Abstract
CCN5, a member of the CCN family of growth factors, inhibits the proliferation and migration of smooth muscle cells in cell culture and animal models. Expressed in both embryonic and adult tissues, CCN5 exhibits a matricellular localization pattern characteristic of secreted proteins that are closely associated with the cell surface. In addition to this observed expression pattern, immunohistochemical evidence suggests the presence of nuclear CCN5 in some cells. To determine if CCN5 localizes to the nucleus we performed immunofluorescence, confocal imaging, and cell fractionation to corroborate the immunohistochemical observations. After confirming the presence of nuclear CCN5 using four independent experimental methods, we identified a single putative nuclear localization signal in the von Willebrand factor C domain of mouse and rat CCN5. Site directed mutagenesis of the three basic amino acids in the putative nuclear localization sequence did not prevent nuclear localization of CCN5 in four different cell types, suggesting that CCN5 nuclear transport is not mediated by the only canonical nuclear localization signal present in the primary amino acid sequence. Future work will address the mechanism of nuclear localization and the function of nuclear versus secreted CCN5.
Keywords: CCN5, Nuclear localization, WISP-2, CCN family, Matricellular proteins
Introduction
CCN5 belongs to the cysteine rich 61/connective tissue growth factor/nephroblastoma overexpressed (CCN) protein family. These proteins play a role in numerous disease states and in diverse cellular functions including development, wound healing, cell proliferation, migration, and invasiveness. The CCN protein family consists of six cysteine-rich, matricellular proteins—proteins that are secreted but remain closely associated with the cell surface. All CCN proteins are composed of a homologous modular structure. Five of the six family members—CCN1, CCN2, CCN3, CCN4 and CCN6—contain 4 distinct functional domains: an insulin-like growth factor-binding protein domain (IGFBP), a von Willebrand Factor-C domain (VWC), a thrombospondin-1 domain (TSP-1), and a carboxy terminal domain (CT). In contrast, CCN5 (WISP-2) lacks the carboxy-terminal domain.
CCN5 is a heparin-induced growth arrest specific gene whose protein inhibits the proliferation and migration of smooth muscle cells in both cell culture and animal models (Lake et al. 2003; Lake and Castellot 2003; Mason et al. 2003). In these studies, adenoviral overexpression of CCN5 protein in cultured vascular smooth muscle cells inhibits cell proliferation, motility, and invasiveness resulting in maintenance of cells in a G0 growth state (Lake et al. 2003). Knockdown of CCN5 using RNAi results in morphological changes and disorganization of the actin cytoskeleton (Lake and Castellot 2003). Furthermore, CCN5 protein can attenuate the intimal smooth muscle proliferation seen following vascular injury in a mouse carotid ligation model (unpublished data).
While CCNs are secreted proteins, reports indicate that CCN proteins exist in the nucleus where they may perform additional cellular functions. For example, truncated forms of CCN3 lacking the secretory signal peptide are directed to the nucleus where they exhibit negative transcriptional activity (Planque et al. 2006). Also, the secreted form of CCN2 can be transported via endocytosis to the nucleus after binding at the cell surface (Wahab et al. 2001). We, too, recently observed putative nuclear staining of CCN5 in adult mouse tissues and in cell models via immunohistochemistry, however this evidence is suggestive rather than conclusive (Gray et al. 2007; Wei et al. 2009). To unambiguously determine if CCN5 exists in the nucleus, we used four independent approaches: immunohistochemistry, immunofluorescence, confocal imaging, and subcellular fractionation. Analysis of the data demonstrates that CCN5 is both a nuclear and secreted protein. Furthermore, the sole putative nuclear localization signal (NLS) identified in the VWC domain does not facilitate nuclear localization of CCN5.
