Abstract
Gold nanoparticle (AuNP)-DNA conjugates can enhance in vitro translation of a protein. Enhancement occurs via a combination of non-specific adsorption of translation-related molecules and the ribosome to the AuNP-DNA and specific binding to the mRNA of interest. AuNP-DNA conjugates enhanced protein production of fluorescent proteins (mCherry, eGFP) in retic lysate mixes by 65–100%. Gel electrophoresis was used to probe non-specific adsorption of the AuNP-DNA conjugates to the translation machinery. It was determined that non-specific adsorption is critical for enhancement, and if it was eliminated, expression enhancement did not occur. The interaction of the mRNA with the DNA on the AuNP surface influenced the amount of enhancement, and was probed by expression in the presence of RNase H. These results suggest that higher translation enhancement occurs when the DNA on the AuNP forms an incomplete duplex with the mRNA. Tuning the balance between non-specific adsorption and specific binding of the AuNP-DNA conjugates could result in the translation enhancement of a specific gene in a mixture.
Keywords: nanoparticle-DNA conjugates, non-specific adsorption, in vitro translation, selective enhancement, antisense, DNA-mRNA hybridization, nanoparticle-mPEG
Due to their unique properties, nanoparticles (NPs) are attractive for numerous biological and therapeutic applications.1–10 One of the biggest barriers for utilizing NPs is non-specific adsorption, where biomolecules non-covalently adsorb to NPs, obscuring biological function and leading to denaturation and undesirable effects.11–14 Unfortunately, non-specific adsorption is complex, where an enormous number of non-covalent bonds between biomolecules and NP surfaces or ligands can form. Non-specific adsorption is difficult to not only prevent but also directly probe, and thus remains poorly understood.15–16 Despite the fact that gold NPs (AuNPs) have versatile surface chemistry, efforts to simply eliminate non-specific adsorption via surface modification17 with inert molecules18–19 have met limited success.20–23 Consequently, non-specific adsorption is a major hindrance for nanobiotechnology.
Here we adopt a different perspective of non-specific adsorption and demonstrate that it is ideal for enhancing the efficiency of a biological reaction, in vitro translation. Translation, the synthesis of a protein encoded in mRNA, is complex and involves the ribosome, mRNA, and hundreds of other species.24 It can potentially be enhanced by recruiting and coordinating translation machinery and mRNA.25–26 Because AuNP-DNA conjugates are approximately the same size as proteins, they can act as artificial scaffolds to bring proteins into proximity by non-specifically adsorbing to them (Figure 1a). Numerous weak bonds are what make non-specific adsorption problematic, but this very property is uniquely suited for dynamic and repeating reactions, where multiple species must be fluxional.27 In this case, strong and specific binding to translation machinery would not only be impossible to design since reactions involve numerous species, but it would also be detrimental, preventing turnover of species necessary for dynamic and repeating reactions. In addition, the DNA on the AuNP can have a sequence such that it can bind specifically to the mRNA of interest, increasing enhancement. We show that non-specific adsorption to AuNP-DNA can be exploited synergistically with specific binding to enhance in vitro translation as high as 100%. Expression enhancement by conjugates of AuNP and DNA (AuNP-DNA) depends on the mRNA-DNA interaction and the AuNP surface charge. Finally, AuNP-DNA can be used to enhance specific translation of a target gene in a mixture.
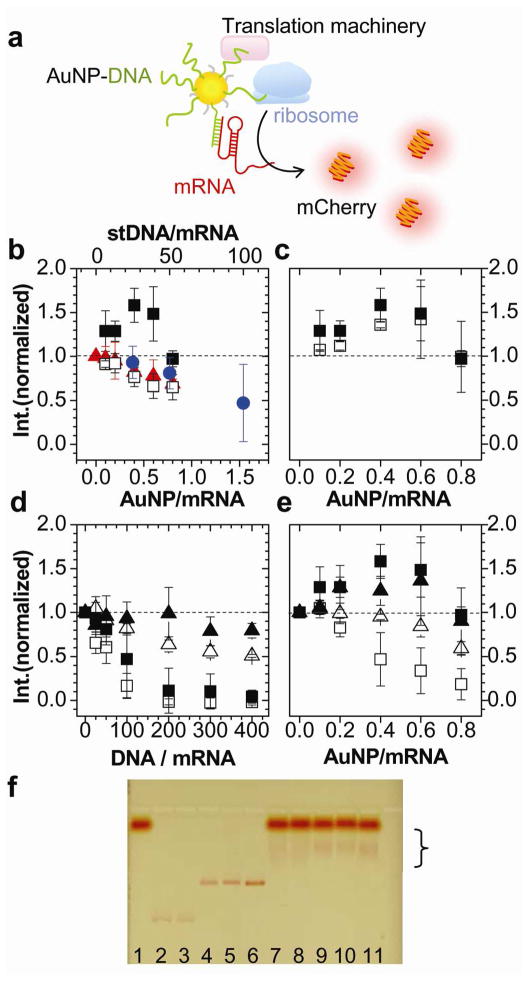
Figure 1.
