Abstract
Silk production has independently evolved in numerous arthropod lineages, such as Lepidoptera, the moths and butterflies. Lepidopteran larvae (caterpillars) synthesize silk proteins in modified salivary glands and spin silk fibers into protective tunnels, escape lines, and pupation cocoons. Molecular sequence data for these proteins are necessary to determine critical features of their function and evolution. To this end, we constructed an expression library from the silk glands of the ghost moth, Hepialus californicus, and characterized light chain fibroin and heavy chain fibroin gene transcripts. The predicted H. californicus silk fibroins share many elements with other lepidopteran and trichopteran fibroins, such as conserved placements of cysteine, aromatic, and polar amino acid residues. Further comparative analyses were performed to determine site-specific signatures of selection and to assess whether fibroin genes are informative as phylogenetic markers. We found that purifying selection has constrained mutation within the fibroins and that light chain fibroin is a promising molecular marker. Thus, by characterizing the H. californicus fibroins, we identified key functional amino acids and gained insight into the evolutionary processes that have shaped these adaptive molecules.
Keywords: Lepidoptera, Trichoptera, Silk, Fibroin, cDNA, Gene tree, Purifying selection
Introduction
Silks are ideal for the study of adaptive evolution because they have independently arisen in numerous arthropod lineages. Typically, silks are primarily made of proteins that are dominated by non-essential amino acids, such as glycine, alanine, and serine (Gatesy et al. 2001). Arthropods use silk for a broad range of ecological functions, such as prey capture, protection, reproduction, and dispersal (Craig 1997). Silk proteins that perform similar tasks tend to possess particular attributes that have been maintained over long periods of evolutionary time. These conserved protein traits can range from the uniform positioning of a single cysteine residue, to more complex features, such as long regions of high sequence identity. For spider silks, studying these conserved features has helped determine important sequence elements and molecular evolutionary patterns of silk proteins (Guerette et al. 1996; Garb and Hayashi 2005; Ittah et al. 2006).
Like spiders, Trichoptera (caddisflies) is a lineage with silk proteins that have highly conserved elements (Yonemura et al. 2009). Caddisfly larvae synthesize silk proteins in paired labial glands, whose original function was the secretion of saliva. The larvae spin silk underwater to construct aquatic capture nets, domiciles, and pupation cocoons. Silk proteins have been studied in species sampled from each of the three trichopteran suborders (Yonemura et al. 2006, 2009). In these exemplars, the core of the larval silk fiber is formed from two proteins, heavy chain fibroin (H-fibroin) and light chain fibroin (L-fibroin). Across trichopteran suborders, there is ~50% sequence identity within the L-fibroin sequence and 56% sequence identity in the amino terminal region of H-fibroin (Yonemura et al. 2009).
H- and L-fibroin genes and proteins are also conserved in Lepidoptera (moths and butterflies). Larval lepidopterans (caterpillars) use silk for diverse functions that in most species include constructing pupal cocoons. Lepidoptera is the sister group to Trichoptera, and together they form the supraorder Amphiesmenoptera (Kristensen 1999; Whiting 2002). Unlike caddisfly larvae that spin their silk underwater, caterpillars spin silk in terrestrial environments. Except for this difference, Lepidoptera produce silk in a homologous manner to Trichoptera. Specifically, silk proteins are secreted in paired labial glands (Grimaldi and Engel 2005). Regions of sequence similarity have been observed throughout their L-fibroins and in the terminal regions of their H-fibroins (Sehnal and Žurovec 2004; Yonemura and Sehnal 2006; Yonemura et al. 2009). The use of L- and H-fibroins in amphiesmenopteran silk production has been hypothesized to be conserved for over 250 million years (Yonemura et al. 2009). However, unlike trichopteran silk, most previously studied moth silks include an additional protein, P25 (Inoue et al. 2000; Sehnal and Žurovec 2004). L-fibroin, H-fibroin, and P25 are thought to have formed the core of lepidopteran silk fibers for over 150 million years (Yonemura and Sehnal 2006).
