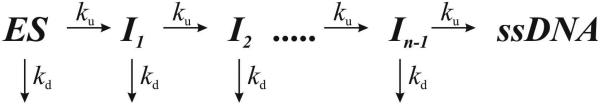Abstract
Helicases catalyze the separation of double-stranded nucleic acids to form single-stranded intermediates. Using transient state kinetic methods we have determined the kinetic properties of DNA unwinding under conditions that favor a monomeric form of the Dda helicase as well as conditions that allow multiple molecules to function on the same substrate. Multiple helicase molecules can align like a train on the DNA track. The number of base pairs unwound in a single binding event for Dda is increased from ~19 bp for the monomeric form to ~64 bp when as many as four Dda molecules are aligned on the same substrate, while the kinetic step-size (3.2 ± 0.7 bp) and unwinding rate (242 ± 25 bp s−1) appear to be independent of the number of Dda molecules present on a given substrate. The data support a model in which the helicase molecules bound to the same substrate move along the DNA track independently during DNA unwinding. The observed increase in processivity arises from the increased probability that at least one of the helicases will completely unwind the DNA prior to dissociation. These results are in contrast to previous reports in which multiple Dda molecules on the same track greatly enhanced the rate and amplitude for displacement of protein blocks on the track. Therefore, only when the progress of the lead molecule in the train is impeded by some type of block, such as a protein bound to DNA, do the trailing molecules interact with the lead molecule in order to overcome the block. The fact that trailing helicase molecules have little impact on the lead molecule in the train during routine DNA unwinding suggests that the trailing molecules are moving at similar rates as the lead molecule. This result implicates a step in the translocation mechanism as contributing greatly to the overall rate-limiting step for unwinding of duplex DNA.
The nature of nucleic acid metabolism requires enzymes that are able to catalyze the separation of double stranded helices to transiently form single stranded intermediates for the purpose of cellular events such as replication, recombination, repair, transcription, translation, and splicing. Helicases fulfill such a requirement by coupling energy associated with NTP hydrolysis to the manipulation of nucleic acid structure (1-5). They are ubiquitous in nature and function in coordination with highly regulated macromolecular complexes (1). The manner in which helicases achieve strand separation appears to be a variation upon a common theme, in which molecular motors power translocation and separation in a directionally biased manner. Different helicases are likely to have different mechanistic features. For some helicases, translocation has been described in terms of an inch-worm mechanism, in which strand-separation may take a form analogous to a ‘snow-plow’ or ‘wire-stripper’ (6). Other helicases are proposed to actively interact with the duplex region at a single-strand/double-strand junction to melt the DNA (7). At least one helicase, RecBCD, utilizes two molecular motors to translocate along each strand of the duplex during DNA unwinding (8-10). One motor moves 5′-to-3′ while the second motor moves 3′-to-5′, thereby ensuring very robust separation of the DNA. Variation in helicase mechanisms are also revealed by the oligomeric state of the functional enzyme, which can include monomers (11;12), dimers (13;14), and hexamers (15-19).
Unwinding of duplexes of varying length has led to several descriptors of the kinetic and physical constants associated with helicases. One of the most confusing values relates to the ‘step size’. The kinetic step size refers to the number of base pairs unwound prior to a rate limiting kinetic step. The physical step size refers to the number of base pairs that are unwound simultaneously. A helicase might unwind one base pair at a time (physical step of one), but then proceed through a slow conformational change that occurs every three base pairs, resulting in a kinetic step size of 3 bp. The chemical step size refers to the number of base pairs unwound per ATP hydrolyzed. In the simplest case, all of these values are equal to one.
A fundamental component of the kinetic mechanism describing helicase action is the number of base pairs that are unwound during a rate-limiting kinetic step. A method to measure this number, termed the kinetic step size, was developed by the Lohman laboratory, which has reported kinetic step sizes of 4 bp for the UvrD helicase and 6 bp for the RecBCD helicase (20-22). Recently, the large kinetic step size for UvrD was proposed to be made of smaller kinetic steps of ~ 1 bp, at least in regards to translocation on ssDNA (23;24). The kinetic step size of the hexameric helicase DnaB was reported as 1 bp (25), whereas the gene4 helicase from bacteriophage T7 exhibited a kinetic step size of 1.4 bp (26). Studies of the NS3 helicase indicated a large kinetic step size of 18 bp (27). More recent single molecule experiments have shown that the large kinetic step size consists of smaller steps of 3-4 bp (28), which are further made up of even smaller physical steps of one bp (29). Indeed, physical models for PcrA and UvrD helicases have been proposed based on multiple x-ray crystal structures in which the number of base pairs unwound per ATP hydrolysis event is one (7;30). The Type I restriction modification enzymes contain a helicase-like motor which moves along dsDNA with one bp step size with one ATP consumed per step (31;32). Hence, it appears that the fundamental, physical step size of many helicases and translocases is one bp.
Dda is one of three helicases encoded by the bacteriophage T4 genome and is believed to play a role in the initiation of DNA replication forks at a DNA origin of replication (33) as well as during replication fork progression (34;35). Biochemical characterization of Dda indicates that it can function as a monomeric molecular motor that does not readily form higher-order oligomeric species in solution (11;36). Dda translocates with a 5′-to-3′ directional bias and can remove several different types of protein blocks from its path (37-39). A ‘cooperative inchworm’ model was proposed to explain how multiple Dda molecules line up along ssDNA and function together to enhance displacement of streptavidin from biotin-labeled oligonucleotides (40). The enhanced activity of multiple Dda molecules does not appear to result from specific protein-protein interactions. Rather, the presence of multiple motors moving in the same direction on ssDNA is thought to increase force production and prevent movement backwards on the ssDNA, thereby driving streptavidin displacement. Multiple Dda molecules align along the DNA to enhance many enzymatic activities including DNA unwinding (41), displacement of tryp repressor from dsDNA (42), and translocation of Dda past chemically modified DNA (43). For some activities, such as displacement of streptavidin, it is clear that one Dda molecule ‘pushes’ another to produce greater force in the direction of translocation. However, it is not clear whether this mechanism applies to unwinding of dsDNA. A model whereby multiple helicase molecules can enhance DNA unwinding by simply increasing the probability that unwinding will occur may also account for the observed increase in activity. Such a model, termed functional cooperativity, has been proposed for the helicase domain of the Hepatitis C viral helicase, NS3 (44). The monomeric form of Dda exhibits a kinetic step size of ~ 3 bp per step and unwinds DNA at a rate of ~250 bp/s, albeit with low processivity (45). The objective of the current study was to determine how the processivity of Dda is increased when multiple molecules bind to the same substrate. The unwinding rate and step size were measured under conditions that favor binding of more than one helicase molecule per DNA substrate and models that allow for functional cooperativity were explicitly considered to analyze the data.
EXPERIMENTAL PROCEDURES
Materials
ATP (disodium salt) and Sephadex (G-25) were obtained from Sigma. HEPES, Na4EDTA, BME, BSA, Mg(OAc)2, KOAc, SDS, xylene cyanol, bromophenol blue, NaCl, glycerol, and KOH were obtained from Fisher. T4 polynucleotide kinase was purchased from New England Biolabs. [γ32P]ATP was purchased from New England Nuclear. DNA oligonucleotides (IDT) were purified by preparative polyacrylamide gel electrophoresis and stored in 10 mM HEPES (pH 7.5) and 1 mM EDTA. Recombinant Dda was overexpressed and purified from E. coli as previously described (36).
Helicase Substrates
Purified oligonucleotides were 5′-radiolabeled with T4 polynucleotide kinase at 37 °C for 1 hour. The kinase was inactivated by heating to 70 °C for 10 minutes, and unincorporated [γ32P]ATP was removed by passing the reaction mixture over two Sephadex G-25 columns. Helicase substrates were prepared by adding 1.2 equivalents of complement to the 5′-radiolabeled oligos, followed by heating to 95 °C for 5 minutes, and then slow cooling to room temperature.