In vitro gene expression with DNA, AuNP and AuNP-DNA. a. Schematic diagram of enhanced mCherry gene expression with AuNP-DNA. AuNP recruits mRNA and translation related molecules into its proximity. AuNP D = 9.6 ± 0.6nm. b. Normalized peak fluorescence intensity of expressed mCherry with AuNP-stDNA of coverage 1:65 (filled squares), AuNP (red triangles), and mixture of 1:65 free AuNP and stDNA (open squares) as a function of AuNP/mRNA ratio (lower axis), and expression with free stDNA (blue circles) as a function of stDNA/mRNA ratio (upper axis). For all translation experiments, the amount of mRNA used was fixed at 0.25μg. Error bars denote 95% confidence intervals with n ≥ 4. Upper axis matches lower axis at NP/mRNA = stDNA/mRNA = 0, and NP/mRNA = 1 and stDNA/mRNA=65, as the coverage of stDNA on the NP is 65 DNA/NP. c. High (1:65, filled squares) or low (1:29, open squares) coverage of AuNP-stDNA with mCherry expression. d. Effect of wkDNA on mCherry (filled triangles) and eGFP (open triangles), and stDNA on mCherry (filled squares) and eGFP (open squares) translation. e. AuNP-wkDNA (1:59) on mCherry (filled triangles) and eGFP (open triangles), and AuNP-stDNA (1:65) on mCherry (filled squares) and eGFP (open squares). f. 1.5% agarose gel electrophoresis in 0.5× TBE at 3.8V/cm for 1.5hrs. Lane 1: retic lysate kit mixture, 2: AuNP, 3: AuNP-mPEG (reaction ratio 1:200), 4: AuNP-wkDNA (coverage 1:59), 5: AuNP-stDNA (1:65), 6: AuNP-stDNA (1:29), 7: mixture of 1&2, 8: 1&3, 9: 1&4, 10: 1&5, and 11: 1&6.
Results and Discussion
AuNPs (D=9.6nm, Supporting Information, Figure S1) coated with bis(p-sulfonatophenyl) phenylphosphine (BPS) were conjugated covalently to stDNA or wkDNA (Table 1, See Methods). Fluorescence measured mCherry expression (Figure 1b) from fixed amounts of mRNA (0.25μg) in reticular lysate translation kits, and was normalized to reactions that had no AuNP, DNA, or AuNP- DNA added. When AuNP-stDNA (AuNP:DNA=1:65, filled squares) was added to a translation reaction, mCherry expression reached 1.65× the amount for no AuNP-DNA, or an enhancement of 65%. Enhancement depended on AuNP-DNA concentration, peaking at 0.4 AuNP/mRNA molar ratio and switching to inhibition at higher AuNP/mRNA. This suggests that enhancement is strongest when multiple species, including the mRNA, can bind to one scaffold that brings them into proximity. AuNP-stDNA of lower coverage (Figure 1c, AuNP:DNA=1:29, open squares) enhanced expression to a lesser extent (40%), suggesting that more DNA on the AuNP facilitates binding to mRNA and translation-related species.
Table 1.
DNA sequences.
| name | Sequence |
|---|---|
| stDNA | 5′-HS -TTTTT TTTTT CTCGT TGGGG TCTTT -3′ |
| wkDNA | 5′-HS -TTTTT TTTTT GATGT TGACG TTGTA -3′ |
Enhancement did not occur with either free AuNP or DNA. mCherry expression decreased with increasing free AuNP (Figure 1b, red triangles). BPS coated AuNPs are negative, so evidently charge interaction between AuNPs and translation-related molecules interferes with translation. Quenching was not responsible for fluorescence changes,28 as fluorescence was unaffected when AuNPs were added after translation reactions were complete (Supporting Information, Figure S2). Free stDNA only inhibited translation, (Figure 1b, blue circles. Upper axis matches with lower axis at 65:1), acting as antisense DNA to block ribosomal activity via mRNA binding.29–30 This shows that the DNA’s biophysical behavior is completely reversed when on the AuNP surface. AuNPs mixed with unlinked stDNA at a ratio of 1:65 (Figure 1b, open squares) inhibited expression more than free stDNA or AuNP, confirming that stDNA must be covalently bound to AuNPs for the enhancement effect.
