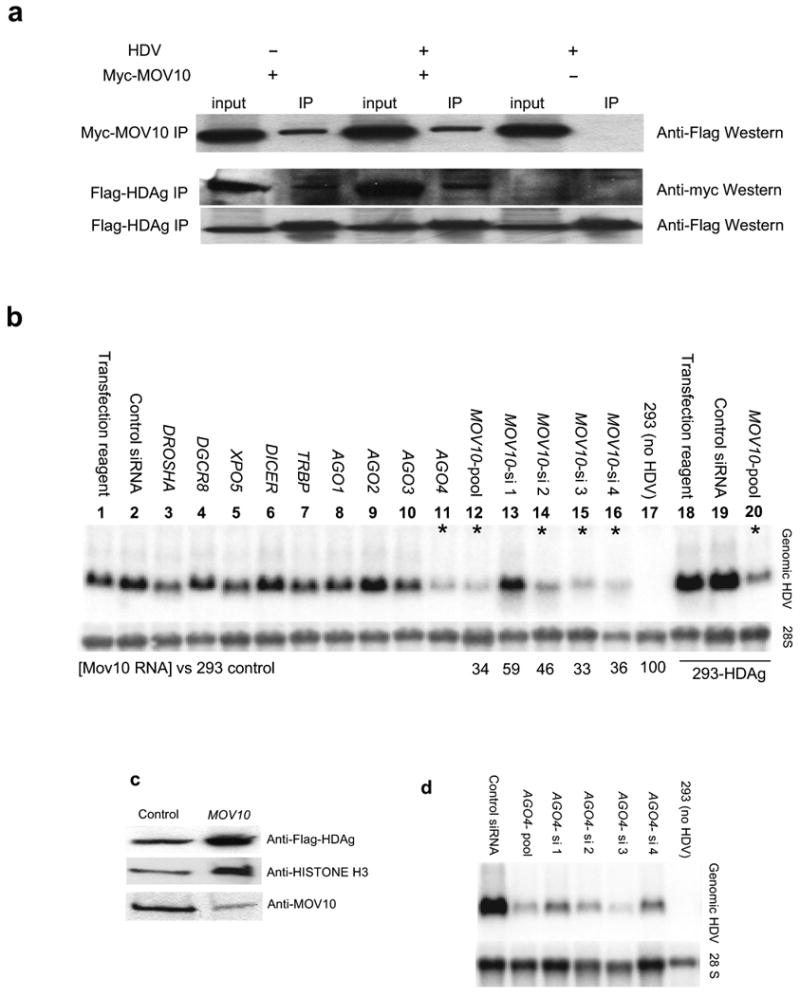
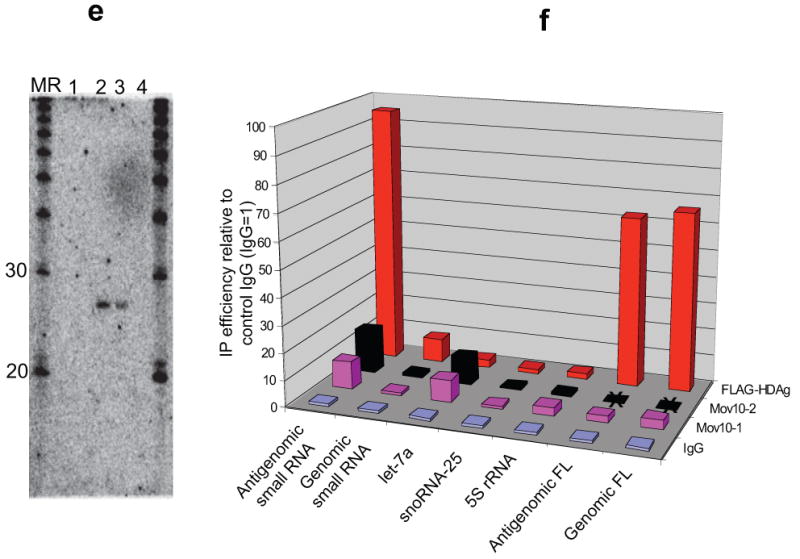
Role for MOV10 in HDV replication. (a) Confirmation of HDAg-interaction with MOV10 (immunoprecipitation analyzed by Western Blot). Flag-HDAg co-precipitated with myc-tagged MOV10, both in the presence and absence of HDV replication, as did myc-MOV10 following Flag-HDAg IP. Due to considerable cross-reactivity with myc-IgG, detection of myc-MOV10 following myc-MOV10 immunoprecipitation was technically not feasible; IP: immunoprecipitation; Myc-MOV10 +/-: with/without co-transfected myc-tagged MOV10; HDV +/-: protein extract from cells expressing Flag-HDAg in the presence or absence of HDV replication. HDV replication of the HDAg-mutant virus was dependent on Flag-HDAg (data not shown). (b) MOV10 knockdown inhibits HDV replication (Northern Blot). MOV10 knockdown with a pool (lane 12) as well as 3 out of 4 individual siRNAs (lanes 14-16) against MOV10 inhibited HDV replication, with the non-inhibitory siRNA (lane 13) showing least effective MOV10 knockdown as determined by qRT-PCR (3 days following siRNA transfection). Targeting of other RNAi-related proteins with pools of siRNAs did not appreciably affect HDV replication, except for AGO4 (lane 11). Viral inhibition by MOV10 knockdown was also observed in cells stably expressing HDAg (lanes 18-20); *: >60% HDV inhibition. (c) Flag-HDAg abundance in the stable cell line is not appreciably altered by MOV10 knockdown (Western Blot); control/MOV10: control/MOV10 siRNA pool. (d) Confirmation of AGO4 knockdown effect on HDV replication using individual AGO4 siRNAs. (e-f) RNA immunoprecipitation. HDV RNA-containing lysate was incubated with rabbit IgG (IgG), two different MOV10 antibodies (MOV10-1, MOV10-2; only MOV10-1 was used for full-length HDV analysis), or antibodies for POL II and Flag and the recovered RNA analysed by Northern blot (e) and stem-loop qRT-PCR (f). 