Single-Turnover RQF Helicase Unwinding Experiments
Unwinding assays were performed with a Kintek rapid chemical quench-flow instrument (Kintek, Austin, TX) maintained at 25 °C with a circulating water bath. All concentrations listed are after mixing, unless otherwise stated. The helicase reaction buffer consisted of 25 mM HEPES pH 7.5, 0.1 mM Na4EDTA, 0.1 mg/ml BSA, 2 mM BME. Dda was diluted into 25 mM HEPES pH 7.5, 1 mM Na4EDTA, 0.1 mg/ml BSA, 2 mM BME, 50 mM NaCl, and 20% glycerol prior to performing the unwinding assays. For the enzyme limiting pre-steady-state experiments, Dda (final concentration of 25 nM) was incubated for 2-5 min with 100 nM radiolabeled DNA substrate and reaction buffer at 25 °C. For the excess enzyme experiments, Dda (final concentration of 100 nM) was incubated for 2-5 min with 10 nM radiolabeled DNA substrate and reaction buffer at 25 °C. Unless otherwise stated, the reaction was initiated by adding 5 mM ATP, 10 mM Mg(OAc)2. In order to prevent ssDNA product from re-forming dsDNA substrate, 300 nM re-annealing trap was placed in the receiving vial for each sample. In the enzyme limiting experiments, which contained 100 nM DNA substrate, 500 nM re-annealing trap was placed in the receiving vial, which was sufficient because only ~ 25 nM ssDNA product was produced under these conditions. The sequence of the annealing trap is complementary to the displaced strand of the substrate so that re-annealing of the displaced strand to the radiolabeled loading strand was prevented. For single-turnover conditions, 5 μM polydT was included in order to prevent Dda from re-binding to the radiolabeled substrate following the first catalytic turnover of substrate. The reaction mixture was rapidly mixed with 400 mM EDTA to quench the reaction following the allotted timeframe. 25 μl of the quenched solution was then added to 5 μl of non-denaturing gel loading buffer (0.1% bromophenol blue, 0.1% xylene cyanol in 6% glycerol). Finally, the dsDNA substrate was separated from ssDNA product on a 20% native polyacrylamide gel. Radiolabeled substrate and product were visualized by using a Molecular Dynamics Phosphorimager system and ImageQuant software. The quantity of radioactivity was used to determine the ratio of double stranded oligonucleotide substrate to single stranded oligonucleotide product as a function of time.
Non-linear Least Squares Analysis of Unwinding Data
Data fitting was performed using the program Scientist (Micromath, St. Louis, MO). The function fss(t) that describes the formation of ssDNA product as a function of n-steps for scheme 1 has been defined previously as equation 1 (21;46).
| equation 1 |
where kobs can be defined as the sum of the forward and dissociative rate constants, ku and kd (equation 2).
| equation 2 |
Processivity, P, is defined by equation 3, where m is the DNA unwinding step size and N is the average number of bp unwound.
| equation 3 |
The method of Laplace transforms has been used previously to solve the system differential equations for reaction schemes similar to those utilized in this study (21;46). The resulting expression Fss(s) is the Laplace transform of equation 1, describing the minimal reaction scheme that describes unwinding for a helicase that dissociates readily from the DNA substrate lengths used for in vitro unwinding assays.
| equation 4 |
where s is the Laplace variable of the fraction of ssDNA product formed over time, fss(t) (equation 1). The inverse Laplace transform, L −1, can be obtained using the numerical integration capabilities of Scientist to obtain fss(t) as shown in equation 5.
| equation 5 |
Scheme 1 assumes that each step in the series along the unwinding pathway is identical. The kinetic step-size (m) is then defined with equation 6.
| equation 6 |
where LT equals the total length of dsDNA in bp, and L0 equals the minimal length of dsDNA that is stable in the presence of an active helicase. The resulting individual kinetic step-size estimates may then be used to obtain an average kinetic step-size for the helicase under investigation.
Scheme 1.
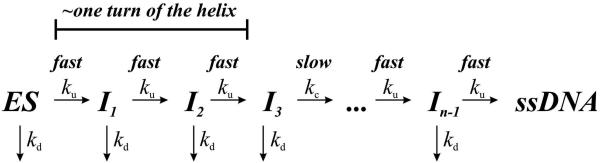
Scheme 2 proposes that a second step occurs h number of times during strand separation as a function of the forward rate constant kc. The inverse Laplace transform fss(t) for scheme 2 was obtained using equation 7, as described previously (21;46).
| equation 7 |
The value h may or may not be involved with strand separation and/or translocation. If it is assumed that h is not involved in either of these two properties then the kinetic step-size is defined only by the number of intermediates, n, that define the step-size (m) using equation 6.
Scheme 2.
Data Analysis by Kintek Global Explorer
The model for functional cooperativity was analyzed by fitting data using Kintek Global Kinetic Explorer (Kintek Corporation, Austin, TX) (47). The scripts used for the data analysis are shown in the supplemental data.
RESULTS
DNA unwinding by monomeric Dda helicase
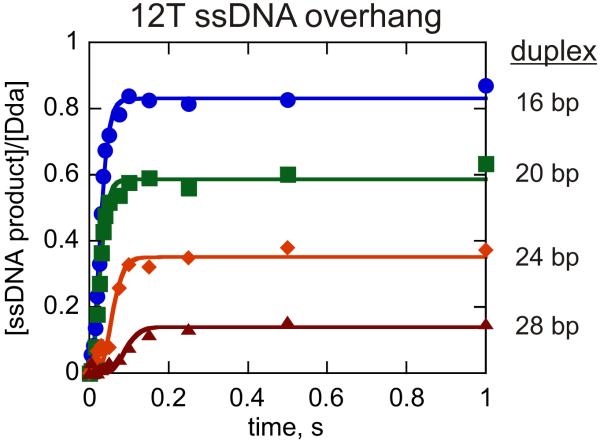
Previous work has evaluated DNA unwinding by monomeric Dda under conditions in which a DNA substrate was provided that was long enough to accommodate only one molecule of Dda (45). However, results indicated that binding of Dda to the duplex portion of the substrate may impede or reduce DNA unwinding under conditions where the helicase concentration exceeds the DNA substrate concentration (41). Here we have performed a kinetic analysis of DNA unwinding under conditions in which the concentration of DNA substrate is four-fold greater than Dda helicase in order to ensure that only one molecule of enzyme is bound to the substrate. Initial experiments were performed with substrate set 1 (Table 1). Previously, a 12 nt ssDNA overhang provided the best results for unwinding of a single length of dsDNA (41), however, a detailed kinetic analysis was not performed, and the effect of increasing duplex length was not evaluated. The DNA substrates used throughout this work are named based on the length of ssDNA overhang and the length of the duplex. For example, the DNA substrate with a 12 nt ssDNA overhang and a 16 bp dsDNA region is labeled as 12T16bp. Four substrates were examined in which the duplex contained 16, 20, 24, or 28 bp in order to measure the kinetic step-size and processivity of monomeric Dda. All four progress curves were fit simultaneously to equation 5 representing Scheme 1 using Scientist (Fig. 1, Table 2).
Table 1. DNA substrates.
| Substrate set 1 : 12T ssDNA overhang | |
| 12T16 | 5′-12t-cg ctg atg tcg cct gg 3′ |
| 3′gc gac tac agc gga cc 5′ | |
| 12T20 | 5′-12t-cg ctg atg tcg cct ggt acg 3′ |
| 3′gc gac tac agc gga cca tgc 5′ | |
| 12T24 | 5′-12t-cg ctg atg tcg cct ggt acg tcg c 3′ |
| 3′gc gac tac agc gga cca tgc agc g 5′ | |
| 12T28 | 5′-12t-cg ctg atg tcg cct ggt acg tcg ctg cc 3′ |
| 3′gc gac tac agc gga cca tgc agc gac gg 5′ | |
The “14T” ssDNA overhang substrates contain 14 thymidine residues with the 16 bp, 20 bp, 24 bp, and 28 bp duplex sequences shown above.
The “21T” ssDNA overhang substrates contain 21 thymidine residues with the 16 bp, 20 bp, 24 bp, and 28 bp duplex sequences shown above
The “28T” ssDNA overhang substrates contain 28 thymidine residues with the 16 bp, 20 bp, 24 bp, and 28 bp duplex sequences shown above.
Figure 1.