Gel electrophoresis confirmed the presence of non-specific adsorption to the translation machinery (Figure 1f). AuNP, AuNP-mPEG, and AuNP-DNA (Lanes 2–6) when added to the translation mix (Lanes 7–11), exhibited lower mobility bands (bracket), which were determined to have protein by blue-staining (Supporting Information, Figure S3). This shows that regardless of whether the particles were modified with DNA, they adsorbed to the translation machinery.
We compared enhancement by AuNP-DNA for two different DNA sequences. DNA was chosen to exhibit strong (stDNA) or weak (wkDNA) antisense inhibition of mCherry or eGFP (Figure 1d). Free stDNA suppressed both eGFP (open squares) and mCherry expression (filled squares). However, stDNA behaved differently on AuNPs (Figure 1e). AuNP-stDNA inhibited eGFP expression strongly (open squares), but enhanced mCherry (filled squares). Antisense inhibition by AuNP-DNA was reported previously31 but we observed that the same DNA strand can either enhance or inhibit translation depending on whether it is conjugated to AuNPs, and is reported for the first time here to our knowledge. Free wkDNA inhibited both genes weakly (Figure 1d, open/filled triangles), and AuNP-wkDNA suppressed eGFP (Figure 1e, open triangles) and enhanced mCherry (filled triangles), but to a lesser extent than AuNP-stDNA. Evidently, enhancement depends not only on the sequence of the DNA on the AuNP-DNA conjugate, but also the gene of interest. Differences in eGFP and mCherry suppression by stDNA or wkDNA could be due to how the oligos interact with the mRNA and the resulting changes in mRNA secondary structure, which would affect ribosomal access and its ability to translate the mRNA. However, the antisense mechanism of oligos is generally not well understood, and difficult to predict.29–30, 32 This shows that enhancement or inhibition of AuNP-DNA can be indirectly checked by the antisense strength of the free DNA, which is correlated with DNA affinity for the mRNA.
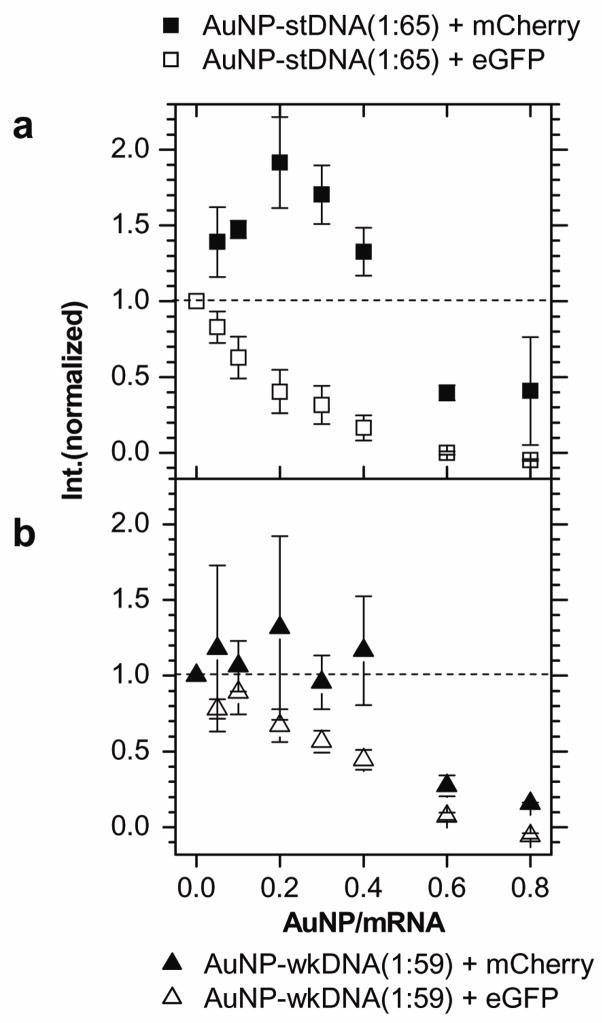
Because enhancement by AuNP-DNA varies for different genes, it is possible to exploit this to selectively enhance a gene in a mixture. AuNP-stDNA incubated with a mixture of mCherry and eGFP mRNA enhanced mCherry expression by ~100% at 0.2 AuNP/mRNA while simultaneously suppressing eGFP (Figure 2a), higher than in the single gene experiments (Figure 1e). AuNP-wkDNA also exhibited selective enhancement of mCherry over eGFP, but to a lesser extent (Figure 2b).