1: rabbit IgG; 2: anti Flag (HDAg); 3: anti POL II; 4: anti MOV10; MR: RNA marker. Average from at least two independent experiments; FL: full-length; X= not analyzed.
Interaction between small RNAs and replication factors
Considering the interaction of MOV10 with HDAg, the replication defect in MOV10 knockdown cells, and the association of its plant homologue with RNAi-related RNA-directed transcription, we were interested in whether Mov10 also interacted with the HDV small RNAs. Northern blotting following immunoprecipitation with MOV10 antibodies did not detect the HDV small RNAs. Employing, however, the more sensitive stem-loop qRT-PCR technique for the detection of small RNAs20, the both of the MOV10 antibodies modestly enriched for the antigenomic small RNA (Fig. 4e,f) to a similar degree as for let-7 microRNA, and may be consistent with an indirect interaction. By contrast, the antigenomic small RNA was readily detected following HDAg immunoprecipitation not only by qRT-PCR, but also by Northern blot (Fig. 4e), strongly supporting it to be part of functional HDV RNP complexes. Similarly, although not as consistent as for HDAg, the antigenomic small RNA could be immunoprecipitated with an antibody against POL II supporting a close involvement in active RNA-directed transcription. The variability in the Pol II interaction could be due to a more transient interaction that is sensitive to the particular stage of viral replication at the time of extract preparation. Little association was detected for HDAg and the genomic small RNA, and none above background between MOV10 and POL II with the genomic small RNA (Fig. 4f and data not shown).