Materials and methods
Cell culture
Vascular smooth muscle cells (VSMC) were obtained from aortae of Sprague-Dawley rats (Charles River Breeding Laboratories, Inc. Wilmington, MA). They were isolated, cultured, and characterized as previously described (Castellot et al. 1982). Briefly, the abdominal segment of the aorta was removed and the fascia cleaned away under a dissecting microscope. The aorta was cut longitudinally, and small pieces of the media were carefully stripped from the vessel wall. Two to three such strips were placed in 60 mm dishes. Within 1–2 weeks, VSMC migrated from the explants; they were capable of being subcultured approximately 1 week after the first appearance of cells. They were identified as VSMC by their characteristic hill-and-valley growth pattern and indirect immunofluorescence staining for SMC specific α-actin. Human aortic smooth muscle cells were purchased from Lonza Group Ltd. All cultures were maintained in RPMI-1640 or DMEM medium containing 10% bovine growth serum at 37°C in a humidified, 5% CO2/95% air atmosphere. Other cell lines NIH 3T3, C2C12, HEK293, and COS7 were maintained in DMEM medium containing 10% bovine growth serum (BGS, Hyclone), 2 mM L-glutamine (GIBCO, Carlsbad, CA), and 100 ug/ml penicillin/ streptomycin (GIBCO) at 37°C in a humidified, 5% CO2/95% air atmosphere.
Immunohistochemistry
Frozen sections of mouse tissues were prepared as described in Gray et al. 2007. Slides were immunostained with an affinity purified polyclonal antibody to a rat peptide of the CCN5 protein and visualized using a rabbit IgG VectaStain Elite ABC reagent kit (Vector Laboratories, Burlingame, CA).
Immunofluorescence
VSMC were plated in 2- and 4- well chamber slides at a low density. Cells were grown according to the cell culture protocol provided above and subsequently growth arrested in medium containing 0.4% serum for 72 h. Cells were fixed in 0.4% paraformaldehyde in PBS, permeabilized when indicated in 2% Triton X-100/PBS, blocked with 3% BSA/PBS, and exposed to the affinity-purified rabbit anti-peptide antibody specific to CCN5 and/or the lamin A+C antibody (Chemicon International, Temecula, CA). Secondary antibody (donkey anti-rabbit (CCN5) or donkey anti-mouse (for lamin A) conjugated to Cy3 and FITC, respectively) was used to visualize the cells. Samples were mounted in Vectashield containing DAPI to visualize nuclei.
Human aortic smooth muscle (hAoSMC) were growth arrested, fixed and stained as described above. HAoSMC and cell nuclei were visualized with confocal imaging using a Leica TCS SP2 AOBS confocal microscope.
Cell fractionation
Subcellular fractionation of human aortic and rat vascular smooth muscle cells previously infected with adenovirus expressing GFP or CCN5 was performed using a modified version of the QIAGEN Qproteome Nuclear Subfractionation kit (QIAGEN, Valencia, CA). Briefly, cell suspensions were centrifuged at 1800 × g to produce a cell pellet. The supernatant was carefully removed and the cell pellet was exposed to a series of buffer incubations and centrifugation (i.e., washing) steps resulting in the isolation of two distinct cellular fractions: a nuclear and non-nuclear fraction. Acetone precipitation of the isolated fractions was performed to prepare fractions for further analysis. Briefly, fractions were incubated with equal volumes of ice cold acetone for 15 min at room temperature. Samples were then centrifuged for 10 min at 12,000 × g in a pre-cooled microcentrifuge at 4°C. Supernatants were discarded, the pellets air dried, and reconstituted in loading buffer containing β-mercaptoethanol for subsequent gel electrophoresis.
Western blot analysis
Cell fractions prepared as described above were boiled in loading buffer, resolved by SDS-polyacrylamide gel electrophoresis (PAGE) using 4–15% gradient gels, and blotted onto 0.2-μm pore size ImmunoBlot polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA) at 100 V for 1 h. The blots were dried and rewetted with methanol. Membranes were blocked for 1 h in PBST (PBS containing 0.2% Tween-20) containing 10% non-fat dried milk. Blots were probed using the following antibodies: anti-HA and anti-lamin A/C. Following PBST washes, blots were probed with the appropriate secondary antibodies (horseradish peroxidase-conjugated anti-mouse IgG and horseradish peroxidase-conjugated anti-rabbit IgG, respectively) diluted in PBST. Bands were visualized using the Western Lighting Chemiluminescence Reagent Plus (Perkin Elmer Life Sciences) reagents.