DNA unwinding by monomeric Dda. Unwinding was measured under conditions in which substrate concentration (100 nM) exceeded the helicase concentration (25 nM) to ensure that only one molecule of Dda was bound per substrate molecule. DNA substrates contained a 12nt overhang of ssDNA and varying lengths of duplex including 12T16bp ( ), 12T20bp (
), 12T20bp ( ), 12T24bp (
), 12T24bp ( ), and 12T28bp (
), and 12T28bp ( ). The experiments were performed in the presence of 5 μM polydT to create single-turnover conditions. The data were fit to equation 5 using Scientist and the resulting kinetic parameters are listed in Table 2.
). The experiments were performed in the presence of 5 μM polydT to create single-turnover conditions. The data were fit to equation 5 using Scientist and the resulting kinetic parameters are listed in Table 2.
Table 2. Kinetic parameters determined from NLLS analysis of data in Fig. 1 for the substrate containing a 12 nt overhang (one molecule of Dda per substrate).
| ku, s−1 | step-size (m), bp | unwinding rate (mku), bp s−1 | Na |
| 107 ± 12 | 2.4 ± 0.7 | 256 ± 74 | 19 bp |
|
| |||
| Individual kd values (s−1) for different lengths of dsDNA | |||
| 12T16bp | 12T20bp | 12T24bp | 12T28bp |
| 6.0 ± 0.5 | 17.0 ± 1.0 | 16.2 ± 1.1 | 19.4 ± 2.6 |
DNA unwinding produced steadily less product as the duplex length increased from 16bp up to 28 bp (Fig. 1). The kd values were allowed to float for each substrate. The rationale for this was based on the idea that longer duplexes may actually present a greater opposing force than shorter duplexes as previously suggested (26). Hence, longer duplexes might exhibit a different dissociation rate than shorter duplexes. All kinetic parameters that use the length of dsDNA in the calculation assumed that the final 8 bp melt spontaneously. For example, in order to calculate the step-size for the 12T16bp substrate, we assume that Dda is catalyzing the separation 8 bp of the initial 16 bp. The 8 bp value is based on previous results with methylphosphonate modified substrates, which indicated that the last few bp melt spontaneously during Dda-catalyzed DNA unwinding (45). The value for the unwinding rate, ku, is the rate constant for each kinetic step, which can be multiplied by the number of base pairs unwound per step to obtain an overall unwinding rate constant ku,bp. The step-size, 2.4 ± 0.7 bp, and the overall unwinding rate constant, 256 ± 74 bp s−1, are similar to values obtained previously with a substrate that can accommodate one molecule of Dda on the ssDNA overhang (45). The small increase in the kd values obtained here for longer duplexes (Table 2) may reflect the greater opposing force presented by the longer regions of duplex DNA (26). We conclude that DNA unwinding by monomeric Dda under conditions in which substrate concentration exceeds enzyme concentration proceeds with similar kinetic constants, ku, kd and kinetic step size (m) as were observed under conditions in which the enzyme concentration exceeded the substrate concentration (45).
Protein-protein interactions are not required to account for increased processivity during DNA unwinding by Dda
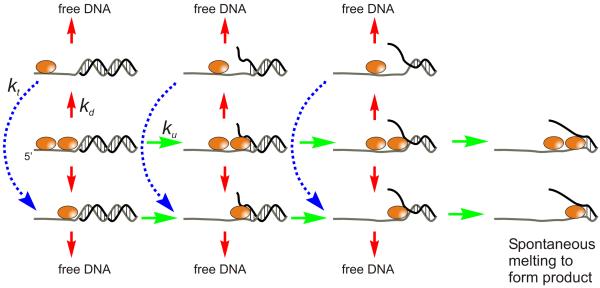
It is known that multiple molecules of Dda bound to the same DNA substrate give rise to increased processivity for DNA unwinding (41), however, the mechanism for the increased processivity is not clear. Processivity (P) is generally controlled by the rate for DNA unwinding, ku, and the rate for dissociation from the DNA substrate, kd according to the equation P = ku/(kd + ku). The increase in processivity could arise from an increase in unwinding, decrease in dissociation, or both. It is also possible that the increase in processivity is due to functional cooperativity, whereby the probability of one molecule of Dda successfully unwinding a DNA substrate is increased when multiple molecules start on the same substrate (44). In this case, processivity is increased, but neither ku nor kd are necessarily altered. Figure 2 illustrates how functional cooperativity can allow multiple opportunities for unwinding to occur, thereby increasing processivity without changing rate constants. The model shows two molecules of helicase bound to the ssDNA overhang. Three kinetic steps lead to unwinding of sufficient base pairs to allow spontaneous melting of the remaining base pairs (25;45). The model allows the leading or trailing molecule to dissociate independently of one another at any step during the reaction. If the leading molecule dissociates, then the trailing molecule must translocate to the ss/dsDNA junction for unwinding to continue. Translocation steps may not occur at the same rate as the steps for unwinding (23;24), but in the case of Dda, translocation rates are very similar to unwinding rates (Byrd, Matlock and Raney, in preparation). Therefore, each stepping rate, unwinding (ku) and translocation (kt), was made equal in this model.
Figure 2.
Model for functional cooperativity for Dda helicase. Two Dda molecules are shown bound to the 14T16bp substrate (14 nt overhang and 16 bp of duplex DNA). Upon addition of ATP and Mg+2, DNA unwinding occurs according to rate constant ku (green arrows). Three kinetic steps are shown for unwinding of the 16 bp substrate (11). Either enzyme molecule can dissociate from the DNA substrate according to rate constant kd (red arrows). If the leading enzyme molecule dissociates, then the trailing molecule can translocate to the ss/ds junction according to rate constant kt. The blue dotted lines and arrows indicate two kinetic steps that are required for the trailing enzyme to move to the ss/ds DNA junction. The final base pairs can melt spontaneously giving rise to ssDNA product.
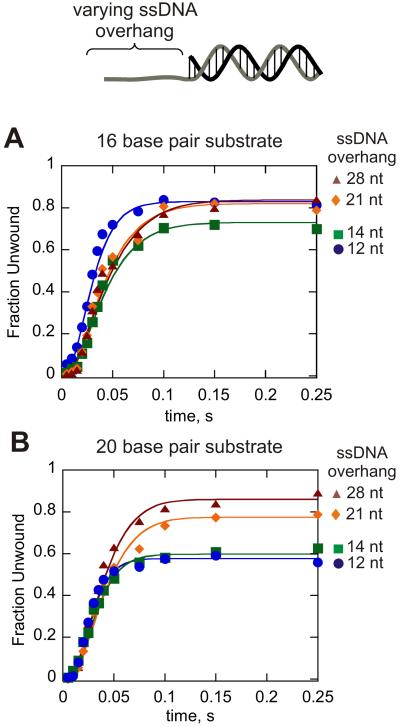
DNA unwinding was conducted with substrates containing varying length ssDNA overhangs of 12, 14, 21, and 28 nt and duplex lengths of 16 and 20 bp (Figure 3). Longer ssDNA overhang lengths were largely based on fluorescence titration data suggesting that a single Dda molecule occupies 6-7 nt (40). Also, substrates with an overhang of less than 6 nt do not exhibit significant unwinding in vitro (41). Kinetic mechanisms were defined in which one, two, three, or four molecules of Dda were bound to the substrates containing 12, 14, 21, and 28 nt overhangs, respectively. Three or four unwinding steps were designated for the 16 bp or 20 bp substrates, respectively based on the average kinetic step size of 3.2 bp and the number of base pairs that spontaneously melt. For example, 8 base pairs melt spontaneously, so only 8 bp from the 16 bp substrate and 12 bp from the 20 bp substrate are considered in the mechanism. Progress curves for each substrate were fit individually to their respective kinetic models by using Kintek Global Kinetic Explorer (47), allowing the rate constants ku and kd to float. The resulting kinetic parameters are listed in Table 3. The dissociation rate constants vary by four-fold (5.6 ± 1.2 s−1 to 25 ± 3.4 s−1). It has been suggested that longer duplexes may present a greater force opposing the movement of a helicase (26), which might account for the small variation observed here when comparing a 16bp to a 20bp duplex. The rate constant for each kinetic step, ku, ranged from 65 ± 5 s−1 to 116 ± 6 s−1. The small increase in ku may relate to the fact that the exact number of kinetic steps may be slightly less than 3 for the 16 bp substrate and slightly more than 4 steps for the 20 bp substrate. However, the overall consistency in the unwinding rate constants within each substrate set (16 bp duplexes and 20 bp duplexes) indicates that ku does not increase as more molecules of Dda are added to the substrate.