Figure 2.
Selective gene expression enhancement. a. Normalized peak fluorescence intensity of mCherry (filled squares) and eGFP (open squares) when a mixture of both genes in equal amounts (0.25μg each) are translated with AuNP-stDNA (coverage 1:65). b. Repeated for AuNP-wkDNA (coverage 1:59). mCherry (filled triangles) and eGFP (open triangles). Note that the AuNP-DNA in the mixtures was ~2× used in single gene experiments.
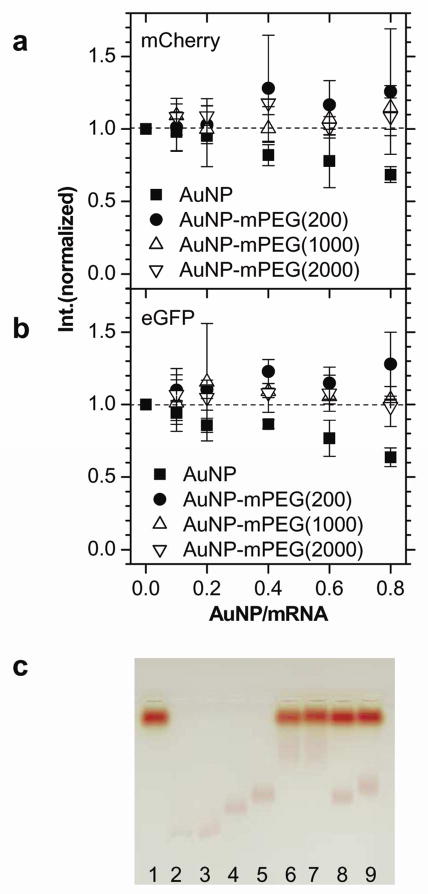
To understand how non-specific adsorption and translation enhancement are related, AuNPs conjugated with different amount of mPEG (MW=356.5) were used. BPS coated AuNPs became less negative with increasing mPEG conjugation while their hydrodynamic size did not change,15 retarding their mobility (Figure 3c, Lanes 3–5. Supporting Information, Figure S3). AuNP-mPEG 1:1000 and 1:2000 exhibited minimal mobility shifts when added to the translation mix (Lanes 4 vs. 8, and 5 vs. 9), indicating negligible non-specific adsorption to translation machinery with high mPEG coverage. AuNP-mPEG 1:1000 and 1:2000 had little effect on both mCherry and eGFP expression (Figure 3a and 3b, triangles and inverted triangles). Furthermore, AuNP and AuNP-mPEG 1:200 differ only slightly in surface charge (Figure 3c, Lanes 2, 3) and adsorption behavior (Lanes 6, 7), but showed opposite translation behavior, where AuNP-mPEG 1:200 enhanced (circles) while AuNP inhibited (squares). Free mPEG affected translation negligibly (Supporting Information, Figure S4). Thus for enhancement, AuNPs need to be charged to enable non-specific adsorption, but not too highly charged or inhibition results. Furthermore, non-specific adsorption alone cannot enhance selectively, as both eGFP and mCherry are affected similarly by AuNP or AuNP-mPEG. Thus, specific enhancement requires AuNPs conjugated to DNA designed for the gene. Also, the enhancement effect is lower if there is no DNA on the AuNP, where AuNP-mPEG 1:200 enhances only 25%, while AuNP-stDNA enhances 65%.
Figure 3.
Translation enhancement by AuNP-mPEG. a. Normalized peak fluorescence intensity of mCherry when AuNP (squares), AuNP-mPEG (reaction ratio 1:200, circles), AuNP-mPEG (1:1000, triangles), and AuNP-mPEG (1:2000, inverted triangles) are added. b. Repeated for eGFP translation. c. 1.5% agarose gel electrophoresis in 0.5× TBE at 3.8 V/cm for 1.5hrs. Lane 1: retic lysate kit mixture, Lane 2: AuNP, Lane 3: AuNP-mPEG (reaction ratio 1:200), Lane 4: AuNP-mPEG (1:1000), 5: AuNP- mPEG (1:2000), Lane 6: mixture of 1 & 2, Lane 7: 1 & 3, Lane 8: 1 & 4, and Lane 9: 1 & 5.