To date, all silk studies of Lepidoptera have focused on members of Ditrysia, which includes most moth and butterfly species (Mita et al. 1994; Žurovec and Sehnal 2002; Yonemura and Sehnal 2006). However, Ditrysia is a highly derived clade (Friedlander et al. 1996) and so to provide better insight into the composition and evolution of lepidopteran silk, we focused on the ghost moth, Hepialus californicus. Hepialus is a member of Exoporia, a basal lepidopteran lineage of moths with primitive appearances (Nielsen et al. 2000; Wiegmann et al. 2002). H. californicus larvae feed on the roots of lupines (Strong et al. 1996) and live in silk lined subterranean tunnels that they build around the host plant’s roots (Nielsen et al. 2000).
In this study, we describe H- and L-fibroin transcripts in a cDNA library constructed from the silk glands of H. californicus. We compare the fibroin sequences to those known in other Lepidoptera and Trichoptera. We then determine the rate of molecular evolution at each amino acid site and assess the potential phylogenetic utility of the silk genes across Amphiesmenoptera. One of our aims is to reconstruct the silk groundplan for Lepidoptera by ascertaining whether H. californicus utilizes three core silk proteins like previously studied Lepidoptera, or only two silk proteins as found in Trichoptera. In addition, by comparing the sequences of H. californicus silk to other known fibroins, we shed light on the evolution and function of these molecules of adaptive significance.
Methods
Tissue Collection
Hepialus californicus (Lepidoptera: Hepialidae) larvae were collected from the Bodega Marine Reserve (38°32′N 123°07′W) in Sonoma County, CA, USA, and placed in individual culture trays (3.5-cm diameter) with moistened filter paper and organic carrot chunks. The larvae were kept at ambient room temperature until dissection. At that time, 18 final instar larvae were anesthetized with CO2 gas and then individually placed in a dissection dish filled with 0.15 M sodium chloride, 0.015 M sodium citrate buffer. Once submerged within the solution, the head was gently disconnected from the thorax so as not to rupture the paired labial silk glands. For each larva, the silk gland pair was carefully separated from the head by delicately pulling on the anterior gland regions, placed in a 1.5 ml microfuge tube, immediately flash frozen in liquid nitrogen, and stored at −80°C.
cDNA Library Construction and Sequencing
Frozen silk glands were pulverized under a small volume of liquid nitrogen with mortar and pestle and the resulting powder was added to TRIzol reagent (Invitrogen, Carlsbad, CA, USA) for RNA extraction. Total RNA was purified using the RNeasy mini kit (Qiagen, Valencia, CA, USA). After precipitation with isopropanol, the total RNA was dissolved in 0.5% SDS containing 20 mM sodium acetate (pH 5.3) and sent to Takara Bio (Shiga, Japan) for cDNA library construction. cDNA was made with the Stratagene (La Jolla, CA, USA) cDNA Synthesis Kit then directionally cloned into pBluescript II SK(+) (Stratagene, La Jolla, CA, USA) that had been digested with EcoRI and XhoI. The resulting cDNA library size was 1.1 × 107 cfu, and the average insert size was 1.58 kb, which was estimated from 16 randomly chosen clones. Nucleotide sequences were determined from the 5′ (T3) and 3′ (T7) ends of 4,000 randomly chosen clones using an ABI3730XL (Applied Biosystems, Foster City, CA, USA).
Sequence Characterization
Vector sequences were trimmed away and short reads of <100 bp were removed from the data set using Sequencher (Gene Codes, Ann Arbor, MI, USA). Batch BLASTX and BLASTN searches were performed on all sequences against the nr database (www.ncbi.nlm.nih.gov/BLAST). Both BLAST searches were performed twice, once with the low complexity filter enabled (default) and once with the filter disabled. The filter disabled searches were necessary to identify the H-fibroin gene, which is almost entirely highly repetitive (i.e., low complexity) sequence. All BLAST results were compared and all hits were visually inspected. Contiguous sequences (contigs) were generated using Phrap (Green 1994) employing the default settings with the exception of minimum base match, which was set to 60 instead of 20 to increase stringency. Contig assemblies were refined and checked for chimeras with Lasergene’s SeqMan (DNASTAR, Madison, WI, USA). To verify the accuracy of the H- and L-fibroin contigs, a second set of these contigs was assembled using Sequencher 4.2 (settings were 90% minimum mismatch, 60 bp minimum overlap) from the sequences annotated as H- or L-fibroin based on BLAST searches.
