Abstract
Purpose
To replicate the previous association of single nucleotide polymorphisms (SNPs) with risk of intracranial aneurysm (IA) and to examine the relationship of smoking with these variants and the risk of IA.
Methods
White probands with an IA from families with multiple affected members were identified by 26 clinical centers located throughout North America, New Zealand, and Australia. White controls free of stroke and IA were selected by random digit dialing from the Greater Cincinnati population. SNPs previously associated with IA on chromosome 2, 8, and 9 were genotyped using a TaqMan assay or were included in the Affymetrix 6.0 array that was part of a genome-wide association study of 406 IA cases and 392 controls. Logistic regression modeling tested whether the association of replicated SNPs with IA was modulated by smoking.
Results
The strongest evidence of association with IA was found with the 8q SNP rs10958409 (genotypic P = 9.2 × 10-5; allelic P = 1.3 × 10-5; OR = 1.86, 95% CI: 1.40−2.47). We also replicated association with both SNPs on chromosome 9p, rs1333040 and rs10757278, but were not able to replicate the previously reported association of the two SNPs on chromosome 2q. Statistical testing showed a multiplicative relationship between the risk alleles and smoking with regard to the risk of IA.
Conclusion
Our data provide complementary evidence that the variants on chromosome 8q and 9p are associated with IA and that the risk of IA in patients with these variants are greatly increased with cigarette smoking.
Keywords: Intracranial aneurysm, genome-wide association studies, familial, smoking
Introduction
Recent genome-wide association studies (GWAS) have identified several common sequence variants on chromosome 9p21 that are associated with myocardial infarction, coronary artery disease, abdominal aortic aneurysms, and intracranial aneurysms (IA).1-6 In addition, variants on chromosome 2q33 and 8q11 have been associated with IA in distinct populations that attain genome wide-significance.1 Smoking is the most powerful environmental risk factor for ruptured and unruptured IA, with 70-80% of patients reporting a past history of smoking, similar to the strength of the relationship between smoking and lung cancer.7-12 The relationship between sequence variants associated with the risk of IA and smoking has yet to be explored.
We sought to replicate the association of six variants identified in the aforementioned studies with IA in an independent case-control sample from the Familial Intracranial Aneurysm Study (FIA; www.FIAStudy.com) as well to examine the relationship of smoking with these variants and risk of IA.
Methods
Probands with an IA were identified by 26 clinical centers (41 recruitment sites) located throughout North America, New Zealand, and Australia. To be eligible for inclusion in the FIA study,13-15 the proband was required to have additional family members who also had an IA. Exclusion criteria were employed to eliminate subjects who had an IA due to a known genetic cause, such as Ehlers Danlos or polycystic kidney disease, or as a secondary phenotype, such as an association with an arteriovenous malformation. One case was selected from each of the Caucasian multiplex IA families.
White controls free of stroke and known IA were selected from the Greater Cincinnati/Northern Kentucky population. The methodology for control identification and enrollment has been previously published.8, 16 In short, the University of Cincinnati Institute for Policy Research used random-digit-dialing telephone survey techniques to identify control subjects of the same sex, race, and age for comparison with cases of subarachnoid and intracerebral hemorrhage in an ongoing NINDS-funded study. After informed consent was obtained, each control subject, or a proxy was interviewed face to face in a highly structured and identical manner. In addition to the interview, which included detailed questions about past and present cigarette smoking, blood pressure measurements and blood samples for DNA extraction were obtained. FIA cases were interviewed in an identical manner.
The cases (N = 410) and controls (N = 393) were previously genotyped using the Affymetrix 6.0 array (data not shown). These data were used to test for cryptic relatedness among the reportedly unrelated cases and controls and to ensure that the association analysis is not confounded by the effect of population substructure. A principal component-based analysis was performed in PLINK17 to cluster these samples, along with HapMap reference samples (CEU, YRI, CHB, and JPT), to verify that the samples used in this study were derived from European ancestry (Supplemental Figure online). Five subjects (4 cases and one control) who did not cluster with the Caucasian samples and a CEU reference sample were excluded from further analysis. The final analysis sample consisted of 406 IA cases and 392 controls.