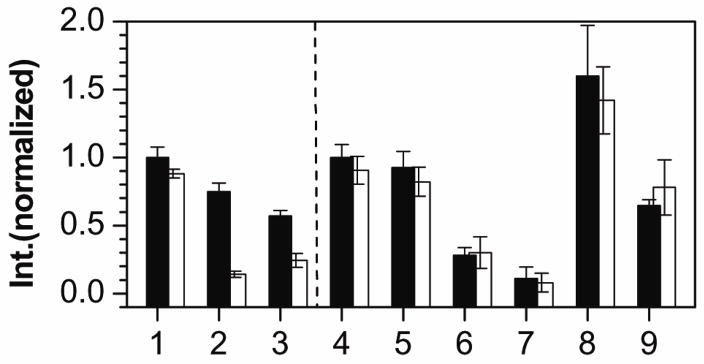
Ribonuclease H (RNase H) was used to probe the DNA-mRNA interaction. RNase H recognizes RNA-DNA duplexes to cleave the RNA, reducing expression levels.29–30 eGFP and mCherry expression was measured with (white) and without (black) RNase H (Figure 4). RNase H had negligible effect on eGFP or mCherry mRNA alone (Samples 1 and 4). eGFP expression decreased with stDNA (Sample 2, black), and dropped further with RNase H (Sample 2, white). This indicates that stDNA binds to eGFP mRNA to form a DNA-mRNA duplex which not only inhibits translation via the antisense effect but also can be recognized by RNase H. AuNP-stDNA with eGFP behaved similarly, where RNase H increased the inhibition of eGFP by AuNP-stDNA (Sample 3, black/white) but to a lesser extent compared with Sample 2. This suggests that AuNP-stDNA also can form a duplex with eGFP mRNA, but RNase H accessibility to mRNA is limited due to the conjugated AuNP.
Figure 4.
Translation with RNase H. Normalized peak fluorescence intensity of eGFP and mCherry expression (black columns), and with identical amount of RNase H (white columns). Sample 1: eGFP, 2: eGFP with stDNA (stDNA/mRNA=50), 3: eGFP with AuNP-stDNA (coverage 1:65, AuNP/mRNA=0.4), 4: mCherry, 5–7: mCherry with stDNA (stDNA/mRNA=50, 75, and 100, respectively), 8–9: mCherry with AuNP-stDNA (coverage 1:65, AuNP/mRNA=0.4 and 0.8, respectively).
However, RNase H did not significantly change mCherry expression in the presence of several free stDNA concentrations (Samples 5–7) and AuNP-stDNA (Samples 8 and 9). Because stDNA exhibited antisense inhibition of mCherry, it somehow binds to mCherry mRNA, but not in a manner suitable for RNase H activity. Similarly, RNase H did not reduce the enhanced mCherry expression of AuNP-stDNA. wkDNA and AuNP-wkDNA exhibited both weak antisense and RNase H activity (Supporting Information, Figure S5). Antisense inhibition by an oligo is related to its ability to sterically block ribosomes from reading and translating the gene, which can result from both non-specific and specific binding of DNA to the mRNA. eGFP and mCherry mRNA used in the experiments are ≥700 bases, with multiple sites for partial or complete binding of the DNA. Also, the poly-T spacers inserted into stDNA and wkDNA can form non-Watson-Crick pairs with mRNA.33 However, RNase H activity requires the DNA-mRNA duplex to be not only well-formed but also sterically accessible, which can result from DNA-induced changes to secondary structure of mRNA.32 Evidently, translation enhancement occurs when the DNA in the free form binds to mRNA strongly enough for antisense inhibition, but not RNase H activity. Interestingly, AuNP-DNA does not enhance RNase H activity. We believe that this is due to the fact that the RNase H mechanism is significantly different from translation. Unlike the ribosome, it does not require the numerous translation factors, amino acids, and tRNAs for activity, and thus does not benefit from non-specific adsorption.
Conclusion
Based on these observations, specific translation enhancement occurs via a combination of non-specific adsorption to translation machinery and specific binding to mRNA by AuNP-DNA. AuNP-DNA brings the related species to within nanometer proximity, and permits species to come on and off. AuNP-DNA may also remove mRNA secondary structure upon binding, facilitating ribosome access and thus enhancing expression. mPEG functionalization also enhances translation by protecting the AuNP surface and reducing particle charge, but to a lesser extent and without gene specificity. Strong binding of AuNP-DNA to mRNA results only in inhibition, as it probably sterically hinders the ribosome from reading the mRNA. With this information, future work can explore how to improve enhancement to even higher levels, or apply it to other biological reactions. This study shows that AuNP non-specific adsorption can be beneficially exploited for their use as nanoscale platforms to enhance protein synthesis.