Amino acid translations were estimated from cDNA transcripts using SeqMan (DNASTAR, Madison, WI, USA). Protparam (ExPASy tools: Gasteiger et al. 2005) was used to compute amino acid composition and isoelectric point (pI).
Comparative Analyses
The H. californicus H- and L-fibroin transcripts were compared to lepidopteran and trichopteran sequences obtained from the NCBI database. Specifically, coding sequences for the H-fibroins of Ephestia kuehniella (accession number AY253535), Antheraea yamamai (AF410906), Bombyx mandarina (DQ459410), Bombyx mori (NM_001113262), Galleria mellonella (AF095240), Hydropsyche angustipennis (AB214507), Limnephilus decipiens (AB214509), Rhyacophila obliterata (AB354588), and Yponomeuta evonymellus (AB195979); and the L-fibroins of Bombyx mandarina (AB001820), Bombyx mori (X17291), Dendrolimus spectabilis (AB001822), Galleria mellonella (S77817), Hydropsyche angustipennis (AB214508), Limnephilus decipiens (AB214510), Papilio xuthus (AB001824), Rhyacophila obliterata (AB354590), and Yponomeuta evonymellus (AB195977) were downloaded. Sequence alignments were performed with ClustalW (Thompson et al. 1994) and refined by eye.
Signatures of selection on the protein coding regions of L-fibroin and the carboxyl-terminal region of H-fibroin were examined through maximum likelihood analyses using the codeml package of PAML (Yang 2007). PAML determined ω, the ratio of non-synonymous substitutions per non-synonymous site to synonymous substitutions per synonymous site, for each codon. We used an input tree based on recent studies of higher-level lepidopteran relationships (Kjer et al. 2002; Whiting 2002; Wiegmann et al. 2002) and analyzed the data with four different models: M1a, neutral model; M2a, selection model; M7, beta distribution neutral model; and M8, beta distribution with selection. Models M1a versus M2a and M7 versus M8 were then compared using likelihood ratio tests (Yang et al. 2005).
Phylogenetic analyses were conducted in PAUP* v.4.0b10 (Swofford 1998). In parsimony analyses, all character transformations were weighted equally. In maximum likelihood analyses, models of DNA evolution were chosen by the Akaike information criterion implemented in MODELTEST v.3.6 (Posada and Crandall 1998) and PAUP*. Parsimony searches utilized the branch and bound method and maximum likelihood searches were heuristic. Both parsimony and maximum likelihood searches were performed with random taxon addition and tree-bisection and reconnection. Nodal support values were assessed using bootstrap analyses (Felsenstein 1985). Bootstrap replicates were heuristic with tree-bisection and reconnection branch swapping. For parsimony searches, 10,000 iterations with 100 random taxon additions were done and 1,000 iterations with 10 random taxon additions were executed in maximum likelihood searches.
Results and Discussion
Identification of Silk cDNAs
The H. californicus silk gland cDNA library was constructed and 4,000 clones were sequenced from both ends. From these reads, 600 contigs that contained between two to 80 sequences were assembled with Phrap (Green 1994). In addition to contigs of cellular and tissue maintenance genes, we identified several contigs of two major silk genes, H- and L-fibroin.
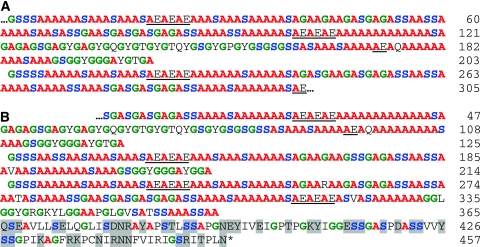
There were 27 clones that encoded long repetitive regions. The top BLAST hit for each of these clones was the H-fibroin from the Japanese oak silkmoth, Antheraea yamamai (Genbank AF410906). The similarity was largely due to the presence of frequent stretches of poly-alanine in the repetitive region. A representative clone containing 305 codons for a portion of the repetitive region of H. californicus H-fibroin (Genbank GU144521) is shown in Fig. 1a. The longest clone with both repetitive region and a complete carboxyl-terminal region (Genbank GU144520) encoded 457 amino acid residues (Fig. 1b).