Discussion
Our studies have uncovered a new type of capped small RNAs. We consider it highly unlikely that these represent merely non-functional turnover intermediates of larger HDV RNA because: a) they map to corresponding genomic and antigenomic hairpin structures reported to harbor HDV promoter activity15; b) they exhibit a distinct cellular distribution that mirrors that of their complementary full-length counterparts, possibly mediated by base-pairing; c) in the case of the antigenomic small RNA, the association with HDAg and POL II; and d) abortive transcription initiation without further functional relevance seems at odds with the antigenomic small RNA being of similar abundance in the cytoplasm as in the nucleus (Fig. 3d). Based on qRT-PCR analyses (data not shown), there was approximately one antigenomic small RNA for every five full-length antigenomic RNAs per cell (∼63,000 antigenomic small RNAs, ∼326,000 antigenomic full-length copies per cell 3 days post replication initiation in 293 cells). This is a rather high ratio considering the stability of circular full-length HDV RNA21. There were relatively few genomic small RNAs, ∼6,400 per cell in contrast to very abundant full-length genomic RNAs (∼1.68 × 106 copies per cell). The more heterogeneous nature of the genomic small RNA as well as its limited enrichment following HDAg immunoprecipitation (Fig. 4e) could mean that it functions, consistent with its absence outside of the nucleus, as a more transient transcription initiation intermediate.
Since post-transcriptional capping of Pol II transcripts is unknown, the capped HDV small RNAs likely reflect RNA-directed transcription initiation. Notably, the 5′ ends of the antigenomic small RNA and HDAg mRNA coincide, and both the antigenomic and genomic small RNAs fall within equivalent hairpin structures. Consequently, a single mode of transcription initiation determined by hairpin structures and marked by the presence of capped small RNAs, may account for essentially all HDV RNAs present during an infection and suggests that antigenomic HDAg mRNA and full-length HDV RNA share a common transcription initiation site. In this model, HDAg polyadenylation would be incompletely coupled to transcription termination, with POL II continuing downstream to generate full-length antigenomic HDV RNA by rolling-circle transcription. Efforts, including our own (data not shown), to determine the 5′ end of full-length genomic RNA have failed. One explanation for this may be the predominantly circular nature of the genomic RNA at steady-state and the challenge of analyzing highly structured RNA. It may also relate to our finding that while the 18-19 nucleotide species was recovered in the 5′ cap-enriched fraction only, the 20-21 nucleotide species, which represent 2-3 nucleotide 3′ extensions, must have been 5′ phosphorylated in order to be cloned (Supplementary Table 1 online). Cap-dependent 5′ RACE analyses would have therefore failed to uncover the genomic transcription initiation site if the nascent 5′ capped genomic RNA were to be converted into a 5′ phosphorylated transcription initiation intermediate. The 5′ capped small RNA therefore represents the best evidence so far for the elusive genomic HDV RNA transcription initiation site.
POL II had previously been shown in vitro to bind to the HDV RNA hairpins12. Another study reported in vitro HDV RNA template-dependent transcription by POL II which was more dependent on the hairpin structure than the primary sequence22. In that study, the hairpin was cleaved next to a bulge proximal to the loop with the newly generated 3′ end serving as a primer for Pol II transcription thus resulting in an RNA with both genomic and antigenomic sequence. As this activity, in the absence of HDAg, occurred on the antipode, not the pode hairpin which is thought to harbor the HDV promoter15 and from which we detected the capped small RNAs, hairpins may have a general ability to direct the initiation of RNA-templated transcription, and additional factors, such as HDAg and interacting proteins determine the fidelity of transcription initiation site selection during HDV replication. We note that by RT-PCR and sequencing we could detect a similar antigenomic pode-genomic RNA hybrid from the pode region (Supplementary Fig.2 online). This hybrid co-immunoprecipitated with POL II and was the only hybrid that we detected among the four possible HDV hairpin combinations tested. Nevertheless, since nicked RNA cannot serve as a template for rolling-circle replication and efforts to visualize the hybrid by means other than PCR failed, cleavage-extension may be an alternative mechanism for HDV transcription initiation, but probably does not account for the majority of HDV transcription.