Six SNPs (rs1429412, rs700651 on 2q33; rs10958409, rs9298506 on 8q11; rs1333040, rs10757278 on 9p21) previously associated with arterial diseases1, 2 were genotyped in our sample of cases and controls. The rs10958506 SNP, included in the Affymetrix 6.0 array, was genotyped as part of our previously completed GWAS; the other five SNPs were genotyped using the TaqMan (fluorogenic 5’ nuclease) assay, and the end-point results were scored on the ABI 7900HT Sequence Detection System. Blind duplicates and known control samples were included in test plates for quality assurance. Quality control metrics and SNP descriptive statistics were computed for each of the six SNPs. Completeness of genotyping and allele frequencies were calculated using all genotyped subjects; Hardy-Weinberg equilibrium (HWE) was assessed using only the data from the control subjects. Genotype proportions at all SNPs conformed to the expectation of HWE (data not shown). Two tests of association for each SNP with IA were performed in PLINK; a 2 degree of freedom genotypic test for differences between cases and controls with any of the three observed genotypes, and an allelic test (1 degree of freedom) comparing the minor allele frequency between cases and controls. Pairwise linkage disequilibrium (LD) between all pairs of SNPs in the three chromosomal regions was computed based on the r2 statistic. We examined six SNPs in 3 regions, suggesting that a conservative correction for multiple testing would require 0.05/3 (0.017) threshold for replication. We had 80% power to detect odds ratios of 1.42-1.45 across the range of minor allele frequencies typed at the p=0.017 significance threshold.
For SNPs for which we were able to replicate evidence of association, we augmented the available data with the SNP genotypes generated as part of the previously completed GWAS in the same samples. We added to the dataset all SNPs within 250 kilobases (kb) upstream and downstream of a replicated SNP. These SNPs underwent similar quality review to ensure genotypic completeness and lack of deviation from HWE.
Since smoking is such an important risk factor in IA, we next performed logistic regression analyses to test whether the association of the replicated SNPs on chromosomes 8 and 9 were modulated by smoking. We used the same case-control design and a logistic regression model to test each SNP. Each model included the presence of SNP risk alleles, scored as 0 = no risk allele, 1 = 1 risk allele (heterozygous), and 2 = 2 risk alleles (homozygous). The risk allele was defined as the allele more common in cases than controls. Log of pack years smoked was used to evaluate the effect of smoking. For purposes of the logistic regression, persons without any history of smoking were defined as having 0.05 pack years. Each model was adjusted for age and the data presented as odds ratio (OR) and 95% confidence intervals (CI). An explicit interaction between log of pack years and the risk allele score was tested to determine whether there was a deviation from the multiplicative effect on risk that is modeled by the logistic regression (i.e. closer to additive effects on risk or greater than multiplicative interaction). We also compared the geometric mean of the log of pack years smoked for those subjects with one IA as compared to those with multiple Ias.
Results
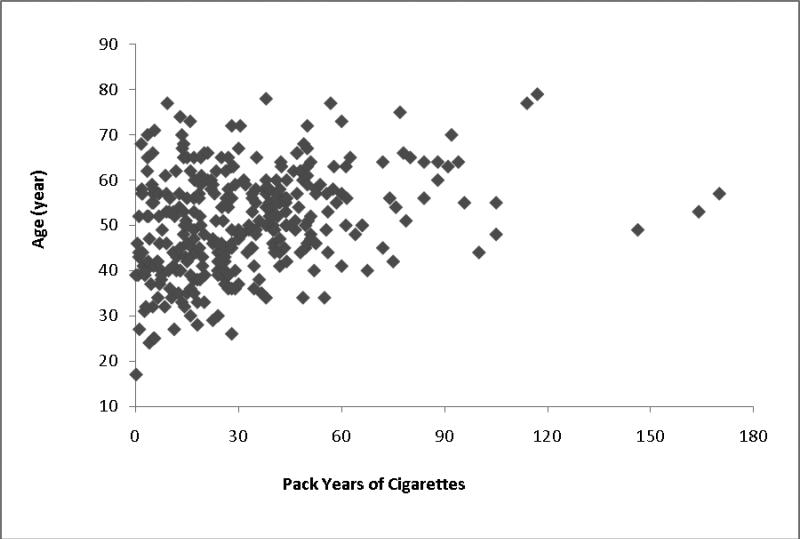
Of the 406 cases, 46.7% were male, compared with 54.3% of the 392 controls (p = .0004). The mean ± SD age of the cases at time of diagnosis was 50.5 ± 11.6 versus 63.4 ± 15.1 at time of interview for the controls (p < .0001). At diagnosis, 47.3% of cases were current smokers and 35.2% were prior smokers versus 16.6% and 35.7%, respectively, at interview for the controls (p < .0001). Figure 1 shows the plot of the all cases who were smokers by pack-years of smoking and age at diagnosis of IA. Of the 406 cases of IA, 159 were ruptured IAs.