Mutation of putative Nuclear Localization Signal (NLS)
Putative NLS sequences were determined via bioinformatic analysis of the rat, mouse, and human amino acid sequences. Only one possible sequence, PRPKRI, was found in the VWC domain of rat CCN5. This sequence was mutated using the Stratagene QuikChange Site-Directed Mutagenesis (SDM) Kit according to the manufacturer’s protocol. The three basic amino acid residues in the putative NLS were mutated to neutral amino acids: PRPKRI to PLPMGI. Following site-directed mutagenesis, NIH3T3, HEK293, C2C12, and COS7 cells were transfected with the mutated construct. These cell types transfect easily and are routinely used in subcellular localization studies (Thakur et al. 1997; Lee et al. 2004). The construct contains an N-terminal Flag tag and a C-terminal myc tag. Localization of the mutated CCN5 construct was observed in the four cell types via immunofluorescence using the anti-FLAG M2 monoclonal antibody Cy3 conjugate (Sigma, St. Louis, MO) and the monoclonal anti-c-Myc-FITC conjugate (Sigma, St. Louis, MO).
Results
Immunohistochemistry and immunofluorescence suggest possible nuclear localization of CCN5
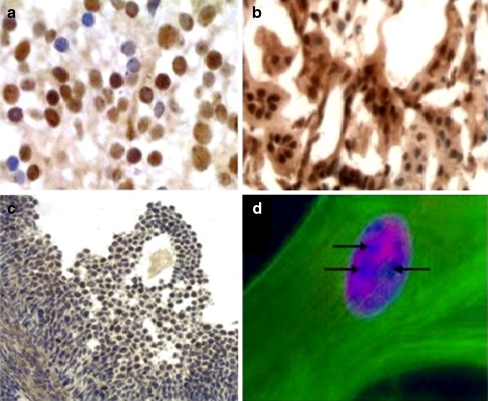
Earlier work from this laboratory showed that CCN5 protein expression is widely distributed in both adult and embryonic rodent tissues (Gray et al. 2007; Jones et al. 2007). These studies focused primarily on the expression patterns of CCN5 and another family member, CCN2. Closer analysis of tissue sections revealed nuclei that appeared positive for CCN5 in many cells and tissues including heart, brain, lung, intestine, spinal cord, ovary, and kidney (Gray et al. 2007; Jones et al. 2007; Wei et al. 2009). Figure 1a–c includes representative images of CCN5 nuclear staining in mouse embryonic spinal cord (GD16), adult rat kidney, and adult mouse ovary sections (Graafian follicle). A human uterine smooth muscle cell triple stained with phalloidin, Hoeschst, and an anti-CCN5 antibody is included in panel 1D, with arrows indicating apparent nucleolar exclusion.
Fig. 1.
Immunohistochemistry and immunofluorescence of tissues and cells from rodents and humans. Immunohistochemistry and immunofluorescence using specific anti-CCN5 antibodies as a probe was carried out as described in “Materials and methods”. Immunohistochemical images of frozen sections of mouse embryonic spinal cord, GD16 (a), adult rat kidney (b), and adult mouse ovary (Graafian follicle) (c) are shown. d shows an immunofluorescence image of cultured human uterine myometrial SMC triple-stained with Hoecsht (nucleus), phalloidin (actin), and the anti-CCN5 antibody. Arrows indicate areas of apparent nucleolar exclusion
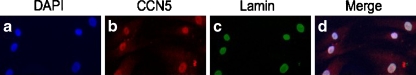
After observing putative CCN5 nuclear staining patterns in adult and embryonic rodent tissue sections, we examined the possibility that CCN5 might localize to the nucleus of cultured, growth arrested vascular smooth muscle cells. Growth arrest of VSMC via serum deprivation robustly increases CCN5 protein expression (Delmolino et al. 2001). To determine if CCN5 could be detected in the nucleus of growth arrested VSMC in culture we performed immunofluorescence using the well-characterized, highly specific antibody to CCN5 protein described above and costained with lamin A/C. As lamins A and C specifically localize to the nuclear compartment, lamin serves as a useful nuclear marker. Both lamin (Fig. 2c) and CCN5 (Fig. 2b) co-localized to the nuclei of human aortic SMC (Fig. 2d merged image).
Fig. 2.