Fig. 1.
Hepialus californicus heavy chain fibroin. a Middle segment clone depicting a full-length and partial-length ensemble repeat (GU144521). b Carboxyl-terminal clone with upstream non-repetitive region (GU144520). Single letter abbreviation for amino acids, residue position number on the right, and ellipsis (…) for additional, undetermined sequence. Highlights for glycine (green), serine (blue), alanine (red), acidic motifs in the repetitive region (underlined), and polar residues in the non-repetitive region (gray shading)
In addition to the 27 H-fibroin transcripts, 537 clones of our cDNA library contained L-fibroin transcripts. Many of these clones were full-length, as determined from the bidirectional sequencing reads (Genbank GU180666–GU180675; Fig. 2). The top BLAST hit for all these clones was the L-fibroin of the greater wax moth, Galleria mellonella (Genbank S77817). L-fibroin was by far the most numerous transcript identified within the cDNA library, reflecting a high expression level of the L-fibroin gene in the silk glands.
Fig. 2.
Alignment of Hepialus californicus light chain fibroin (LCF) alleles (GU180666-GU180675). Shading indicates 100% amino acid identity. Dots and arrow denote cysteines involved in bonding within L- and with H-fibroin, respectively. Dashes are alignment gaps. Position number appears above each block
Unlike previous lepidopteran silk studies (Inoue et al. 2000; Tanaka and Mizuno 2001; Sehnal and Žurovec 2004; Yonemura and Sehnal 2006), we did not detect a H. californicus transcript that corresponded to the P25 protein. This absence may indicate that the P25 transcript was present in much lower abundance than those of L-, H-fibroin, and the ~300 other genes that were identified in the cDNA library. Such rarity would have made P25 transcripts unlikely to be sampled from our cDNA library. However, it would be unexpected for an integral component of silk to be so scarce in an active silk gland. Hence, it is more likely that H. californicus lacks P25, which has only been identified within the lepidopteran clade Ditrysia (Inoue et al. 2000; Sehnal and Žurovec 2004; Yonemura and Sehnal 2006). P25 is also not known from Trichoptera, despite characterization of silk gland cDNA and N-terminal sequencing of silk gland proteins close to the expected size (Yonemura et al. 2006, 2009). Thus, P25 may be restricted to Ditrysia. Genomic characterization of H. californicus would provide more definitive evidence as to whether the lack of P25 in the silk gland cDNA library was due to absence or non-expression of a P25 gene.
A third explanation for the absence of P25 in our silk gland expression library is that H. californicus may have secondarily lost P25. There is precedence for losing this silk component. P25 has been lost along with L-fibroin in Saturniidae, a family within the Ditrysia. Silk fibers of these silkmoths are constructed solely from H-fibroins (Tamura et al. 1987; Tanaka and Mizuno 2001). Secondary loss of P25 in H. californicus, however, is not the most parsimonious given the absence of P25 in Trichoptera. Analysis of silks in more basal lepidopteran clades, such as Micropterygidae, is needed to more definitively address this possibility.
Heavy Chain Fibroin
From the 27 H. californicus H-fibroin clones, we obtained 12 middle fragments that encompassed only the repetitive region (described below) and 20 end fragments that included repetitive region and the carboxyl-terminal region. No nucleotide variation was found in the 3′ sequences that encoded the 92 amino acid long, non-repetitive carboxyl-terminal region (Fig. 1b). In the repetitive region, however, the sequences from both middle (Fig. 1a) and end fragments (Fig. 1b) varied in subrepeat length and arrangement. These differences were likely due to recombination and insertion/deletion mutations because highly iterated sequences, such as the genes for moth H-fibroins are prone to slip strand mispairing and unequal crossover (Ueda et al. 1985).