Figure 1.
Plot of all cases who were smokers at any time by pack-years of smoking and age of diagnosis.
The association analyses, which include the genotype frequencies as well as the frequency of the risk allele, are presented in Table 1. The strongest evidence of association with IA was found with the 8q SNP rs10958409 (genotypic P = 9.2 × 10-5; allelic P = 1.3 × 10-5; OR = 1.86, 95% CI: 1.40−2.47). We also found evidence of association with both SNPs on chromosome 9p, rs1333040 and rs10757278, with rs1333040 meeting our corrected level of significance. We were not able to replicate the association of the two SNPs on chromosome 2q reported by Bilvugar and colleagues.1
Table 1.
Association of six SNPs on chromosomes 2, 8, and 9 with familial intracranial aneurysm
| Chr. | SNP | Pos. (Mb) | Genotype Frequencies (%) | P-value Genotypic | RAF* | P-value Allelic | OR (95% CI) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 2q33 | rs1429412 | 197.9 | GG | GA | AA | G | ||||
| Case | 13.2 | 44.6 | 42.2 | 0.423 | 0.355 | 0.605 | 1.06 (0.85–1.32) | |||
| Control | 10.3 | 47.9 | 41.9 | 0.342 | ||||||
| 2q33 | rs700651 | 198.3 | GG | GA | AA | G | ||||
| Case | 12.5 | .43.0 | 44.5 | 0.178 | 0.340 | 0.973 | 1.0 (0.81–1.24) | |||
| Control | 9.5 | 48.9 | 41.6 | 0.339 | ||||||
| 8q11 | rs10958409 | 55.5 | AA | AG | GG | A | ||||
| Case | 4.3 | 31.3 | 64.4 | 9.2 × 10-5 | 0.199 | 1.3 × 10-5 | 1.86 (1.40–2.47) | |||
| Control | 1.3 | 20.9 | 77.7 | 0.118 | ||||||
| 8q11 | rs9298506 | 55.6 | GG | GA | AA | A | ||||
| Case | 1.7 | 27.0 | 71.3 | 0.522 | 0.848 | 0.268 | 1.18 (0.88–1.56) | |||
| Control | 2.4 | 30.1 | 67.5 | 0.826 | ||||||
| 9p21 | rs1333040 | 22.1 | CC | CT | TT | T | ||||
| Case | 17.0 | 40.0 | 43.0 | 0.017 | 0.630 | 0.039 | 1.24 (1.01–1.52) | |||
| Control | 17.6 | 48.9 | 33.4 | 0.579 | ||||||
| 9p21 | rs10757278 | 22.1 | AA | AG | GG | G | ||||
| Case | 19.8 | 49.5 | 30.7 | 0.021 | 0.554 | 0.005 | 1.33 (1.09–1.62) | |||
| Control | 26.9 | 49.5 | 23.6 | 0.484 | ||||||
RAF = Frequency of the more common allele in cases compared to controls.