Immunofluoresence of permeabilized growth arrested human aortic smooth cells. Immunofluorescence was performed on growth arrested human aortic smooth muscle cells using anti-CCN5 (b) and anti-lamin A/C antibodies (c). Nuclei were visualized using DAPI (a). The merged image (d) shows overlapping nuclear staining
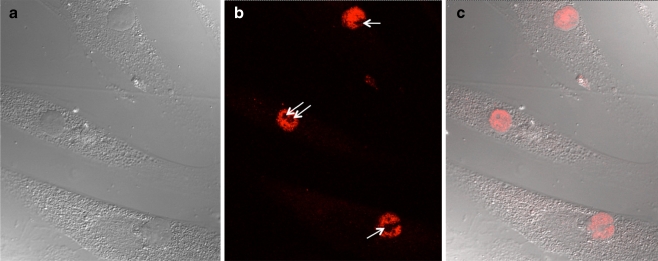
Confocal image analysis demonstrates the presence of CCN5 protein in the nucleus
Though highly suggestive, immunohistochemistry and standard immunofluorescence methods do not definitively demonstrate nuclear localization, since the observed staining could be concentrated above or below the nucleus in the cytoplasm. To more rigorously assess the presence of CCN5 within the nucleus, we used confocal microscopy to optically dissect the cell and nucleus. Confocal analysis of growth arrested hAoSMC show clear evidence of nuclear CCN5 (Fig. 3). Figure 3a shows a typical differential interference contrast micrograph of several growth arrested hAoSMC in culture. A 1 micron section through the center of the nuclei of these cells indicates the presence of nuclear CCN5 protein (Fig. 3b); the pattern of nuclear expression is reminiscent of that expected for nuclear matrix proteins. Superimposing the confocal image from the optical plane through the center of the nuclei with the differential interference contrast micrograph clearly demonstrates the presence of CCN5 in the nuclei (Fig. 3c). It is also interesting to note that the nucleoli in all of the cultured cells we have examined appear to be excluded from CCN5 staining.
Fig. 3.
Confocal image analysis. Confocal and light microscopy images of CCN5-stained, growth arrested human aortic smooth muscle cells were produced as described in “Materials and methods”. a. Differential Interference Contrast micrograph. b. Confocal microscopy of a 1 micron optical section through the center of the nuclei of the cells shown in a. White arrows denote the nucleoli. c. Superimposed Differential Interference Contrast and confocal micrographs from a and b, respectively
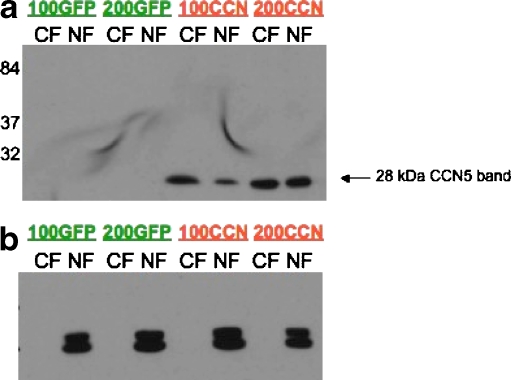
Cell fractionation demonstrates the presence of nuclear CCN5 protein
To definitively establish the presence of CCN5 in the nucleus, we corroborated the immunohistochemical, immunofluorescence, and confocal imaging analysis data using a biochemical method—cell fractionation (Fig. 4). We infected human aortic smooth muscle cells with adenovirus expressing CCN5 or green fluorescent protein (GFP), as previously described in our laboratory. The CCN5 adenovirus contains an HA-tag that allows monitoring of adenovirus infection; >90% of the cells are infected (Lake et al. 2003). Infected smooth muscle cells were separated into nuclear and non-nuclear fractions as described in “Materials and methods”. Western blot analysis of subcellular fractions with an anti-HA antibody indicates the presence of both nuclear and non-nuclear CCN5 protein (Fig. 4a). In contrast, CCN5 could not be detected in GFP-infected cells. Additionally, lamin—a highly specific nuclear marker—is detected only in the nuclear fractions of both the GFP- and CCN5-infected cells, indicating efficient separation of the nuclear and cytoplasmic compartments (Fig. 4b).
Fig. 4.