Methods
RNase Free treatment
RNase-free water was either purchased or made by incubation with 0.1% Diethyl Pyrocarbonate and autoclaving. All the samples used for this work were prepared with RNase-free water.
AuNP synthesis
AuNPs (D=9.6nm, s.d.=0.6nm) were synthesized by reduction of HAuCl4 according to literature methods.34 Average size of the particles was obtained by analysis of TEM images (JEOL 2011, Supporting Information, Figure S1) with ImageJ software. AuNPs were functionalized with the negatively charged ligand BPS (bis(p-sulfonatophenyl) phenylphosphine) by incubating with excessive amount of BPS for ~12 hrs. Unnecessary reaction residues were excluded by taking a narrow electrophoretic band of AuNP from 1% agarose gel and letting the particles diffuse into fresh 0.5× TBE (45 mM tris, 45 mM boric acid, and 1 mM EDTA). Concentration of AuNP solution was calculated from the peak of absorption spectra at 520nm.
AuNP–mPEG functionalization
BPS coated AuNPs were incubated in mPEG-SH (Methoxypolyethylene glycol thiol, MW = 356.5) bath for ~24 hrs with different ratios of AuNP:mPEG-SH (1:200, 1:1000 and 1:2000, [AuNP] = 5×10−7 M) to allow formation of thiol linkages between AuNPs and mPEG. The solution was centrifuged with a bench-top micro-centrifuge at 10 krpm for 30 min, and then the thick-colored bottom layer was collected and re-suspended in 0.5× TBE. This step was repeated at least 3 times to wash off free mPEG molecules.
AuNP–DNA conjugation
DNA was modified with a 5′ thiol that attaches covalently to AuNP, and has poly-T spacers to reduce self-adsorption of DNA to the AuNP.16, 35 AuNPs were lyophilized with thiol-functionalized DNA (AuNP:DNA = 1:160 for high coverage, 1:80 for low coverage) and incubated in ~1× TBE for 2 days for further conjugation. Free DNA strands were washed off by the same way free mPEG molecules were washed away. Coverage (average # DNA strands per particle) was measured by displacing the DNA completely from AuNP in concentrated MCH solutions (6-Mercapto-1-hexanol, ~1–100mM) for extended time (>24 hrs), excluding aggregated bare NP’s by centrifugation, and staining supernatant with SYBR gold (Invitrogen) to measure fluorescence intensity.20, 35 Concentration of DNA was interpolated from fluorescence intensity of DNA solutions with known concentration.
In vitro transcription/translation
Genes used encode the proteins eGFP and mCherry,36–37 which have distinct emission and excitation fluorescence peak wavelengths, and are encoded in peGFP-C1 plasmid (GenBank accession #: U55763, Clontech) or pmCherry-C1 (GenBank accession # not available, Clontech). Standard T7 promoter was inserted during DNA replications using Taq DNA polymerase. Replicated DNA was amplified by PCR and the products were purified with PCR Purification Kit (Qiagen). Concentration of DNA was determined by measuring optical absorbance at 260nm. mRNA was then transcribed from the DNA using PROTEINscript II T7 Kit (Applied Biosystems) and cleaned with a RNA cleaning kit (Qiagen). Template DNA remaining in the solution was degraded by DNase I Kit (Qiagen). Achieved mRNA was quantified by optical absorbance at 260nm and stored at −80°C. mRNA was used as a template for translation reaction using Retic Lysate IVT™ Kit (Applied Biosystems). 0.25μg of eGFP and/or 0.25 μg of mCherry mRNA, together with DNA, Au NP-DNA or Au NP-mPEG were mixed with a batch amount of the translation kit and incubated at 30°C for 1hr. Note that the amount of AuNP-DNA used in experiments of gene mixtures is ~2× the amount used in single gene experiment since 0.25μg of eGFP and/or 0.25μg of mCherry mRNA were put into the reaction, and the amount of AuNP was based on the total mass of mRNA, where both genes have similar molecular weight. After the incubation was finished, the samples were cooled down to 4°C and maintained at that temperature for > 12hrs. All the translation processes were performed using manufacturers’ protocols. Fluorescence of the samples were measured at λemission ~ 507nm (λexcitation = 488nm) for eGFP, and at λemission ~ 610nm (λexcitation = 587nm) for mCherry. Fluorescence spectra of the sample mixture that lacked mRNA substrate were identically subtracted from the data to collect the actual spectra of eGFP or mCherry protein only. Data were normalized with the fluorescence intensity of the resultant which was translated with only mRNA. When RNase H is applied to translation reaction, 1 unit amount of the enzyme as defined by the manufacturer (Applied Biosystems) was used for each experiment.