We propose a model for HDV replication (Supplementary Fig. 3 online) in which following infection, the incoming viral RNA encounters the largely cytoplasmic RNA helicase MOV10 which re-models the HDV RNA into a transcription initiation-competent RNP. Once in the nucleus, POL II binds to the pode hairpin and initiates antigenomic HDV RNA synthesis. Given that primed, but not unprimed RNA-directed transcription initiation is readily observed in vitro22,23, it is possible that an annealed small RNA would facilitate this process by serving as a small priming RNA (spRNA). This could increase the odds for a successful viral infection, particularly since early HDAg mRNA transcription is thought to be critical for replication24.
The HDV small RNAs themselves are likely generated by de novo transcription initiation and subsequent pausing, possibly regulated by the loop. While a fraction of the paused complexes would get elongated, some may be exported into the cytoplasm either for viral packaging and/or MOV10-mediated remodeling for subsequent rounds of replication. This would be consistent with the experimental HDV replication system that we have used which does not include a bona fide viral infection step and might explain the nuclear-cytoplasmic shuttling of HDV RNP particles25. The exact nature of the relationship between MOV10 and plant SDE3 with RNA-directed transcription in general and the associated small RNAs in particular remain to be determined.
It has been debated whether HDV-related RNA-directed transcription by a human RNA polymerase evolved independently, or whether it taps into an ancient, either dormant or still active capacity for RNA-directed transcription harking back to an ‘RNA World’. Our findings of capped HDV small RNAs marking the initiation sites of HDV transcription, the requirement for the SDE3-related MOV10, and the observation that RNA-directed transcription by (S. cerevisiae) Pol II in vitro is possible even in the absence of HDAg and solely dependent on the template structure23, suggests that HDV might indeed harness such a conserved mechanism. Interestingly, the RdRP-dependent secondary small RNAs in C. elegans RNAi amplification were 5′ triphosphorylated14 and very likely generated by non-processive de novo transcription initiation26. The use of Pol II rather than a dedicated RdRP may be due to co-evolution with the response of the mammalian innate immune system to dsRNA and triphosphorylated 5′ ends. These are recognized as foreign and trigger innate immune responses27.
The existence of so-called mirror-spliced antisense transcripts (MSATs) is consistent with the notion that non-viral RNA-directed transcription occurs in mammalian cells28. MSATs are transcripts that are the reverse complement of spliced mRNA and are best explained as having been generated by RNA-directed transcription from their corresponding mRNA. We propose that HDV may be a suitable model system for such RNA-directed transcription studies. Moreover, the discovery of non-viral capped small RNAs that can be mapped to hairpin structures and transcripts sensitive to MOV10 inhibition may offer an opportunity to identify related spRNAs and RNA-directed transcription in mammals.
Methods
Tissue culture
To induce HDV replication by DNA transfection, we transfected Huh-7 and 293 cells with plasmid pCMV3DCHDV×1_2ag, or variants containing either a replication-deficient HDAg mutant or a construct capable of only expressing the small isoform of HDAg. For RNA-induced replication, RNA from T7 polymerase in vitro transcription off BamHI-linearised pCMV3DCHDV×1_2ag and HDAg mRNA generated with the mMessage mMachine kit (Ambion) was transfected. The template for the HDAg mRNA was obtained by XbaI linearization of pcDNA3 into which the HDAg ORF from pCMV3DCHDV×1_2ag was cloned downstream of the T7 promoter. RNA was harvested 3-6 (293) or 11 (Huh) days after transfection with Trizol reagent (Gibco) for total RNA, or the mirVana kit (Ambion) for low-molecular weight RNA. Medium containing 5×10e8 HDV particles/ml was kindly provided by R. Lanford (Southwest Foundation for Biomedical Research, San Antonio)30. 2.5ml of medium was cleared by 2min centrifugation at 4000×g and 4°C and RNA isolated by adding 22.5ml Trizol. The ratio of the amount of small RNAs in the virion versus the cellular RNA samples was 10 to 100-fold higher for the antigenomic small RNA compared to control cellular 5S rRNA, let-7 microRNA, and sno25 RNA (1.2 versus 0.142, 0.019, and 0.037, respectively.