Western blot analysis of fractionated cells. Human aortic smooth muscle cells were infected with adenovirus expressing HA-tagged CCN5 or GFP (without an HA tag) and subjected to cell fractionation as described in “Materials and methods”. The nuclear and cytoplasmic fractions were examined by Western blot analysis. a shows a 28 kDa band revealed by the HA-tag antibody in both nuclear and cytoplasmic fractions of cells infected with adenovirus making CCN5, but not in adenovirus-GFP infected cells. b shows that only the nuclear fractions contain lamin A/C, a specific nuclear marker protein
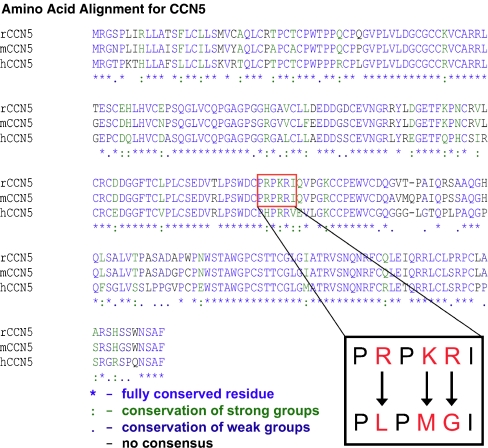
Analysis of the amino acid sequence of CCN5 reveals a putative, but weak, NLS
Using the PSORT II program (http://psort.ims.u-tokyo.ac.jp; Horton and Nakai 1997), we analyzed the rat, mouse, and human CCN5 protein primary amino acid sequence to predict potential nuclear localization signals (NLS). We identified a putative monopartite NLS in the VWC domain of rat and mouse CCN5 (Fig 5); notably, the human sequence does not contain a putative NLS in its primary amino acid sequence. The identified NLS in rat is PRPKRI, while in mouse the sequence is PRPRRI. Since NLS typically have 4 or 5 basic amino acids, and the putative NLS in the rodent CCN5 sequences have only 3 basic amino acids, this NLS would be considered rather weak. Nonetheless, we tested the ability of this sequence to mediate nuclear localization, as described below. In contrast, the human sequence (PHPRRV) has only 2 basic amino acid sequences, making it extremely unlikely to be an NLS.
Fig. 5.
Alignment of the rat, mouse and human CCN5 protein sequences. The rat, mouse and human amino acid sequences for CCN5 are aligned. Bioinformatic analysis using the PSORT II program predicted a weak putative NLS in the VWC domain as indicated by the red box. The inset shows the putative rat NLS sequence, PRPKRI. The three basic amino acids indicated in red were mutated to neutral amino acids to produce the sequence PLPMGI. The mutated sequence was overexpressed in the four cell types shown in Fig. 6
Mechanism of nuclear localization
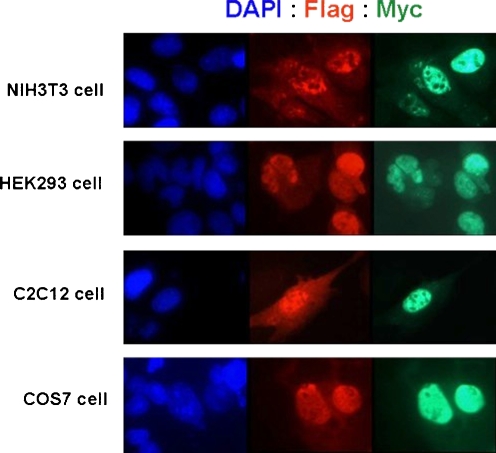
To determine if the putative NLS in rat CCN5 directs the protein to the nucleus, we employed site directed mutagenesis to alter the sequence. The three basic amino acids were mutated to neutral amino acids (Fig. 5). The mutated CCN5 protein was transfected into four cell types (NIH3T3, HEK293T, C2C12, and COS7) as described in “Materials and methods”. The CCN5 construct contains both an N-terminal Flag tag and a C-terminal Myc tag. Transfected cells were visualized using immunofluorescence with anti-Flag Cy3 and anti-myc FITC conjugated antibodies. However, we still observed CCN5 in the nucleus of all four cell types transfected with the mutated CCN5 protein (Fig. 6), indicating that this sequence alone is not sufficient to direct CCN5 protein to the nucleus.
Fig. 6.