Gel electrophoresis of AuNP and AuNP-DNA
Agarose gel electrophoresis (1.5%) was performed in 0.5× TBE at E = 3.8 V/cm for 90 min. Gel was stained with Coomassie blue for ~2 hrs and de-stained for ~12 hrs.
Supplementary Material
Acknowledgments
We thank NIH NIBIB (R21 EB008156-01) for funding the project. We are grateful to A. Rich group in the Department of Biology at M.I.T. for use of their fluorometer, and to M.I.T. Center for Materials Science and Engineering for use of transmission electron microscopy.
Footnotes
SUPPORTING INFORMATION. Nanoparticle sizing, effects of fluorescence quenching, and free mPEG on expression, and additional experiments exploring the effect of RNase H is included in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.De M, Ghosh PS, Rotello VM. Applications of Nanoparticles in Biology. Adv Mater. 2008;20:4225–4241. [Google Scholar]
- 2.Alivisatos P. The Use of Nanocrystals in Biological Detection. Nat Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 3.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum Dot Bioconjugates for Imaging, Labelling and Sensing. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 4.Thomas M, Klibanov AM. Conjugation to Gold Nanoparticles Enhances Polyethylenimine’s Transfer of Plasmid DNA into Mammalian Cells. Proc Natl Acad Sci USA. 2003;100:9138–9143. doi: 10.1073/pnas.1233634100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CC, Lin YP, Wang CW, Tzeng HC, Wu CH, Chen YC, Chen CP, Chen LC, Wu YC. DNA-Gold Nanorod Conjugates for Remote Control of Localized Gene Expression by Near Infrared Irradiation. J Am Chem Soc. 2006;128:3709–3715. doi: 10.1021/ja0570180. [DOI] [PubMed] [Google Scholar]
- 6.Loo C, Lowery A, Halas N, West J, Drezek R. Immunotargeted Nanoshells for Integrated Cancer Imaging and Therapy. Nano Lett. 2005;5:709–711. doi: 10.1021/nl050127s. [DOI] [PubMed] [Google Scholar]
- 7.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum Dots for Live Cells in Vivo Imaging, and Diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barhoumi A, Huschka R, Bardhan R, Knight MW, Halas NJ. Light-Induced Release of DNA from Plasmon-Resonant Nanoparticles: Towards Light-Controlled Gene Therapy. Chem Phys Lett. 2009;482:171–179. [Google Scholar]
- 9.Braun GB, Pallaoro A, Wu G, Missirlis D, Zasadzinski JA, Tirrell M, Reich NO. Laser-Activated Gene Silencing via Gold Nanoshell-siRNA Conjugates. ACS Nano. 2009;3:2007–2015. doi: 10.1021/nn900469q. [DOI] [PubMed] [Google Scholar]
- 10.Lee SE, Liu GL, Kim F, Lee LP. Remote Optical Switch for Localized and Selective Control of Gene Interference. Nano Lett. 2009;9:562–570. doi: 10.1021/nl802689k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parak WJ, Pellegrino T, Micheel CM, Gerion D, Williams SC, Alivisatos AP. Conformation of Oligonucleotides Attached to Gold Nanocrystals Probed by Gel Electrophoresis. Nano Lett. 2003;3:33–36. [Google Scholar]
- 12.Aubin-Tam ME, Hwang W, Hamad-Schifferli K. Site-Directed Nanoparticle Labeling of Cytochrome c. Proc Natl Acad Sci. 2009;106:4095–4100. doi: 10.1073/pnas.0807299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nel AE, Madler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding Biophysicochemical Interactions at the Nano-Bio Interface. Nat Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 14.Kimura-Suda H, Petrovykh DY, Tarlov MJ, Whitman LJ. Base-Dependent Competitive Adsorption of Single-Stranded DNA on Gold. J Am Chem Soc. 2003;125:9014–9015. doi: 10.1021/ja035756n. [DOI] [PubMed] [Google Scholar]
- 15.Park S, Hamad-Schifferli K. Evaluation of Hydrodynamic Size and Zeta-potential of Surface-Modified Au Nanoparticle-DNA Conjugates via Ferguson Analysis. J Phys Chem C. 2008;112:7611–7616. [Google Scholar]
- 16.Brown KA, Park S, Hamad-Schifferli K. Nucleotide-Surface Interactions in DNA-Modified Au-Nanoparticle Conjugates: Sequence Effects on Reactivity and Hybridization. J Phys Chem C. 2008;112:7517–7521. [Google Scholar]