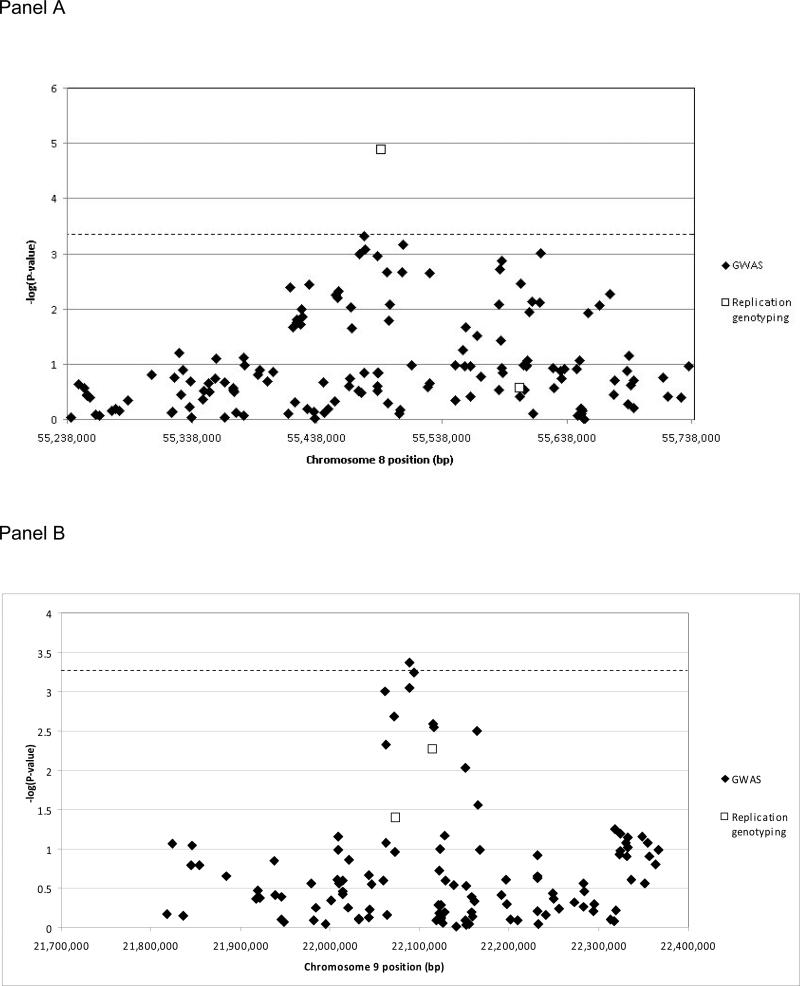
As shown in Figure 2 panel A, substantial support for the association to chromosome 8 was provided by the SNPs genotyped in the GWAS. Six of these SNPs, located on both sides of the index replication SNP rs10958409, achieved association p-values less than 0.001. These results suggest the presence of a substantial linkage disequilibrium block near the 55.5 megabase position on chromosome 8 that contains a variant associated with IA. The significance of the p-value obtained for rs10958409, as compared with that of the surrounding GWAS SNPs, suggests the frequency of the IA-predisposing allele is near that of rs10958409 (MAF=0.199 in cases and 0.118 in controls). In contrast, as shown in Figure 2 panel B, we were able to improve the evidence for association via examination of the results from GWAS SNPs surrounding rs10757278 on chromosome 9p. P-values as small as 4 × 10-4 were observed in this region in the GWAS, compared with p=0.005 for rs10757278, the SNP reported in the previous studies. The GWAS SNP providing strongest support for association to chromosome 9p in our study, rs2891168, surpassing the α=0.05 significance threshold corrected by the simpleM method22 for the SNPs in the 500 kb region considered. These results clearly illustrate the benefit of denser SNP coverage over a range of allele frequency values.
Figure 2.