Localization of CCN5 containing a mutated putative NLS. Site-directed mutagenesis of the putative NLS sequence in rat CCN5 and subsequent localization of the mutated CCN5 was performed as described in “Materials and methods”. Immunofluorescence of NIH3T3, HEK293, C2C12, and COS7 cells transfected with the mutated CCN5 construct shown in Fig. 5 (inset) and stained with DAPI (to reveal the nucleus), and anti-Flag and anti-Myc antibodies (to reveal CCN5) is shown
We also considered the possibility that CCN5 might diffuse into the nucleus through the nuclear pores, since its mass (27.5 kDa) is less than the theoretical maximum protein mass (approximately 50 kDa) for diffusion through the nuclear pores. To rule out diffusion, we prepared a dual-tagged expression plasmid containing the IGFBP-TSP1 domains of rat CCN5 and transfected this construct into several different cells types. Despite having a molecular mass of only 18.4 kDa, this partial CCN5 peptide was excluded from the nucleus in all cell lines tested strongly suggesting that the larger full-length CCN5 protein does not enter the nucleus via diffusion, but rather through a regulated process (data not shown, manuscript in preparation). Moreover, the exclusion of the IGFBP-TSP1 construct from the nucleus suggests that nuclear import of CCN5 may require the VWC domain.
Discussion
In this communication, we provide strong evidence that CCN5 is both a secreted and a nuclear protein using four independent methods: immunohistochemistry, immunofluorescence, confocal image analysis, and subcellular fractionation. In addition, we show that the putative nuclear localization signal identified in the primary amino acid sequence of rat CCN5 does not mediate nuclear translocation.
Preliminary immunohistochemical analysis of adult mouse tissues including kidney, ovary, spinal cord, brain, heart, lung, and gut indicates the potential presence of nuclear CCN5. Additionally, immunofluorescence of growth arrested, permeabilized human aortic smooth muscle cells with a specific, affinity purified CCN5 antibody suggested nuclear CCN5 localization, with apparent nucleolar exclusion. Confocal microscopy provided direct evidence for nuclear localization with apparent nucleolar exclusion. Biochemical fractionation and analysis of the nuclear and cytoplasmic compartments of smooth muscle cells corroborated the presence of CCN5 in the nucleus. Furthermore, antibodies to the nucleus-specific protein lamin revealed colocalization of CCN5 and lamin, suggesting that the nuclear localization was not an artifact.
With respect to the apparent exclusion of CCN5 from the nucleolus, we cannot formally rule out the possibility of sequestering of the binding epitope or technical artifacts such as quenching of the fluorescent signal. Nonetheless, exclusion of CCN5 from the nucleolus is highly likely to be real for the following reasons: 1) we see exclusion with three different antibodies, including a monoclonal, against three different epitopes within the CCN5 protein; 2) we have used several different cell fixation methods in cultured cells that generally do not require epitope unmasking, and have never observed CCN5 in the nucleus; 3) sequence analysis via the databases previously mentioned do not indicate any similarity or motifs associated with any known ribonuclear protein; and 4) we see exclusion in many different cell types.
How CCN5 reaches the nucleus is not known. Using the PSORTII program, we analyzed the protein sequence of rat, mouse, and human CCN5 and identified a putative, albeit weak, nuclear localization sequence [PRPKRI] in the VWC domain of the rat CCN5 protein. A nearly identical sequence is observed in the mouse CCN5 protein [PRPRRI]. Careful analysis of several databases including NucPred, NLStradamus, and PredictNLS did not reveal the presence of any other possible nuclear localization signals, such as the M9 sequence found in some ribosomal proteins, within the primary amino acid sequence of CCN5. The NetNES Program for predicting nuclear export signals does predict two possible nuclear export signals, but as yet we have no evidence to support the possibility that CCN5 is able to move out of the nucleus.