Figure 3.
Dda helicase-catalyzed unwinding of 16 bp (A) and 20 bp (B) substrates containing varying length ssDNA overhang. For all substrates containing 12 nt of ssDNA, the DNA substrate concentration (100 nM) exceeded the concentration of Dda (25 nM) and the quantity of product was divided by the enzyme concentration to obtain the fraction unwound. These conditions were chosen to ensure that only one molecule of Dda was bound to the substrate. For all other substrates, the concentration of Dda (100 nM) exceeded the DNA substrate concentration (10 nM) in order to ensure that multiple molecules were bound to the ssDNA overhangs. All experiments were performed in the presence of 5 μM poly dT to create single-turnover conditions with respect to the DNA substrate. The lengths of the ssDNA overhangs are listed in the figure. Data were fit to a model as depicted in Figure 2 by using the program Kintek Global Kinetic Explorer (Kintek Corp.). Three or four kinetic steps for unwinding were used for the unwinding step with the 16 bp substrate and 20 bp substrate, respectively. One, two, three, or four molecules of Dda were pre-bound to the DNA substrate for the 12 nt, 14 nt, 21 nt, and 28 nt substrates, respectively. The rate constant for translocation was set equal to that for DNA unwinding. The rate constants for DNA unwinding, ku, and dissociation, kd, were allowed to float for each substrate to obtain the best fit for each mechanism. The resulting rate constants are shown in Table 3.
Table 3. Kinetic parameters determined from fitting data in Figure 3 to the model for functional cooperativity as depicted in Figure 2.*.
| substrate | number of Dda molecules bound$ |
ku, s−1 | kd, s−1 |
|---|---|---|---|
| 12T16bp | 1 | 87 ± 6 | 5.6 ± 1.2 |
| 14T16bp | 2 | 65 ± 5 | 13 ± 2.0 |
| 21T16bp | 3 | 65 ± 5 | 9.5 ± 2.2 |
| 28T16bp | 4 | 76 ± 9 | 23 ± 7.3 |
| 12T20bp | 1 | 114 ± 5 | 17 ± 1.4 |
| 14T20bp | 2 | 110 ± 4 | 24 ± 1.5 |
| 21T20bp | 3 | 103 ± 6 | 23 ± 3.1 |
| 28T20bp | 4 | 116 ± 6 | 25 ± 3.4 |
Data were fit by using Kintek Global Kinetic Explorer (Kintek Corp.). Kinetic schemes were based on the model from Figure 2, which depicts two molecules bound to the substrate. Three or four unwinding steps were used for the 16bp and 20bp substrates, respectively. More complex kinetic mechanisms were used for substrates containing three or four Dda molecules (Supplementary data). Errors are standard errors obtained from the best fit of the data.
The number of Dda molecules bound per substrate is estimated from the binding site size and the length of the ssDNA overhang. In the case of the 12T substrates, the concentration of DNA substrate was in four-fold excess over the concentration of enzyme to ensure binding of one molecule of Dda per substrate molecule.
We conclude that the model depicted in Figure 2 for functional cooperativity readily accounts for the increased processivity observed when multiple molecules of Dda unwind the same DNA substrate. The significance of this model is in the fact that protein-protein interactions are not required to account for the observed increase in product formation. This result is in contrast to previous results for streptavidin-displacement activity exhibited by Dda, which clearly requires trailing helicase molecules to ‘push’ the lead molecule in order to push streptavidin from biotin-labeled DNA (40).
The kinetic step size for multiple Dda molecules is similar to that of monomeric Dda
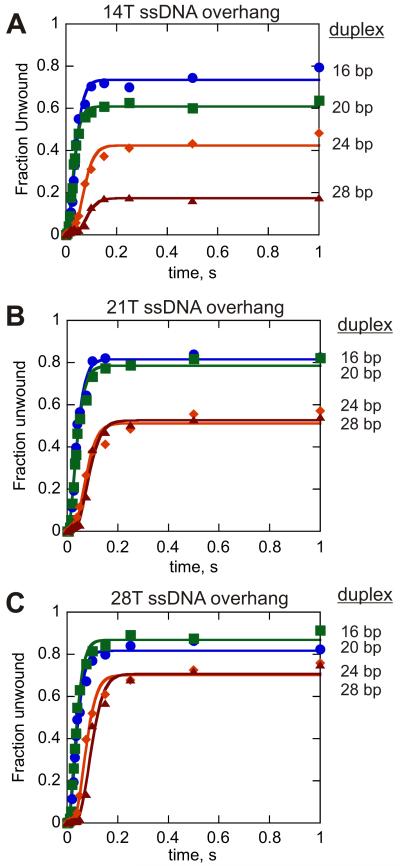
In addition to the ku and kd values, the kinetic step size, m, also defines the kinetic mechanism for helicase-catalyzed unwinding. The kinetic step size can be determined by measuring DNA unwinding with DNA substrates of increasing duplex length (20;21). In previous work, we found that the kinetic step size varied from 3.5 to 7.0 bp unless one accounted for spontaneous melting of the last few base pairs of the substrate (45). The final 8 bp of the DNA were observed to melt spontaneous, and when this was taken into account, the step size varied from 2.5 to 3.5 bp for different substrate lengths. Therefore, spontaneous melting of the last 8 bp was included in the analysis of all substrates in this work. Single-turnover RQF experiments were performed with substrates possessing a 14 nt ssDNA overhang (substrate set 2). Enzyme concentration greatly exceeded the substrate concentration so that more than one molecule of Dda binds to the substrate prior to initiation of the reaction. Two molecules of Dda should be able to bind to the 14nt overhang based on the binding site size of 6-7 nt. Four substrates containing a 14 nt ssDNA overhang and increasing lengths of dsDNA were examined (Fig. 4A). All four data sets were fit simultaneously to equation 5 describing reaction Scheme 1. The rate constant ku was constrained to be identical for all data sets, while the kd value and number of steps, n, were allowed to float. The resulting kinetic parameters are listed in Table 4. The step-size, 3.1 ± 0.9 bp, and the value for ku, 66 ± 7, were similar to those obtained under enzyme-limiting conditions (45). Small variations in the kd values were also observed, although no clear trend was evident with the different substrates (Table 4).
Figure 4.
Single-turnover results for Dda-catalyzed unwinding of DNA substrates possessing 14, 21, and 28 nt ssDNA overhangs. A. Results for 100 nM Dda-catalyzed unwinding of 10 nM 14T16bp ( ), 14T20bp (
), 14T20bp ( ), 14T24bp (
), 14T24bp ( ), and 14T28bp (
), and 14T28bp ( ) DNA substrates, respectively. The data were fit to equation 5 using Scientist and the resulting kinetic parameters are listed in Table 4. B. Results for 100 nM Dda-catalyzed unwinding of 10 nM 21T16bp (
) DNA substrates, respectively. The data were fit to equation 5 using Scientist and the resulting kinetic parameters are listed in Table 4. B. Results for 100 nM Dda-catalyzed unwinding of 10 nM 21T16bp ( ), 21T20bp (
), 21T20bp ( ), 21T24bp (
), 21T24bp ( ), and 21T28bp (
), and 21T28bp ( ) DNA substrates, respectively. The experiments were performed in the presence of 5 μM polydT to create single-turnover conditions. The data were fit to equation 7 using Scientist and the resulting kinetic parameters are listed in Table 5. C. Results for 100 nM Dda-catalyzed unwinding of 10 nM 28T16bp (
) DNA substrates, respectively. The experiments were performed in the presence of 5 μM polydT to create single-turnover conditions. The data were fit to equation 7 using Scientist and the resulting kinetic parameters are listed in Table 5. C. Results for 100 nM Dda-catalyzed unwinding of 10 nM 28T16bp ( ), 28T20bp (
), 28T20bp ( ), 28T24bp (
), 28T24bp ( ), and 28T28bp (
), and 28T28bp ( ) DNA substrates, respectively. The experiments were performed in the presence of 5 μM polydT to create single-turnover conditions. The data were fit to equation 7 using Scientist and the resulting kinetic parameters are listed in Table 6.