RNAi
293 cells plated in 24-wells were knocked down with 12 (pools)-48nM (individual) siRNA (Dharmacon). 24 hours later, HDV replication was induced by transfecting cells with 0.4μg of pCMV3DCHDV×1_2ag. Total RNA was harvested 48 hours thereafter with Trizol (Invitrogen) and analyzed by Northern Blot. Actin-normalized Mov10 RNA knockdown was assessed from the same RNA by qRT-PCR as described31. For primers and siRNA sequences, see Supplementary Methods.
Northern blot
For the detection of small RNAs, mirVana (Ambion) low-molecular weight RNA was separated by 15-20% (v/v) urea-polyacrylamide gel electrophoresis, transferred onto Hybond-N (Amersham) nitrocellulose, and hybridized to T4 PNK end-labeled oligo probes (see Supplementary Methods) at 32°C with PerfectHyb Plus (Sigma). Blots were washed and exposed to phosphorimager. For the detection of full-length HDV RNAs, Trizol total RNA was separated on 1% (w/v) denaturing formaldehyde-agarose gels, transferred onto Hybond-N (Amersham) nitrocellulose, and hybridized to either alpha-UTP labeled, T7 polymerase transcribed RNA from BamHI-linearized pCMV3DCHDV×1_2ag for the detection of genomic HDV RNA, or T4 PNK (NEB) end-labeled ggcggcagtcctcagtactcttactctt for the detection of antigenomic HDV RNA.
Primer extension
The primer extension oligo was end-labeled with T4 PNK (NEB) and γ-32P ATP (PerkinElmer). For the extension, Superscript II Reverse Transcriptase (SS2RT, Invitrogen) was employed. mirVana (Ambion) low molecular weight RNA was annealed to the end-labeled oligo primer. Following the addition of buffer, dNTP, and rRNasin (Promega), the reaction was incubated at 42°C for 5 minutes at which point 2μl Superscript II Reverse Transcriptase (SS2RT, Invitrogen) was added for 90 minutes. The reaction was terminated by heating and run out on a 20% polyacrylamide gel. PE-small: gcggcagtcctcagta; PE-NegControl: gactcggaccggctcatct.
Analysis of small RNA 3′ and 5′ ends
β-elimination was performed essentially as described16. Enzyme treatments were performed by heat-denaturing 10μg mirVana (Ambion) RNA followed by the addition of enzyme buffer, rRNasin (Promega; except for the Terminator Exonuclease treatment) and finally enzyme. 100μl reactions (Tobacco Acid Pyrophosphatase: 40μl) were incubated at 37°C (Terminator Exonuclease: 30°C) for 45 minutes (Terminator Exonuclease: 1 hour), acid phenol/chloroform extracted, ethanol precipitated, and analyzed by Northern blot.
RNA immunoprecipitation
For the immunoprecipitation of capped RNAs, 20μl of K121 beads were incubated with 10μg of low-molecular weight RNA by rotation in IP buffer [0.01% (w/v) SDS, 1% (v/v) Triton-X-100, 1.2mM EDTA pH8.0, 16.7mM Tris-HCl PH8.0, 167mM NaCl]. The beads were washed and the eluted RNA analyzed by Northern blot. For the detection of the antigenomic-genomic hybrid RNA, small RNA, protein-protein immunoprecipitations, and MOV10 RNAi, a stable 293 cell line was established expressing functional Flag-tagged HDAg (Cao, Haussecker, Kay, unpublished data). HDV replication was induced by transfecting these cells with the HDAg mutant version of pCMV3DCHDV×1_2ag or T7 in vitro transcribed RNA from the same BamHI-linearized plasmid. Lysate was prepared as described32, and 5μg antibodies added (see Supplementary Methods).