All GWAS SNPs 250 kb upstream and downstream of the replicated SNPs are shown in each panel, chromosome 8 in panel A, chromosome 9 in panel B. The dashed horizontal line in each panel indicates the □=0.05 significance threshold correcting for the SNPs in each region by the simpleM method22
Statistical testing was consistent with a multiplicative relationship between the at-risk alleles and smoking (Table 2). For example, a non-smoker with one risk allele for SNP rs10958409 on chromosome 8 would have an OR of 1.48 for having an IA, a 20 year smoker with no copies of this risk allele would have an OR of 5.04, and 20-pack year smoker with one at risk allele would have an OR of 7.46 (see footnote in Table 2 for explanation). We tested for interactions of each SNP with the log of pack years of smoking to determine if they differed from the multiplicative relationship inherent in the logistic regression models. No test reached statistical significance indicating a multiplicative relationship provided a good fit to the data (or no evidence for supra-multiplicative or less than multiplicative relationship).
Table 2.
Logistic Regression Models for Three SNPs and Smoking (pack years) on Chromosome 8 and 9 (Models Adjusted for Age)
| SNP | Odds Ratio | 95% Confidence Interval | P value |
|---|---|---|---|
| rs10958409 (chr 8q) | |||
| Score (per risk allele) | 1.48 | 1.06-2.07 | 0.023 |
| 20 pack years of smoking | 5.04 | 3.50-7.61 | <0.001 |
| Age (per year) | 0.93 | 0.92-0.94 | <0.001 |
| rs10757278 (chr 9p) | |||
| Score (per risk allele) | 1.40 | 1.10-1.78 | 0.007 |
| 20 pack years of smoking | 5.75 | 3.99-8.29 | <0.001 |
| Age (per year) | 0.93 | 0.92-0.94 | <0.001 |
| rs1333040 (chr 9p) | |||
| Score (per risk allele) | 1.37 | 1.08-1.74 | 0.012 |
| 20 pack years of smoking | 5.16 | 1.88-2.71 | <0.001 |
| Age (per year) | 0.93 | 0.92-0.94 | <0.001 |
Footnote: The OR for any given number of K pack-years can be calculated using the following equation: OR = exp(β(Ln(K) + 2.99)) where β = the regression coefficient for log(pkyrs). β=0.270 for rs10958409, β=0.292 for rs10757278, and β=0.278 for rs1333040. For example, the odds ratio for 40 pack years of smoking for subjects in the model of the rs10757278 risk allele = exp(0.292(Ln40) + 2.99)) = 7.03. To determine the odds ratio for presence of two risk alleles of rs10757278 (homozygous state) and 40 pack years of smoking, one would multiply (1.40)2 =1.96 (two risk alleles) × 7.03 which equals an odds ratio of 13.78.
There was a significantly greater geometric mean of log(pkyrs) for smoking among the 147 subjects with > 1 aneurysm (11.19) as compared to the 254 subjects with one aneurysm (5.93; t=2.59, p=0.010). We did not find a significant difference in prevalence of risk alleles for chromosome 8 (rs10958409)and 9 (rs1333040, rs10757278) in those subjects with one aneurysm and those with > 1 aneurysm.
Discussion
This study replicates the associations of SNPs on chromosome 8 and 9 and demonstrates the powerful effect of smoking on the risk of IA in persons with these risk variants. For example, in the logistic regression model, a non-smoker with two rs10757278 alleles on chromosome 9 has an OR of 1.96 for the presence of IA, whereas someone with 40 pack- years would have an OR of 13.78. Our replication of SNPs on chromosome 8 and 9 show that we are getting close to identifying the causal variants associated with IA. Even though the exact gene variants that are associated with IA have yet to be found, it is clear that smoking greatly enhances their effect and that cessation of smoking would have a tremendous impact in prevention of IA, particularly in those at increased genetic risk. The fact that 82.5% of our IA cases were smokers at some point and 47% were current smokers speaks strongly to the opportunity for prevention.