- 17.Lee SE, Sasaki DY, Perroud TD, Yoo D, Patel KD, Lee LP. Biologically Functional Cationic Phospholipid–Gold Nanoplasmonic Carriers of RNA. J Am Chem Soc. 2009;131:14066–14074. doi: 10.1021/ja904326j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W, Howarth M, Greytak AB, Zheng Y, Nocera DG, Ting AY, Bawendi MG. Compact Biocompatible Quantum Dots Functionalized for Cellular Imaging. J Am Chem Soc. 2008;130:1274–1284. doi: 10.1021/ja076069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng M, Li Z, Huang X. Ethylene Glycol Monolayer Protected Nanoparticles: Synthesis, Characterization, and Interactions with Biological Molecules. Langmuir. 2004;20:4226–4235. doi: 10.1021/la035981i. [DOI] [PubMed] [Google Scholar]
- 20.Park S, Brown KA, Hamad-Schifferli K. Changes in Oligonucleotide Conformation on Nanoparticle Surfaces by Modification with Mercaptohexanol. Nano Lett. 2004;4:1925–1929. [Google Scholar]
- 21.Aubin-Tam ME, Hamad-Schifferli K. Gold Nanoparticle-Cytochrome c Complexes: The Effect of Nanoparticle Ligand Charge on Protein Structure. Langmuir. 2005;21:12080–12084. doi: 10.1021/la052102e. [DOI] [PubMed] [Google Scholar]
- 22.Otsuka H, Nagasaki Y, Kataoka K. PEGylated Nanoparticles for Biological and Pharmaceutical Applications. Adv Drug Deliver Rev. 2003;55:403–419. doi: 10.1016/s0169-409x(02)00226-0. [DOI] [PubMed] [Google Scholar]
- 23.Choi HS, Liu W, Liu F, Nasr K, Misra P, Bawendi MG, Frangioni JV. Design Considerations for Tumor-Targeted Nanoparticles. Nat Nanotechnol. 2010;5:42–47. doi: 10.1038/nnano.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser CS, Doudna JA. Structural and Mechanistic Insights into Hepatitis C Viral Translation Initiation. Nat Rev Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 25.Fraser CS, Hershey JW, Doudna JA. The Pathway of Hepatitis C Virus mRNA Recruitment to the Human Ribosome. Nat Struct Mol Biol. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gingras AC, Raught B, Sonenberg N. eIF4 Initiation Factors: Effectors of mRNA Recruitment to Ribosomes and Regulators of Translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh PS, Kim CK, Han G, Forbes NS, Rotello VM. Efficient Gene Delivery Vectors by Tuning the Surface Charge Density of Amino Acid-Functionalized Gold Nanoparticles. ACS Nano. 2008;2:2213–2218. doi: 10.1021/nn800507t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubertret B, Calame M, Libchaber A. Single-Mismatch Detection Using Gold-Quenched Fluorescent Oligonucleotides. Nat Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 29.Jason TLH, Koropatnick J, Berg RW. Toxicology of Antisense Therapeutics. Toxicol Appl Pharmacol. 2004;201:66–83. doi: 10.1016/j.taap.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 30.Patil SD, Rhodes DG, Burgess DJD. NA-Based Therapeutics and DNA Delivery Systems: A Comprehensive Review. AAPSJ. 2005;7 doi: 10.1208/aapsj070109. Article 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 32.Walton SP, Stephanopoulos GN, Yarmush ML, Roth CM. Thermodynamic and Kinetic Characterization of Antisense Oligodeoxynucleotide Binding to a Structured mRNA. Biophys J. 2002;82:366–377. doi: 10.1016/S0006-3495(02)75401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 4. W. H. Freeman and Company; New York: 2005. pp. 288–289. [Google Scholar]
- 34.Beesley JE. Colloidal Gold: A New Perspective for Cytochemical Marking. 1. Oxford University Press; Oxford: 1989. [Google Scholar]
- 35.Demers LM, Mirkin CA, Mucic RC, Robert A, Reynolds I, Letsinger RL, Elghanian R, Viswanadham G. A Fluorescence-Based Method for Determining the Surface Coverage and Hybridization Efficiency of Thiol-Capped Oligonucleotides Bound to Gold Thin Films and Nanoparticles. Anal Chem. 2000;72:5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 36.Shaner NC, Steinbach PA, Tsien RY. A Guide to Choosing Fluorescent Proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 37.Chalfie M. GFP: Lighting up Life. Proc Natl Acad Sci USA. 2009:106. doi: 10.1073/pnas.0904061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.