Like other lepidopteran H-fibroins and some spider silk proteins (Gatesy et al. 2001), the H. californicus H-fibroin is highly repetitive in sequence and dominated by alanine, glycine, and serine (48, 17, 17%, respectively, of amino acid composition; Fig. 1). This composition resembles the 43% alanine, 27% glycine, and 11% serine of the H-fibroin of the saturniid silkmoths, A. yamamai and A. pernyi (Sezutsu and Yukuhiro 2000; Hwang et al. 2001). In other moth H-fibroins, alanine is typically much lower (e.g., 26% in the H-fibroin of Y. evonymellus; Yonemura and Sehnal 2006). The H-fibroins of caddisflies contain dramatically less alanine; in Limnephilus decipiens, the repetitive region of H-fibroin is completely devoid of alanine and instead is composed of glycine (25%), serine (17%), and non-polar residues (Yonemura et al. 2006).
H. californicus H-fibroin resembles moth H-fibroins in that it consists of a series of repetitive motifs that form a higher-level repeat unit known as an ensemble (Fig. 1). The ensemble repeat in H-fibroin of H. californicus is ~203 amino acids in length and includes three distinct motifs, poly-alanine, glycine–X (where X is usually alanine, serine, or tyrosine), and alanine–glutamic acid. Each of these motifs is likely to form sequence-specific secondary structures that contribute to the formation of the silk filament. The poly-alanine, glycine–alanine, and glycine–serine repeats are known to form crystalline β-sheets that provide strength and stiffness to the silk fiber (Gosline et al. 1986; Guerette et al. 1996). H. californicus H-fibroin also contains glycine–glycine–X (where X is either alanine, serine, or threonine), which is part of the α-helical structures thought to reorient the amino acid chain in many spider fibroins (Hayashi et al. 1999; Hayashi and Lewis 2001; Ashida et al. 2003).
Polar amino acids are interspersed among the prevalent glycine and alanine residues in H. californicus H-fibroin. The acidic amino acids, such as glutamic acid, increase the hydrophilicity and decrease the pI of the H-fibroin, both of which may be important for stability of the silk dope and subsequent filament formation (Wong Po Foo et al. 2006). The overall pI (the pH at which a molecule has zero net charge) of the H. californicus repetitive region (Fig. 1a) is 3.98, which is close to the 4.03 of B. mori H-fibroin (Wong Po Foo et al. 2006). The pI of the carboxyl-terminal region (last 92 residues) of the H. californicus H-fibroin (Fig. 1b) is 8.10, and in B. mori it is 10.53. The high pI of this region is offset by the low pI of the L-fibroin (see below).
The large number of poly-alanine repeats within H. californicus H-fibroin may be required to provide structural stiffness to the silk fibers. H. californicus larvae live underground in silk lined tunnels and feed on the root systems of plants. In preparation for metamorphosis, the larvae burrow into the main tap root and migrate into the plant stalk, where they spin pupation cocoons (Strong et al. 1996; Nielsen et al. 2000). As the larvae live underground, high tensile strength, which is associated with prey capture or escape line functions, is unnecessary. Instead, the stiffness provided by the poly-alanine repeats may improve tunnel stability. Keeping tunnels clear and open could facilitate the maintenance of easy escape routes and access to food resources. Stiff silk may also prevent tunnel failure in sandy or sandy loam soil, such as at the Bodega Bay Marine reserve where our H. californicus were collected.
While the repetitive region is important for the structural characteristics of H-fibroin, the carboxyl-terminal region is implicated in fiber formation. In H. californicus, this region contains 42 polar amino acids (indicated by gray shading, Fig. 1b). In silk proteins it is common to have a higher proportion of polar amino acids in the carboxyl-terminal region than in the repetitive region (Bini et al. 2004; Sehnal and Žurovec 2004). The polarity of these amino acids increases hydrophilicity and thus can aid solubility of H-fibroin within the glandular lumen (Bini et al. 2004). Also potentially contributing to solubility is the longer length of the H. californicus H-fibroin carboxyl-terminal region (92 amino acids) compared to the homologous region in other lepidopteran silks. For example, in B. mori the equivalent region is 50 amino acids in length and in G. mellonella it is 60 amino acids long (Bini et al. 2004). By having a larger number of hydrophilic polar amino acids, the longer non-repetitive region in H. californicus H-fibroin may increase silk protein solubility, and possibly compensate for lacking a homolog to the hygroscopic glycoprotein P25.