) DNA substrates, respectively. The experiments were performed in the presence of 5 μM polydT to create single-turnover conditions. The data were fit to equation 7 using Scientist and the resulting kinetic parameters are listed in Table 6.
Table 4. Kinetic parameters determined from NLLS analysis of data in Fig. 4A for the substrate containing a 14 nt overhang (two molecules of Dda per substrate).
| ku, s−1 | step-size (m), bp | unwinding rate (mku), bp s−1 | Na (bp per binding event) |
| 66±7 | 3.1±0.9 | 206 ± 58 | 18 bp |
|
| |||
| Individual kd values (s−1) for different lengths of dsDNA : | |||
| 14T16bp | 14T20bp | 14T24bp | 14T28bp |
| 6.9 ± 0.4 | 13.4 ± 0.7 | 10.5 ± 0.6 | 17.7 ± 1.8 |
Dda-catalyzed unwinding of DNA substrates with ≥ 21 nt ssDNA overhang lengths
Next, single-turnover RQF experiments were performed with substrates possessing 21 nt and 28 nt ssDNA overhangs (substrate sets 3 and 4, respectively) under excess-enzyme conditions (Fig. 4B, 4C). These conditions should favor binding of 3 or 4 molecules of Dda to the 21nt and 28nt substrates, respectively. One of the most obvious differences between the data for longer ssDNA overhangs and those obtained under conditions that favor a monomer is the increased amplitude of product formation, especially for the longer duplexes (compare unwinding for the 28 bp substrate in Figure 4C to the same length in Figure 4A).
Another intriguing observation resides in the fact that product curves for substrates containing long ssDNA overhangs exhibit a discontinuous change in the lag phases of different lengths of dsDNA. For example, there is a similar lag phase for the 21T16 bp and 21T20 bp substrates, but the lag phase for the 21T24 bp and 21T28 bp substrates is much longer (Fig. 4B and 4C). Hence, the four substrates containing the 21nt overhang appear to fall into two distinct populations, as do the four substrates containing the 28nt overhang. If the helicase reaction scheme is defined by a series of ‘fast’ steps followed by a rate-limiting ‘slow’ step, then as the rate of each ‘fast’ step approaches infinity, unwinding curves for substrates with the same number of ‘slow’ steps will become super-imposable (21). Such a phenomenon has been described for the HCV NS3 helicase using a combinatorial, time-resolved unwinding assay with single bp resolution (48), and may explain the data here for Dda. Substrates that can accommodate 3-4 Dda molecules (i.e. 21 nt and 28 nt ssDNA overhang) clearly exhibit a trend in which the unwinding curves for substrates of different physical lengths become super-imposable as the formation of ssDNA product becomes limited by a ‘slow’ step. Consequently, the reaction scheme defining DNA unwinding by multiple Dda molecules was expanded to include a non-identical step along the reaction pathway (Scheme 2), where the “slow” step is defined by the rate constant kc. The unwinding results for the 21T and 28T ssDNA overhang substrates were fit to equation 7 representing Scheme 2 and the resulting kinetic parameters are listed in Tables 5 and 6, respectively.
Table 5. Kinetic parameters determined from NLLS analysis of data from Fig. 4B for the substrate containing a 21nt overhang (three molecules of Dda per substrate).
| ku, s−1 | kc, s−1 | step-size (m), bp | unwinding rate (mku), bp s−1 | Na (bp per binding event) |
| 63±8 | 20±7 | 4.1±0.9 | 261 ± 59 | 47 bp |
|
| ||||
| Individual kd values (s−1) for different lengths of dsDNA | ||||
| 21T16bp | 21T20bp | 21T24bp | 21T28bp | |
| 4.8 ± 0.5 | 5.6 ± 0.5 | 6.9 ± 0.8 | 6.0 ± 0.6 | |
Table 6. Kinetic parameters determined from NLLS analysis of data from Fig. 4C for the substrate containing a 28 nt overhang (four molecules of Dda per substrate).
| ku, s−1 | kc, s−1 | step-size (m), bp | unwinding rate (mku), bp s−1 | Na (bp per binding event) |
| 72±9 | 34±13 | 3.4 ± 0.7 | 248 ± 48 | 64 bp |
|
| ||||
| Individual kd values (s−1) for different lengths of dsDNA | ||||
| 28T16bp | 28T20bp | 28T24bp | 28T28bp | |
| 4.8 ± 0.5 | 3.6 ± 0.5 | 4.1 ± 0.4 | 3.2 ± 0.3 | |
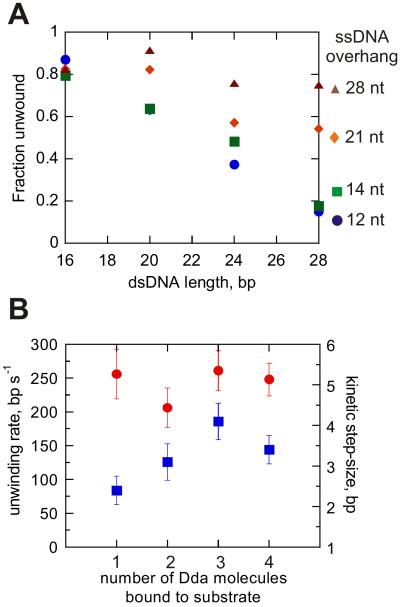
The amplitudes for product formation are plotted for each of the substrate sets in Figure 5A. The trend in the data clearly shows how the substrates with longer ssDNA overhangs which can accommodate more Dda molecules give rise to more product. The number of base pairs unwound per binding event can be estimated by fitting the data to a linear function and then solving for y = 0. Unwinding per binding event is increased from ~30 bp for the monomeric enzyme to ~110 bp for multiple-molecules bound to the 28 nt ssDNA overhang (calculated based on the 28 bp duplex).
Figure 5.
Multiple Dda molecules increase the amplitude of ssDNA formed, but do not change the kinetic step size, or the rate for unwinding. A. Re-plot of product amplitudes as a function of dsDNA length for the 12T ( ), 14T (
), 14T ( ), 21T (
), 21T ( ), and 28T (
), and 28T ( ) substrate sets. B. Re-plot of the unwinding rate constant (
) substrate sets. B. Re-plot of the unwinding rate constant ( ) and the kinetic step-size (
) and the kinetic step-size ( ) as a function of the estimated number of Dda molecules bound to the different substrates at the start of the reaction. One, two, three, or four Dda molecules were bound to substrate 12T, 14T, 21T, and 28T, respectively. Error bars are the errors in the fits to the data.
) as a function of the estimated number of Dda molecules bound to the different substrates at the start of the reaction. One, two, three, or four Dda molecules were bound to substrate 12T, 14T, 21T, and 28T, respectively. Error bars are the errors in the fits to the data.
Figure 5B shows the unwinding rate constants and step-size values plotted as a function of the number of Dda molecules bound to substrate. The unwinding rate constants remain similar, regardless of the number of Dda molecules that are initially bound to the substrate. No clear trend is observed in the kinetic step-size values as a function of the number of Dda molecules bound. Hence, the major role of multiple Dda molecules in DNA unwinding is to provide increased processivity, by increasing the probability for the helicase to complete unwinding prior to dissociation.