The associations of the two 8q SNPs with IA were initially found in two European cohorts from Finland and the Netherlands. However, in a Japanese sample, only rs10958409 was replicated; rs9298506 was not. In our study cohort, comprised only of Caucasian samples, the strongest evidence of association was with rs10958409 (genotypic P = 9.2 × 10-5; allelic P = 1.3 × 10-5; OR = 1.86, 95% CI 1.40-2.47). As in the Finnish, Dutch and the Japanese cohorts, we found the association of the same risk allele in this SNP in our sample of IA cases (Table 1). We did not find evidence of association with rs9298506. However, with our relatively small sample size, it is premature to exclude the involvement of this SNP influencing the risk of IA. As described by Bilguvar et al,1 SOX17 is the closest gene within the interval of the 8q variants, which is involved in endothelium formation and maintenance.
A series of genome-wide association studies has reported association of sequence variants on 9p21 with myocardial infarction (MI), coronary artery disease (CAD), abdominal aortic aneurysm (AAA), and IA in populations of European ancestry followed by replication in a Japanese cohort.1-6 This locus was also implicated in type 2 diabetes18 although the same SNPs were not associated with IA.1 Thus, the chromosome 9p21 locus has emerged as a potentially important region involved in the risk of arterial diseases. We genotyped the two SNPs rs1333040 and rs10757278, reported as significantly associated with CAD, AAA and IA.1-3 We found significant association of the T allele of rs1333040 and the G allele of rs10757278 with IA, thus confirming the previous findings. These two SNPs are 41kb apart and at modest level of LD (r2 = 0.529) in our sample. The mechanism by which the 9p21 locus influences risk to IA and other arterial diseases remains unknown. Two cyclin-dependent kinase genes, CDKN2B and CDKN2A, as well as a non-coding RNA transcript ANRIL, are in close proximity of these sequence variants. CDKN2A and CDKN2B encode the cyclin-dependent kinase inhibitors p16INK4a and p16INK4a, which are involved in senescence and apoptosis of cell types including ameliorating age-related physiological effect on stem cells and repair of aged tissue.19, 20 ANRIL is the closest known gene from the 9p SNPs, 38kb from rs1333040 and 3kb from rs10757278. ANRIL was shown to be expressed cells and tissues involved in atherosclerosis.21
The chromosome 2q SNPs, rs1429412 and rs700651, were novel variants associated with IA in the two European cohorts noted above; rs700651 was replicated in the Japanese sample, but rs1429412 was not.1 We did not find association of either SNP with IA in our study. At this time, the evidence of association is inconclusive and requires further evaluation.
The modest sample size of FIA study cases and controls limits the power to detect genes of smaller effect size. Nevertheless, our cases represent a unique group in that all are from families with strong familial aggregration of IA as compared with prior GWAS studies of IA, where the majority of subjects did not have a positive family history.1 We believe that such a cohort may provide greater ability to detect genetic risk factors for IA and eventually will facilitate identification of potentially causal gene variants within our larger FIA families.
Another limitation of our analysis is that our control group was not perfectly matched to our IA cases. Consequently, the controls were older than the cases and there was a slightly greater proportion of men among our controls. While the control group was identified by random-digit dialing from the entire population as part of an ongoing NINDS-funded study, cases of intracerebral hemorrhage comprised about 2/3 of the cases of intracranial hemorrhage that were used for identification of matched controls. People with intracerebral hemorrhage are more likely to be older and more likely to be male than those with subarachnoid hemorrhage, which accounts for the older age of available controls. That older controls make it more likely that these individuals do not have or would not develop an IA can be considered a strength of genetic studies. However, an age difference between cases and controls is a potential disadvantage when environmental covariates are correlated with age. The frequency of current smoking is inversely associated with advancing age because some people stop smoking for health reasons as they enter middle and older age. Pack years and history of smoking, which look at lifetime exposure rather than current smoking state, help ameliorate this concern, as does the adjustment for age in the logistic regression models. Finally, the lack of information about second-hand smoking for cases and controls limits the precision of true exposure to smoking among cases and controls.
In summary, we analyzed six sequence variants reported to be significantly associated with several vascular diseases in a sample of unrelated cases from multiplex families affected with IA and population controls. Our data provide complementary evidence that the variants on chromosome 8q are strongly associated with IA and variants on chromosome 9p are moderately associated with IA and that the associated risks of IA in patients with these variants are greatly increased with cigarette smoking. We did not find significant association of the 2q variants.