Mutation of the three basic amino acids within the rat putative NLS had no effect on CCN5 nuclear localization in four different cell types. All of these cell types are well-characterized with respect to nuclear import and are used regularly by investigators studying this process. While this data, combined with our bioinformatic analysis, makes the presence of a classical nuclear localization signal within the primary amino sequence of CCN5 highly unlikely, it is still possible that the three-dimensional, native conformation of CCN5 produces a nuclear localization signal via “patching” of two or more different regions of the protein. In addition, it is possible that CCN5 is translocated to the nucleus via chaperone proteins. Both of these possibilities are well-recognized mechanisms for nuclear localization of proteins without a canonical nuclear localization signal (Planque 2006). Additional possibilities include internalization of cell surface CCN5 through an endocytic trafficking mechanism, translation of a splice variant with a nuclear localization signal, or post-translational modifications that target CCN5 to the nucleus. Finally, we note that human CCN5 does not contain even a weak NLS motif, nor any other recognized sequence that directs proteins to the nucleus, further suggesting that nuclear translocation does not occur via a classical NLS.
Since it is also theoretically possible that molecules smaller than 50 kDa in mass can localize in the nuclear compartment by diffusion through nuclear pores, we tested this possibility as well. The exclusion from the nucleus of a truncated CCN5 protein indicates that diffusion of the full-length CCN5 protein is highly unlikely (data not shown). This is not surprising, since most peptides >6–8 kDa in mass are transported into the nucleus via regulated mechanisms, not via diffusion though the nuclear pores. Examination of the mechanism(s) involved in translocating CCN5 to the nucleus is currently underway.
Previous studies indicate that other members of the CCN protein family localize to the nucleus, albeit via different mechanisms. Using both immunohistochemistry and subcellular fractionation, Tamura et. al., identified Cyr61 (CCN1) in the nucleus of mechanically stretched bladder smooth muscle cells, while Cyr61 was absent in the unstretched cells (Tamura et al. 2001). Interestingly, they noted that Cyr61 lacks a classic NLS in its primary sequence (Tamura et al. 2001). CCN3 (Nov) contains a lysine-rich NLS in the carboxy-terminal domain; however, as is usually the case the amino-terminus signal peptide overrides the nuclear localization signal and the protein is targeted for secretion. Interestingly, truncated CCN3 protein lacking the amino terminus was observed in HeLa cells and several tumor cell types (Perbal 1999; Planque and Perbal 2003; Cervello et al. 2004; Planque et al. 2006). CCN2 (CTGF) has been identified in the nucleus of human mesangial cells by immunofluorescence and subcellular fractionation. Wahab et. al., suggest that CCN2 reaches the intracellular compartment by endocytosis and is phosphorylated en route to the nucleus (Wahab et al. 2001). Despite these differences in the mechanism of nuclear localization among CCN family proteins, all of these studies associate nuclear CCN proteins with potential transcriptional activity in addition to the traditional receptor mediated signaling expected from a matricellular protein.
While a receptor for CCN5 is not yet known, as a secreted protein we would expect CCN5 to bind membrane receptors and transduce its effects from the cell surface to the nucleus. The presence of nuclear CCN5 presents the possibility of nuclear as well as cell surface-mediated functions, and underscores the complex biology of the CCN protein family. These possibilities are under active investigation in our laboratory.
Acknowledgments
This paper is dedicated to the memory of Mark Gray. The work was support in part by grants HD046251 and HL49973 to JJC and an American Heart Association Fellowship to CB.