The stepwise kinetic mechanism for multiple Dda molecules proceeds through a non-uniform ‘pause’ characterized by an intermediate species
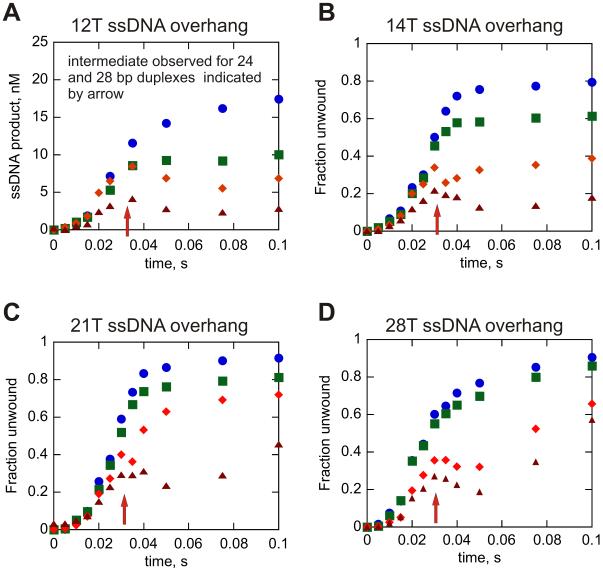
A single-stranded oligonucleotide that is complementary to the strand that is displaced by the helicase must be introduced into the standard in vitro unwinding assay in order to prevent spontaneous re-annealing of the ssDNA products. This oligonucleotide is referred to as a re-annealing trap. By increasing the amount of re-annealing trap present in solution, we were previously able to observe the transient appearance of an intermediate, which was correlated with a ‘pause’ in Dda-catalyzed unwinding occurring as a function of dsDNA length (45). This intermediate species was observed with 24 bp and 28 bp DNA substrates, but not with shorter 16 bp and 20 bp substrates. Therefore, the non-identical step, or pause, takes place after unwinding ≥ 20 bp. Since Dda only catalytically separates ~12 bp of a 20 bp substrate (taking into account spontaneous melting of the final 8 bp), ‘pausing’ was proposed to occur once every ~10 bp.
The appearance of the non-uniform step, as shown by the intermediate, was investigated by using the 12T substrates under conditions that favor monomeric Dda (substrate concentration > enzyme concentration). The concentration of annealing trap was increased from 300 nM to 5 μM and the results clearly show the appearance of the intermediate, but only with the duplex substrates of 24 and 28 bp (Fig. 6A). Since the data with the 12T substrates were obtained under conditions in which the substrate concentration exceeds the enzyme concentration, the appearance of the intermediate clearly shows that monomeric Dda undergoes the non-uniform step in the kinetic mechanism.
Figure 6.
A transient increase in the amount of ssDNA product in the presence of excess re-annealing trap suggests a slow step for unwinding of longer duplexes. All of the experiments shown were performed with 5 μM re-annealing trap in the reaction. The transient peak at around 0.035 s is observed for all of the 24 and 28 bp dsDNA lengths tested. Panel A shows the results of enzyme-limiting conditions (25 nM Dda and 100 nM DNA); all of the other experiments (panels B-D) were performed under excess enzyme conditions (100 nM Dda and 10 nM DNA). All experiments were initiated with 5 mM ATP and 10 mM Mg(OAc)2. Each panel uses the same symbol for each length of dsDNA: 16 bp ( ), 20 bp (
), 20 bp ( ), 24 bp (
), 24 bp ( ), and 28 bp (
), and 28 bp ( ).
).
In order to determine whether the intermediate appeared under conditions in which multiple Dda molecules bind to the same substrate, DNA unwinding assays were performed with the substrates containing longer ssDNA overhangs under conditions that favor binding of multiple Dda molecules per substrate (enzyme concentration > substrate concentration). A large excess of annealing trap was included (500-fold excess) in these experiments and single-turnover RQF experiments were performed. The appearance of the intermediate was observed for all ssDNA overhang lengths tested, but only for dsDNA lengths of 24 bp and 28 bp (Fig. 6B-6D). Thus, multiple Dda molecules aligned on the same substrate exhibit a similar non-uniform step in the unwinding mechanism as monomeric Dda.
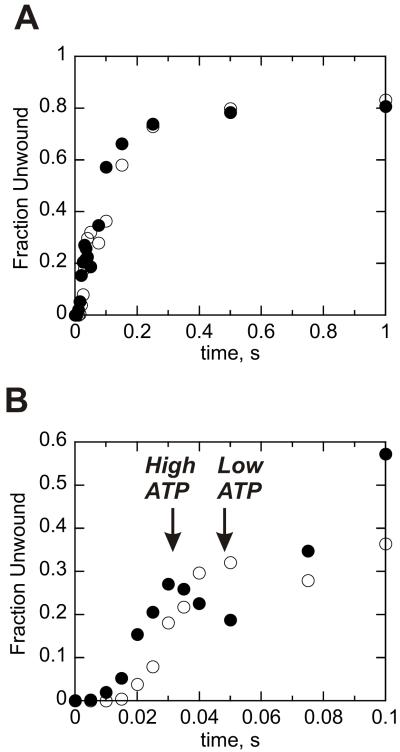
The intermediate appears if duplex lengths are sufficiently long, indicating that Dda must unwind several base pairs prior to undergoing the slower step. If this intermediate is related directly to DNA unwinding, then its appearance should be delayed under conditions in which unwinding is slowed. To determine whether the intermediate appears later in the progress curve under conditions of slower unwinding, the reaction was performed at 100 μM ATP. Unwinding by multiple Dda molecules is slowed under these conditions, but the processivity remains similar based on the amplitude of product formation (Fig. 7A). However, the appearance of the intermediate is delayed when the concentration of ATP is reduced, indicating that the intermediate is directly related to DNA unwinding and to the number of base pairs unwound (Fig. 7B, open circles). The presence of the intermediate provides evidence that the non-uniform step occurs even when multiple Dda molecules are involved in DNA unwinding.
Figure 7.
Dda-catalyzed unwinding of a 28T28bp substrate in the presence of 500-fold excess re-annealing trap results in the appearance of an intermediate species. A. Results for 100 nM Dda-catalyzed unwinding of 10 nM 28T28bp substrate in the presence of 500-fold excess re-annealing trap and ATP concentrations of either 5 mM (●) or 100 μM (○). Appearance of the transient peak in product formation is delayed when ATP concentration is reduced. B. Results in panel 7A are shown to 0.1 seconds.
DISCUSSION
Functional cooperativity without protein-protein interactions can account for the increased processivity exhibited by multiple Dda molecules
Previous work in which multiple Dda molecules were placed on the same substrate molecule indicated that Dda molecules “pushed” one another upon encountering a blockade along the DNA. For example, when biotin/streptavidin blocks are placed in the path of Dda, the rate of streptavidin displacement is increased by ~ one million fold when 5-6 molecules of Dda push together compared to monomeric Dda (38). Similarly, monomeric Dda was unable to displace trp repressor protein bound to DNA under single cycle conditions, whereas two or more Dda molecules readily pushed the protein off of the DNA (42). Perturbations in the DNA structure such as abasic sites blocked DNA unwinding by monomeric Dda, but were overcome by the presence of two or more molecules bound to the same substrate (43). Hence, one molecule of Dda can push another molecule through a block in the path of the helicase.
Adding additional molecules of Dda to the same DNA substrate results in increased product formation, however the kinetic mechanism for this increase was not known until now. Multiple molecules may lead to increased unwinding rates, decreased dissociation rates, or both. It is also possible that the observed increase in product formation results from functional cooperativity (44), whereby the unwinding rates and dissociation rates need not change, but unwinding is increased by the increase in the probability that at least one helicase molecule completes the unwinding process. Data in this report favor a mechanism in which the increase in product is accounted for simply due to the increased probability that at least one molecule completes the unwinding process. This phenomenon has been referred to as ‘functional cooperativity’ (44). Cooperativity is generally associated with protein-protein interactions that give rise to enhanced activity within an enzyme active site. In functional cooperativity, no protein-protein interactions are required because of the nature of the DNA substrate. Helicases can simply align on the same substrate molecule, thereby increasing the likelihood of that DNA duplex being unwound by mass action of the bound helicases. However, protein-protein interactions may contribute to functional cooperativity whereby a trailing helicase molecule engages a leading molecule on the same DNA track. Whether the trailing helicase pushes the lead helicase, or simply prevents the lead helicase from slipping back has not been completely addressed. However, loading of an ATPase-deficient form of Dda on the same track as active molecules failed to enhance the rate of streptavidin displacement (40).