Supplementary Material
Supplemental Figure Online: Multidimensional scaling (MDS) plot for the study subjects with reference to Hapmap population controls. First and second principal component scores for each individual are plotted on the x- and y-axis, respectively, based on the genotypes for the markers from the Affymetrix 6.0 array that passed quality control thresholds. Population reference samples are CEU=Caucasians from Utah, YRI=Nigerians from Yoruba, CHB=Han Chinese from Beijing, and JPT=Japanese from Tokyo.
Sources of Funding
This study was funded by grants from the National Institute of Neurological Diseases and Stroke (NINDS R01 NS39512) (R-01-NS 36695), National Institutes of Health, Bethesda, MD; the State of Ohio TECH 04-042, Ohio Dept. of Development, Wright Centers of Innovation Program Computational Medicine Center for the “Cincinnati Control Cohort Study” and by the Intramural Research Program of the NIH, National Cancer Institute and National Human Genome Research Institute.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bilguvar K, Yasuno K, Niemela M, Ruigrok YM, von Und Zu Fraunberg M, van Duijn CM, van den Berg LH, Mane S, Mason CE, Choi M, Gaal E, Bayri Y, Kolb L, Arlier Z, Ravuri S, Ronkainen A, Tajima A, Laakso A, Hata A, Kasuya H, Koivisto T, Rinne J, Ohman J, Breteler MM, Wijmenga C, State MW, Rinkel GJ, Hernesniemi J, Jaaskelainen JE, Palotie A, Inoue I, Lifton RP, Gunel M. Susceptibility loci for intracranial aneurysm in European and Japanese populations. Nat Genet. 2008;40:1472–1477. doi: 10.1038/ng.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJ, Blankensteijn JD, Ronkainen A, Jaaskelainen JE, Kyo Y, Lenk GM, Sakalihasan N, Kostulas K, Gottsater A, Flex A, Stefansson H, Hansen T, Andersen G, Weinsheimer S, Borch-Johnsen K, Jorgensen T, Shah SH, Quyyumi AA, Granger CB, Reilly MP, Austin H, Levey AI, Vaccarino V, Palsdottir E, Walters GB, Jonsdottir T, Snorradottir S, Magnusdottir D, Gudmundsson G, Ferrell RE, Sveinbjornsdottir S, Hernesniemi J, Niemela M, Limet R, Andersen K, Sigurdsson G, Benediktsson R, Verhoeven EL, Teijink JA, Grobbee DE, Rader DJ, Collier DA, Pedersen O, Pola R, Hillert J, Lindblad B, Valdimarsson EM, Magnadottir HB, Wijmenga C, Tromp G, Baas AF, Ruigrok YM, van Rij AM, Kuivaniemi H, Powell JT, Matthiasson SE, Gulcher JR, Thorgeirsson G, Kong A, Thorsteinsdottir U, Stefansson K. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 3.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 4.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H, WTCCC and the Cardiogenics Consortium Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT. Lund University. Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 7.Longstreth WTJ, Nelson LM, Koepsell TD, van Belle G. Cigarette smoking, alcohol use, and subarachnoid hemorrhage. Stroke. 1992;23:1242–1249. doi: 10.1161/01.str.23.9.1242. [DOI] [PubMed] [Google Scholar]
- 8.Woo D, Khoury J, Haverbusch MM, Sekar P, Flaherty ML, Kleindorfer DO, Kissela BM, Moomaw CJ, Deka R, Broderick JP. Smoking and family history and risk of aneurysmal subarachnoid hemorrhage. Neurology. 2009;72:69–72. doi: 10.1212/01.wnl.0000338567.90260.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kissela BM, Sauerbeck L, Woo D, Khoury J, Carrozzella J, Pancioli A, Jauch E, Moomaw CJ, Shukla R, Gebel J, Fontaine R, Broderick J. Subarachnoid hemorrhage: a preventable disease with a heritable component. Stroke. 2002;33:1321–1326. doi: 10.1161/01.str.0000014773.57733.3e. [DOI] [PubMed] [Google Scholar]