For the detection of antigenomic-genomic hybrid RNA, realtime RT-PCR was performed as described31. Quantitation of small RNAs was done by stem-loop qRT-PCR20. To test the specificity of the assay for the detection of the small RNA, but not other HDV RNA species of the same polarity we a) confirmed the HDV replication-dependent cDNA product by sequencing; and b) included a forward PCR primer designed to anneal upstream of the small RNA 5′ end, which did not yield a PCR product showing that the RT-PCR was not amplifying a longer RNA sharing the same 3′ end with the HDV small RNA. For primers, see Supplementary Methods.
Small RNA quantitation
3 days following induction of 293 cells with pCMV3DCHDV×1_2ag, total RNA was isolated and small RNA quantified by qRT-PCR as described in the preceding section (‘RNA immunoprecipitation’) except that to generate an absolute standard curve, pre-determined amounts of in vitro transcribed HDV small RNAs were added to untreated 293 RNA for RT-PCR.
Nuclear-cytoplasmic RNA fractionation
Cells scraped from a 10cm dish were first pelleted, resuspended in 500μl lysis buffer (0.14M NaCl, 1.5mM MgCl2, 10mM TrisHCl pH 7.5, 0.5% [v/v] NP40) and left on ice for 10 minutes. Nuclei were gently pelleted with 3000×g for 2 minutes at 4°C. For cytoplasmic RNA, the supernatant was underlayered with an equal volume of lysis buffer containing 12% (w/v) sucrose and spun at 15,800×g for 10 minutes. The supernatant (cytoplasmic fraction) was then phenol/chloroform extracted and ethanol precipitated. For nuclear RNA, nuclei were resuspended in 1ml of lysis buffer, gently spun down at 3000×g for 2minutes, and the RNA extracted from the pellet with Trizol.
Small RNA sequencing
mirVana (Ambion) low-molecular weight RNA was adapted as described in Lau et al.33 for the 5′phosphate-dependent cloning, and Pak and Fire14 for the 5′ligation-independent cloning. To preferentially clone capped small RNAs, the RNA was first treated with Antarctic Phosphatase and then Tobacco Acid Pyrophosphatase before 5′-phosphate-dependent cloning. Samples were cloned as part of a barcoded pool and sequenced using the 454/Roche GS20 sequencing platform34.
Western blot
Western Blot was performed according to standard protocols. Antibodies: mAb Flag M2 (Sigma, A8592); rabbit MOV10-2 (Protein Tech Group, 10370-1-AP); rabbit HISTONE H3 dimethyl K4 (Abcam ab7766).
Dual luciferase assay
The Dual-Luciferase® Reporter (DLR™) Assay (Promega) was performed according to the manufacturer's instructions, with modifications as described in the Supplementary Methods.
Supplementary Material
Acknowledgments
We thank J. Glenn (Stanford University) for reagents and R. Lanford (Southwest Foundation for Biomedical Research, San Antonio) for HDV virions. This work was supported by grants DK78424 and AI71068 from the National Institutes of Health (to M.A.K.), a Helen Hay Whitney Foundation Postdoctoral Research Fellowship to D.C., and a Stanford Dean's Postdoctoral Fellowship to D.H.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
For detailed methods, see Supplementary Methods.