The rate constants that control DNA unwinding processivity, as well as the kinetic step size, were determined under conditions in which only one molecule of Dda could bind to the DNA substrate (Fig. 1, Table 2). The kinetic values were then used to construct a model for functional cooperativity in which one, two, three, or four molecules of Dda were bound to the same DNA substrate. The data obtained from these experiments were readily fit by the model for functional cooperativity (Figs. 2 and 3, Table 3). No trends were observed in resulting unwinding rate constants as a function of increasing numbers of Dda molecules on the same DNA substrate. Hence, for regular DNA unwinding, the inherent unwinding activity of Dda is not activated by the presence of multiple Dda molecules. This result is in sharp contrast to the results obtained for protein displacement or for unwinding of DNA that contains chemical perturbations. We conclude that Dda molecules move along the DNA independently, until the lead molecule encounters a block to translocation. At that point, multiple molecules ‘pile up’ and begin to push one another, resulting in displacement of the block or movement passed the block.
The data are also in contrast to helicases such as E. coli Rep (49), and E. coli UvrD (13), and B. stearothermophilus PcrA (50;51) which are highly activated for DNA unwinding upon formation of dimeric or higher order oligomers. Recent work indicates that PcrA helicase can be activated by the presence of a DNA binding protein as well. Processivity for DNA unwinding by PcrA increases dramatically in the presence of the initiator protein for plasmid replication, RepD (52).
We found that small variations in the dissociation constant were observed for monomeric Dda as the length of the duplex increases (Table 1). This may be due to the increased stability exhibited by longer duplexes, which has been described in terms of an opposing force to the helicase (26). Dda is especially sensitive to opposing forces encountered during translocation on DNA as shown by the previously mentioned results for displacement of the trp repressor bound to DNA in which monomeric Dda could not displace the protein. Here we observed a small increase in dissociation constants when the length of the duplex was increased from 16 bp to 28 bp, which may be due to an increased opposing force caused by greater strand rigidity in the longer duplexes (53).
A non-uniform step in the kinetic mechanism is observed for multiple Dda molecules
A feature of the kinetic mechanism for unwinding by Dda is the appearance of an intermediate species that can be observed by increasing the concentration of annealing trap in the reaction (45). Analysis of our data provides two lines of evidence for the existence of a non-uniform step in the kinetic mechanism of Dda, even when multiple molecules are present on the same substrate. First, using a ssDNA overhang of ≥21 nt results in an unusual trend in the amount of time required to unwind substrates (i.e. the lag phase) of different dsDNA lengths. For example, the 21T16 and 21T20 substrates are unwound in a similar amount of time and to a similar extent (Fig. 4B). The lag phases for Dda-catalyzed unwinding of the 21T24 and 21T28 substrates are also very similar, but Dda requires a much longer period of time for product to form relative to the 21T16 and 21T20 substrates. Such a large difference in lag phases for 20 bp and 24 bp substrates strongly suggests that a slower, rate-limiting event takes place somewhere between separation of 20 bp and 24 bp lengths. Second, an intermediate species is clearly observed for Dda-catalyzed separation of ≥24 bp of dsDNA (Fig. 6). Taken together, the two results reported here support a mechanism in which Dda exhibits a non-uniform step that occurs as a function of dsDNA length (Scheme 2).
Others have described non-uniform stepping in the case of DNA unwinding by RecBCD (46), RNA unwinding by NS3 (48), and translocation on ssDNA for UvrD (24). Previously, we suggested that this step for Dda involved a “hand-off” of the displaced strand during DNA unwinding (24;45). Dda may interact with both strands of the duplex during the unwinding reaction, and after unwinding of around one turn of the duplex, the displaced strand is released. Reports on NS3-catalyzed RNA unwinding have proposed a similar mechanism by which the displaced strand remains bound to the enzyme during several sub-steps of unwinding (27). Release of the displaced strand is proposed to limit the overall unwinding cycle, resulting in a slow kinetic step that gives rise to a large kinetic step size. In the case of UvrD, the pause in the kinetic mechanism was observed during translocation on ssDNA, so a different mechanism must be operating to produce the non-uniform step. Movement of UvrD helicase domains during ATP hydrolysis was proposed to “scrunch” the ssDNA between the domains, thereby leading to a slow step during which this ssDNA was released (24). It appears that non-uniform steps in the kinetic mechanisms of helicases occur within several different sub-families of helicases, but different mechanisms may give rise to these steps.
The alignment of Dda molecules on DNA has a greater impact when translocation or DNA unwinding is impeded when compared to routine unwinding of dsDNA
The rate constant for streptavidin displacement increases by 75-fold when comparing one molecule vs. 2 molecules of Dda aligned on the same substrate (40). Therefore, when translocation is impeded, the activity of two or more molecules is clearly enhanced due to the trailing molecule being able to push the lead molecule. Such a synergistic effect is not observed during routine DNA unwinding in going from one molecule to 2 molecules of Dda. The major gain of function for routine DNA unwinding when multiple Dda molecules align is in terms of processivity, which increases by less than four-fold in going from one molecule of Dda to 4 molecules of Dda on the same substrate. Therefore, multiple molecules of Dda aligned on the same substrate impart a much greater affect on displacement of protein blocks than on DNA unwinding. Protein displacement is clearly an important function for some helicases, as illustrated by the role of Srs2 in disassembly of Rad51 protein filaments to regulate homologous recombination (54). It remains to be determined whether helicase-mediated protein displacement in the cell is regulated by the number of helicase molecules that are aligned along the DNA.
DNA unwinding processes in the cell clearly require greatly varying degrees of processivity. Replication requires unwinding of thousands of base pairs per single binding event, whereas some DNA repair processes require unwinding of only a few base pair. Modulation of processivity is likely to be strictly controlled in the cellular environment. This control occurs through the inherent activity of a particular helicase or through protein-protein interactions that can greatly alter helicase processivity. Highly processive helicases typically have multiple sites of interaction with DNA, as with RecBCD helicase, or encircle the DNA, as with replicative hexameric helicases. Translocases such as the Type 1 restriction-modification enzymes contain “helicase” motors that can move along dsDNA with very high processivity, as a result of multiple contacts between protein subunits and DNA (31;32). Helicases with inherently low processivity, such as PcrA helicase, can exhibit very high processivity when bound to accessory factors (52). The role of accessory factors for helicases may be similar as with processivity factors and DNA polymerases. The processivity factor essentially provides additional contacts between the protein complex and the DNA, thereby holding the complex onto the DNA during translocation. Dda can interact with other T4 proteins, such as gp32 and UvsX, which are likely to modulate its processivity during viral replication.
DNA unwinding can be broken down into steps for melting of the duplex and for translocation along the strand. The fact that trailing molecules of Dda do not push the lead molecule during DNA unwinding indicates that the lead molecule is moving along the DNA at similar rates as the trailing molecules. Therefore, the lead molecule is able to unwind duplex DNA and move forward at similar rates as the trailing molecules which only need to translocate on ssDNA. This implies that the rate-limiting step in moving along DNA is the same for DNA unwinding as it is for translocation on ssDNA. Therefore, the likely rate-limiting step for the activity of Dda helicase is contained in the translocation step, and DNA melting does not limit the overall rate of DNA unwinding.
Supplementary Material
Abbreviations
- RQF
Rapid Quench Flow
- SA
streptavidin
- NLLS
non-linear least squares
Footnotes
This work was supported by the National Institutes of Health grant R01 GM059400 (K.D.R) and the UAMS Committee for Allocation of Graduate Student Research Funds (R.L.E).
SUPPORTING INFORMATION AVAILABLE
Scripts describing the data analysis using Kintek Global Kinetic Explorer (Kintek Corporation, Austin, TX) are available free of charge via the internet at http://pubs.acs.org.