- 10.Anderson CS, Feigin V, Bennett D, Lin RB, Hankey G, Jamrozik K, Australasian Cooperative Research on Subarachnoid Hemorrhage Study (ACROSS) Group Active and passive smoking and the risk of subarachnoid hemorrhage: an international population-based case-control study. Stroke. 2004;35:633–637. doi: 10.1161/01.STR.0000115751.45473.48. [DOI] [PubMed] [Google Scholar]
- 11.Anderson C, Ni Mhurchu C, Scott D, Bennett D, Jamrozik K, Hankey G, Australasian Cooperative Research on Subarachnoid Hemorrhage Study Group Triggers of subarachnoid hemorrhage: role of physical exertion, smoking, and alcohol in the Australasian Cooperative Research on Subarachnoid Hemorrhage Study (ACROSS). Stroke. 2003;34:1771–1776. doi: 10.1161/01.STR.0000077015.90334.A7. [DOI] [PubMed] [Google Scholar]
- 12.Isaksen J, Egge A, Waterloo K, Romner B, Ingebrigtsen T. Risk factors for aneurysmal subarachnoid haemorrhage: the Tromso study. J Neurol Neurosurg Psychiatry. 2002;73:185–187. doi: 10.1136/jnnp.73.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foroud T, Sauerbeck L, Brown R, Anderson C, Woo D, Kleindorfer D, Flaherty ML, Deka R, Hornung R, Meissner I, Bailey-Wilson JE, Langefeld C, Rouleau G, Connolly ES, Lai D, Koller DL, Huston J, 3rd, Broderick JP, Familial Intracranial Aneurysm Study Investigators Genome screen in familial intracranial aneurysm. BMC Med Genet. 2009;10:3. doi: 10.1186/1471-2350-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foroud T, Sauerbeck L, Brown R, Anderson C, Woo D, Kleindorfer D, Flaherty ML, Deka R, Hornung R, Meissner I, Bailey-Wilson JE, Rouleau G, Connolly ES, Lai D, Koller DL, Huston J, 3rd, Broderick JP, FIA Study Investigators Genome screen to detect linkage to intracranial aneurysm susceptibility genes: the Familial Intracranial Aneurysm (FIA) study. Stroke. 2008;39:1434–1440. doi: 10.1161/STROKEAHA.107.502930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broderick JP, Sauerbeck LR, Foroud T, Huston J, 3rd, Pankratz N, Meissner I, Brown RD., Jr The Familial Intracranial Aneurysm (FIA) study protocol. BMC Med Genet. 2005;6:17. doi: 10.1186/1471-2350-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo D, Sauerbeck LR, Kissela BM, Khoury JC, Szaflarski JP, Gebel J, Shukla R, Pancioli AM, Jauch EC, Menon AG, Deka R, Carrozzella JA, Moomaw CJ, Fontaine RN, Broderick JP. Genetic and environmental risk factors for intracerebral hemorrhage: preliminary results of a population-based study. Stroke. 2002;33:1190–1195. doi: 10.1161/01.str.0000014774.88027.22. [DOI] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, Wellcome Trust Case Control Consortium (WTCCC) McCarthy MI, Hattersley AT. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT. Stem-cell aging modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 20.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, Clarke R, Collins R, Franzosi MG, Tognoni G, Seedorf U, Rust S, Eriksson P, Hamsten A, Farrall M, Watkins H, PROCARDIS consortium Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 22.Gao X, Becker LC, Becker DM, Starmer JD, Province MA. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol. 2009 doi: 10.1002/gepi.20430. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure Online: Multidimensional scaling (MDS) plot for the study subjects with reference to Hapmap population controls. First and second principal component scores for each individual are plotted on the x- and y-axis, respectively, based on the genotypes for the markers from the Affymetrix 6.0 array that passed quality control thresholds. Population reference samples are CEU=Caucasians from Utah, YRI=Nigerians from Yoruba, CHB=Han Chinese from Beijing, and JPT=Japanese from Tokyo.