References
- 1.Wassenegger M, Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Taylor JM. Structure and replication of hepatitis delta virus RNA. Curr Top Microbiol Immunol. 2006;307:1–23. doi: 10.1007/3-540-29802-9_1. [DOI] [PubMed] [Google Scholar]
- 3.Macnaughton TB, Lai MM. HDV RNA replication: ancient relic or primer. Curr Top Microbiol Immunol. 2006;307:25–45. doi: 10.1007/3-540-29802-9_2. [DOI] [PubMed] [Google Scholar]
- 4.Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–41. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 6.Meister G, et al. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh SY, Chao M, Coates L, Taylor J. Hepatitis delta virus genome replication: a polyadenylated mRNA for delta antigen. J Virol. 1990;64:3192–3198. doi: 10.1128/jvi.64.7.3192-3198.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Modahl LE, Lai MM. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and replication. J Virol. 1998;72:5449–5456. doi: 10.1128/jvi.72.7.5449-5456.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudima S, Dingle K, Wu TT, Moraleda G, Taylor J. Characterization of the 5′ ends for polyadenylated RNAs synthesized during the replication of hepatitis delta virus. J Virol. 1999;73:6533–6539. doi: 10.1128/jvi.73.8.6533-6539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macnaughton TB, Shi ST, Modahl LE, Lai MM. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J Virol. 2002;76:3920–3927. doi: 10.1128/JVI.76.8.3920-3927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang J, Nie X, Chang HE, Han Z, Taylor J. Transcription of Hepatitis Delta Virus RNA by RNA Polymerase II. J Virol. 2007;82:1118–1127. doi: 10.1128/JVI.01758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greco-Stewart VS, Miron P, Abrahem A, Pelchat M. The human RNA polymerase II interacts with the terminal stem-loop regions of the hepatitis delta virus RNA genome. Virol. 2006;357:68–78. doi: 10.1016/j.virol.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nature Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 14.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 15.Beard MR, MacNaughton TB, Gowans EB. Identification and characterization of a hepatitis delta virus RNA transcriptional promoter. J Virol. 1996;70:4986–4995. doi: 10.1128/jvi.70.8.4986-4995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutvagner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 17.Lee CY, Lee A, Chanfreau G. The roles of endonucleolytic cleavage and exonucleolytic digestion in the 5′-end processing of S. cerevisiae box C/D snoRNAs. RNA. 2003;9:1362–1370. doi: 10.1261/rna.5126203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nykaenen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 19.Chendrimada TP, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 20.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Modahl LE, MacNaughton TB, Zhu N, Johnson DL, Lai MM. RNA-dependent replication and transcription of hepatitis delta virus RNA involve distinct cellular RNA polymerases. Mol Cell Biol. 2000;20:6030–6039. doi: 10.1128/mcb.20.16.6030-6039.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filipovska J, Konarska MM. Specific HDV RNA-templated transcription by Pol II in vitro. RNA. 2000;6:41–54. doi: 10.1017/s1355838200991167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehmann E, Brueckner F, Cramer P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature. 2007;450:445–459. doi: 10.1038/nature06290. [DOI] [PubMed] [Google Scholar]
- 24.Sato S, Cornillez-Ty C, Lazinski DW. By inhibiting replication, the large hepatitis delta antigen can indirectly regulate amber/W editing and its own expression. J Virol. 2004;78:8120–8134. doi: 10.1128/JVI.78.15.8120-8134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavanez JP, et al. Hepatitis delta virus ribonucleoproteins shuttle between the nucleus and the cytoplasm. RNA. 2002;8:637–646. doi: 10.1017/s1355838202026432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 28.Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 29.Kuo MY, et al. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and applications. J Virol. 1988;62:1885–1861. doi: 10.1128/jvi.62.6.1855-1861.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sureau C, Guerra B, Lanford RE. Role of the large hepatitis B virus envelope protein in infectivity of the hepatitis delta virion. J Virol. 1993;67:366–372. doi: 10.1128/jvi.67.1.366-372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haussecker D, Proudfoot NJ. Dicer-dependent turnover of intergenic transcripts from the human beta-globin gene cluster. Mol Cell Biol. 2005;25:9724–9733. doi: 10.1128/MCB.25.21.9724-9733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol Biol. 2006;342:33–47. doi: 10.1385/1-59745-123-1:33. [DOI] [PubMed] [Google Scholar]
- 33.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 34.Parameswaran P, et al. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 2007;35:e130. doi: 10.1093/nar/gkm760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