Reference List
- (1).Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: Integration of helicases into cellular processes. Q. Rev. Biophys. 2003;36:1–69. doi: 10.1017/s0033583502003864. [DOI] [PubMed] [Google Scholar]
- (2).Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- (3).Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- (4).Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- (5).Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- (6).von Hippel PH. Helicases become mechanistically simpler and functionally more complex. Nat. Struct. Mol. Biol. 2004;11:494–496. doi: 10.1038/nsmb0604-494. [DOI] [PubMed] [Google Scholar]
- (7).Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Dillingham MS, Spies M, Kowalczykowski SC. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- (9).Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- (10).Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–93. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- (11).Nanduri B, Byrd AK, Eoff RL, Tackett AJ, Raney KD. Pre-steady-state DNA unwinding by bacteriophage T4 Dda helicase reveals a monomeric molecular motor. Proc. Natl. Acad. Sci. U. S A. 2002;99:14722–14727. doi: 10.1073/pnas.232401899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Sikora B, Eoff RL, Matson SW, Raney KD. DNA unwinding by Escherichia coli DNA helicase I (TraI) provides evidence for a processive monomeric molecular motor. J. Biol. Chem. 2006;281:36110–36116. doi: 10.1074/jbc.M604412200. [DOI] [PubMed] [Google Scholar]
- (13).Maluf NK, Fischer CJ, Lohman TM. A Dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J Mol. Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- (14).Maluf NK, Lohman TM. Self-association equilibria of Escherichia coli UvrD helicase studied by analytical ultracentrifugation. J Mol. Biol. 2003;325:889–912. doi: 10.1016/s0022-2836(02)01276-7. [DOI] [PubMed] [Google Scholar]
- (15).Bujalowski W, Klonowska MM, Jezewska MJ. Oligomeric structure of Escherichia coli primary replicative helicase DnaB protein. J Biol Chem. 1994;269:31350–31358. [PubMed] [Google Scholar]
- (16).Dong F, Gogol EP, von Hippel PH. The phage T4-coded DNA replication helicase (gp41) forms a hexamer upon activation by nucleoside triphosphate. J. Biol. Chem. 1995;270:7462–7473. doi: 10.1074/jbc.270.13.7462. [DOI] [PubMed] [Google Scholar]
- (17).Egelman EH, Yu X, Wild R, Hingorani MM, Patel SS. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc Natl Acad Sci U S A. 1995;92:3869–73. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- (19).Patel SS, Picha KM. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- (20).Ali JA, Lohman TM. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- (21).Lucius AL, Maluf NK, Fischer CJ, Lohman TM. General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 2003;85:2224–2239. doi: 10.1016/s0006-3495(03)74648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lucius AL, Lohman TM. Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E.coli RecBCD helicase-catalyzed DNA unwinding. J Mol. Biol. 2004;339:751–771. doi: 10.1016/j.jmb.2004.04.010. [DOI] [PubMed] [Google Scholar]
- (23).Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol. Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- (24).Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol. Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Galletto R, Jezewska MJ, Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: quantitative analysis of the rate of the dsDNA unwinding, processivity and kinetic step-size of the Escherichia coli DnaB helicase using rapid quench-flow method. J Mol. Biol. 2004;343:83–99. doi: 10.1016/j.jmb.2004.07.055. [DOI] [PubMed] [Google Scholar]
- (26).Donmez I, Patel SS. Coupling of DNA unwinding to nucleotide hydrolysis in a ring-shaped helicase. EMBO J. 2008;27:1–9. doi: 10.1038/emboj.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Serebrov V, Beran RK, Pyle AM. Establishing a mechanistic basis for the large kinetic steps of the NS3 helicase. J. Biol. Chem. 2009;284:2512–2521. doi: 10.1074/jbc.M805460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Jr., Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- (31).Seidel R, Bloom JG, Dekker C, Szczelkun MD. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Stanley LK, Seidel R, van der SC, Dekker NH, Szczelkun MD, Dekker C. When a helicase is not a helicase: dsDNA tracking by the motor protein EcoR124I. EMBO J. 2006;25:2230–2239. doi: 10.1038/sj.emboj.7601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Brister JR. Origin activation requires both replicative and accessory helicases during T4 infection. J Mol. Biol. 2008;377:1304–1313. doi: 10.1016/j.jmb.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Formosa T, Alberts BM. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- (35).Ma Y, Wang T, Villemain JL, Giedroc DP, Morrical SW. Dual functions of single-stranded DNA-binding protein in helicase loading at the bacteriophage T4 DNA replication fork. J. Biol. Chem. 2004;279:19035–19045. doi: 10.1074/jbc.M311738200. [DOI] [PubMed] [Google Scholar]
- (36).Morris PD, Tackett AJ, Babb K, Nanduri B, Chick C, Scott J, Raney KD. Evidence for a functional monomeric form of the bacteriophage T4 DdA helicase. Dda does not form stable oligomeric structures. J Biol Chem. 2001;276:19691–19698. doi: 10.1074/jbc.M010928200. [DOI] [PubMed] [Google Scholar]
- (37).Kodadek T, Alberts BM. Stimulation of protein-directed strand exchange by a DNA helicase. Nature. 1987;326:312–314. doi: 10.1038/326312a0. [DOI] [PubMed] [Google Scholar]
- (38).Morris PD, Raney KD. DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- (39).Raney KD, Benkovic SJ. Bacteriophage T4 Dda helicase translocates in a unidirectional fashion on single-stranded DNA. J Biol Chem. 1995;270:22236–22242. doi: 10.1074/jbc.270.38.22236. [DOI] [PubMed] [Google Scholar]
- (40).Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- (41).Byrd AK, Raney KD. Increasing the length of the single-stranded overhang enhances unwinding of duplex DNA by bacteriophage T4 Dda helicase. Biochemistry. 2005;44:12990–12997. doi: 10.1021/bi050703z. [DOI] [PubMed] [Google Scholar]
- (42).Byrd AK, Raney KD. Displacement of a DNA binding protein by Dda helicase. Nucleic Acids Res. 2006;34:3020–3029. doi: 10.1093/nar/gkl369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Eoff RL, Spurling TL, Raney KD. Chemically modified DNA substrates implicate the importance of electrostatic interactions for DNA unwinding by Dda helicase. Biochemistry. 2005;44:666–674. doi: 10.1021/bi0484926. [DOI] [PubMed] [Google Scholar]
- (44).Levin MK, Wang YH, Patel SS. The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J Biol Chem. 2004;279:26005–26012. doi: 10.1074/jbc.M403257200. [DOI] [PubMed] [Google Scholar]
- (45).Eoff RL, Raney KD. Intermediates revealed in the kinetic mechanism for DNA unwinding by a monomeric helicase. Nat. Struct. Mol. Biol. 2006;13:242–249. doi: 10.1038/nsmb1055. [DOI] [PubMed] [Google Scholar]
- (46).Lucius AL, Vindigni A, Gregorian R, Ali JA, Taylor AF, Smith GR, Lohman TM. DNA unwinding step-size of E. coli RecBCD helicase determined from single turnover chemical quenched-flow kinetic studies. J. Mol. Biol. 2002;324:409–428. doi: 10.1016/s0022-2836(02)01067-7. [DOI] [PubMed] [Google Scholar]
- (47).Johnson KA. Fitting enzyme kinetic data with KinTek Global Kinetic Explorer. Methods Enzymol. 2009;467:601–626. doi: 10.1016/S0076-6879(09)67023-3. [DOI] [PubMed] [Google Scholar]
- (48).Serebrov V, Pyle AM. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature. 2004;430:476–480. doi: 10.1038/nature02704. [DOI] [PubMed] [Google Scholar]
- (49).Cheng W, Hsieh J, Brendza KM, Lohman TM. E. coli Rep oligomers are required to initiate DNA unwinding in vitro. J. Mol. Biol. 2001;310:327–350. doi: 10.1006/jmbi.2001.4758. [DOI] [PubMed] [Google Scholar]
- (50).Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J. Biol. Chem. 2007;282:27076–27085. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- (51).Yang Y, Dou SX, Ren H, Wang PY, Zhang XD, Qian M, Pan BY, Xi XG. Evidence for a functional dimeric form of the PcrA helicase in DNA unwinding. Nucleic Acids Res. 2008;36:1976–1989. doi: 10.1093/nar/gkm1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Slatter AF, Thomas CD, Webb MR. PcrA helicase tightly couples ATP hydrolysis to unwinding double-stranded DNA, modulated by the initiator protein for plasmid replication. RepD. Biochemistry. 2009;48:6326–6334. doi: 10.1021/bi900101h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Singh N, Singh Y. Statistical theory of force-induced unzipping of DNA. Eur. Phys. J. E. Soft. Matter. 2005;17:7–19. doi: 10.1140/epje/i2004-10100-7. [DOI] [PubMed] [Google Scholar]
- (54).Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.