Besides affecting solubility, the carboxyl-terminal region of H-fibroin enables intermolecular bonds that are necessary for proper fiber formation (Mori et al. 1995). Comparing the H-fibroins of Lepidoptera and Trichoptera reveals several amino acids that are highly conserved across taxa (Fig. 3). Notably, all the H-fibroins have a cysteine at ~20 residues before the protein end (marked by arrow, Fig. 3). This cysteine forms a disulfide bridge with the cysteine at position 190 of L-fibroin (designated by arrow, Fig. 2; Tanaka et al. 1999). This disulfide bond is necessary for transport of the H- and L-fibroin heterodimers into the glandular lumen (Inoue et al. 2004). The consistent locations of these cysteines across lepidopteran and trichopteran H- and L-fibroins underscore their importance to fiber formation. For example, lacking one of these cysteines (position 190 of L-fibroin) prevents the formation of H- and L-fibroin heterodimers, thus rendering B. mori incapable of spinning a cocoon (the naked pupa mutation; Mori et al. 1995). A similarly positioned cysteine in the carboxyl-terminal region of spider major ampullate (dragline silk) fibroin has also been implicated in silk filament formation (Ittah et al. 2006).
Fig. 3.
Alignment of amphiesmenopteran heavy chain fibroin carboxyl-terminal regions. Shading shows >50% conservation of chemical type: acidic (D, E), hydrophobic (A, G, I, L, V), amine (N, Q), aromatic (F, W, Y), basic (K, R, H), hydroxyl (S, T), proline (P), and sulfur-containing (C, M). Dots and arrow denote cysteines involved in bonding within H- and with L-fibroin, respectively. Underlines at the bottom of columns mark positions that have experienced purifying selection. Dashes are alignment gaps. Positions are numbered in relation to the protein end. Taxon abbreviations and Genbank accessions: Antheraea yamamai (Ant yam; AF410906), Bombyx mandarina (Bom man; DQ459410), Bombyx mori (Bom mor; NM_001113262), Ephestia kuehniella (Eph kue; AY253535), Galleria mellonella (Gal mel; AF095240), Yponomeuta evonymellus (Ypo evo; AB195979), Hepialus californicus (Hep cal; GU144520), Hydropsyche angustipennis (Hyd ang; AB214507), Limnephilus decipiens (Lim dec; AB214509), and Rhyacophila obliterata (Rhy obl; AB354588)
While the H. californicus H-fibroin has the conserved cysteine at −22 (marked by arrow, Fig. 3), it lacks the conserved cysteines that occur in most other lepidopteran H-fibroins at positions −1 and −4 (marked by dots, Fig. 3). In B. mori, it has been shown that these latter two cysteines form an intramolecular bond within the H-fibroin molecule (Tanaka et al. 1999). H. californicus, however, is not unique in missing these cysteines. The lack of these paired cysteines in the H-fibroin of A. yamamai (Saturniidae) is associated with silk filament formation based on H-fibroin homodimers rather than H- and L-fibroin heterodimers (Tamura et al. 1987). Similarly, their absence in the H-fibroin of H. californicus indicates yet another variation of silk fiber formation within lepidopterans.
Light Chain Fibroin
Ten contigs representing unique allelic variants were assembled from the 537 L-fibroin clones (Fig. 2). Nine contigs (LCF-1 to LCF-9) differ from each other by one to five non-synonymous changes. Strikingly, six of the 11 non-synonymous mutations occur at sites that are evolving more rapidly than 80% of the codon sites (see below). The tenth contig, LCF-10, is distinct from the other nine alleles by a sizeable gap that spans 27 amino acids and is likely a deletion mutant. The L-fibroin gene has six highly conserved exon/intron boundaries in the caddisflies H. angustipennis and R. obliterata and also in the moth B. mori, except that in the last species the fifth exon is split into two (Kikuchi et al. 1992; Yonemura et al. 2009). If the H. californicus L-fibroin gene also has this conserved exon arrangement, then a deletion of 81 nucleotides from the central part of exon three would account for LCF-10. Despite the differences present among the 10 L-fibroin alleles, large stretches of amino acids remain constant, suggesting functional importance of the specific sequence in these regions.