Abbreviations
- (CT)
Carboxy terminal domain
- (CCN)
cysteine rich 61/connective tissue growth factor/nephroblastoma overexpressed
- (GFP)
green fluorescent protein
- (hAoSMC)
human aortic smooth muscle cells
- (IGFBP)
insulin-like growth factor-binding protein domain
- (NLS)
nuclear localization signal(s)
- (SMC)
smooth muscle cell
- (TSP-1)
thrombospondin-1 domain
- (VSMC)
vascular smooth muscle cells
- (VWC)
von Willebrand Factor-C domain
References
- Castellot JJ, Jr, Favreau LV, Karnovsky MJ, Rosenberg RD. Inhibition of vascular smooth muscle cell growth by endothelial cell-derived heparin. Possible role of a platelet endoglycosidase. J Biol Chem. 1982;257(19):11256–11260. [PubMed] [Google Scholar]
- Cervello M, Giannitrapani L, Labbozzetta M, Notarbartolo M, D’Alessandro N, Lampiasi N, Azzolina A, Montalto G. Expression of WISPs and of their novel alternative variants in human hepatocellular carcinoma cells. Ann NY Acad Sci. 2004;1028:432–439. doi: 10.1196/annals.1322.051. [DOI] [PubMed] [Google Scholar]
- Delmolino LM, Stearns NA, Castellot JJ., Jr COP-1, a member of the CCN family, is a heparin-induced growth arrest specific gene in vascular smooth muscle cells. J Cell Physiol. 2001;188(1):45–55. doi: 10.1002/jcp.1100. [DOI] [PubMed] [Google Scholar]
- Gray MR, Malmquist JA, Sullivan M, Blea M, Castellot JJ., Jr CCN5 Expression in mammals. II. Adult rodent tissues. J Cell Commun Signal. 2007;1(2):145–158. doi: 10.1007/s12079-007-0013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc Int Conf Intell Syst Mol Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- Jones JA, Gray MR, Oliveira BE, Koch M, Castellot JJ., Jr CCN5 expression in mammals: I. Embryonic and fetal tissues of mouse and human. J Cell Commun Signal. 2007;1(2):127–143. doi: 10.1007/s12079-007-0012-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake AC, Castellot JJ., Jr CCN5 modulates the antiproliferative effect of heparin and regulates cell motility in vascular smooth muscle cells. Cell Commun Signal. 2003;1(1):5. doi: 10.1186/1478-811X-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake AC, Bialik A, Walsh K, Castellot JJ., Jr CCN5 is a growth arrest-specific gene that regulates smooth muscle cell proliferation and motility. Am J Pathol. 2003;162(1):219–231. doi: 10.1016/S0002-9440(10)63813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Lanca AJ, Cheng R, Nguyen T, Ji XD, Gobeil F, Jr, Chemtob S, George SR, O’Dowd BF. Agonist-independent nuclear localization of the Apelin, angiotensin AT1, and bradykinin B2 receptors. J Biol Chem. 2004;279(9):7901–7908. doi: 10.1074/jbc.M306377200. [DOI] [PubMed] [Google Scholar]
- Mason HR, Nowak RA, Morton CC, Castellot JJ., Jr Heparin inhibits the motility and proliferation of human myometrial and leiomyoma smooth muscle cells. Am J Pathol. 2003;162(6):1895–1904. doi: 10.1016/S0002-9440(10)64323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. Nuclear localisation of NOVH protein: a potential role for NOV in the regulation of gene expression. Mol Pathol. 1999;52(2):84–91. doi: 10.1136/mp.52.2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planque N. Nuclear trafficking of secreted factors and cell-surface receptors: new pathways to regulate cell proliferation and differentiation, and involvement in cancers. Cell Commun Signal. 2006;4:7. doi: 10.1186/1478-811X-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planque N, Perbal B. A structural approach to the role of CCN (CYR61/CTGF/NOV) proteins in tumourigenesis. Cancer Cell Int. 2003;3(1):15. doi: 10.1186/1475-2867-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planque N, Long Li C, Saule S, Bleau AM, Perbal B. Nuclear addressing provides a clue for the transforming activity of amino-truncated CCN3 proteins. J Cell Biochem. 2006;99(1):105–116. doi: 10.1002/jcb.20887. [DOI] [PubMed] [Google Scholar]
- Tamura I, Rosenbloom J, Macarak E, Chaqour B. Regulation of Cyr61 gene expression by mechanical stretch through multiple signaling pathways. Am J Physiol Cell Physiol. 2001;281(5):C1524–C1532. doi: 10.1152/ajpcell.2001.281.5.C1524. [DOI] [PubMed] [Google Scholar]
- Thakur S, Zhang HB, Peng Y, Le H, Carroll B, Ward T, Yao J, Farid LM, Couch FJ, Wilson RB, Weber BL. Localization of BRCA1 and a splice variant identifies the nuclear localization signal. Mol Cell Biol. 1997;17(1):444–452. doi: 10.1128/mcb.17.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahab NA, Brinkman H, Mason RM. Uptake and intracellular transport of the connective tissue growth factor: a potential mode of action. Biochem J. 2001;359(Pt 1):89–97. doi: 10.1042/0264-6021:3590089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, McKeon F, Russo JW, Lemire J, Castellot J. Domain-and species-specific monoclonal antibodies recognize the Von Willebrand Factor-C domain of CCN5. J Cell Commun Signal. 2009;3(1):65–77. doi: 10.1007/s12079-009-0054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]