Comparison of the H. californicus L-fibroin to the other lepidopteran and trichopteran L-fibroins shows extensive conservation of amino acid sequence and biochemical properties (shading, Fig. 4). The conserved residues include aspartic acid and glutamic acid (red and bold, Fig. 4) that render the L-fibroin pI acidic. For example, the pI of B. mori L-fibroin is 5.06 and that of H. californicus is 6.13. Acidic pI values are hypothesized to be important for the conversion of silk proteins from a gel state to a liquid crystalline solution prior to spinning fibers (Wong Po Foo et al. 2006). The negatively charged L-fibroin balances the charge of the carboxyl-terminal region of H-fibroin, which is often basic (see above).
Fig. 4.
Alignment of amphiesmenopteran light chain fibroins. Shading shows >50% conservation of chemical type as shown in Fig. 3. Dots and arrow denote cysteines involved in bonding within L- and with H-fibroin, respectively. Underlines at the bottom of columns mark positions that have experienced purifying selection. Dashes are alignment gaps. Conserved aromatic residues are boxed and acidic residues are red and bold. Position number appears above each block. Taxon abbreviations and Genbank accessions: Bombyx mandarina (Bom man; AB001820), Bombyx mori (Bom mor; X17291), Dendrolimus spectabilis (Den spe; AB001822), Papilio xuthus (Pap xut; AB001824), Galleria mellonella (Gal mel; S77817), Yponomeuta evonymellus (Ypo evo; AB195977), Hepialus californicus (Hep cal; GU180666), Hydropsyche angustipennis (Hyd ang; AB214508), Limnephilus decipiens (Lim dec; AB214510), and Rhyacophila obliterata (Rhy obl; AB354590)
Three cysteine residues in H. californicus L-fibroin are present in similar locations in all known L-fibroins of Amphiesmenoptera (Figs. 2, 4). These conserved residues form the intra- and intermolecular bonds required for protein transport and fiber formation (Takei et al. 1987; Tanaka et al. 1999). Specifically, the first two cysteines (marked by dots, Figs. 2, 4) are known in B. mori to form an intramolecular bond thought to be important for proper protein conformation (Tanaka et al. 1999). Also shown through studies of B. mori, the third conserved cysteine (denoted by an arrow, Figs. 2, 4) forms a disulfide bond with the cysteine located in position −22 of H-fibroin (marked by an arrow, Fig. 3). Assembly of the L-fibroin–H-fibroin complex is essential for the transport of both components out of the endoplasmic reticulum and into the Golgi complex (Takei et al. 1987).
In addition to the conserved cysteine residues, there are five sites with aromatic amino acids within all of the lepidopteran and trichopteran L-fibroins (boxed, Fig. 4). There are also aromatic sites that while not conserved across Amphiesmenoptera, are consistent within Ditrysia (positions 114, 174) or Trichoptera (positions 84, 98, 158, 168, and 274). These aromatic amino acid sites may be important for forming aromatic–aromatic interactions that stabilize the configuration of L-fibroin. Burley and Petsko (1985) noted that many globular proteins contain amino acids with aromatic side chains. Using high resolution protein structures, they showed that aromatic side chains interact at dihedral angles between 50° and 90° and that these residues are separated by a mean of 38.6 residues (minimum of seven). Within L-fibroin, the mean interval between aromatic amino acid sites present across all Amphiesmenoptera is 42 and the Ditrysia-specific and Trichoptera-specific means are 60 and 45.8, respectively, with every individual interval much greater than seven. The stabilization resulting from these aromatic–aromatic interactions may enhance the micellar and globular conformations, which are required for the solubility of silk proteins within the glandular lumen prior to silk spinning (Jin and Kaplan 2003).
Molecular Evolution
Measuring the ratio of the non-synonymous substitution (replacement) rate versus the synonymous substitution (silent change) rate in coding sequences can provide insights into the evolution and function of a specific protein (Yang and Bielawski 2000). The ratio of replacement substitutions (d N) over silent substitutions (d S) is ω. Fixation of non-synonymous substitutions may reflect functional adaptive change within a molecule. For example, Gatesy and Swanson (2007) analyzed the ACR gene, which encodes a fertilization protein within mammals. Within ACR, they detected 21 rapidly evolving codon sites that may have adaptive significance. In contrast, conservation of amino acids over evolutionary time is thought to reflect a molecule that is undergoing purifying selection, indicating that change of amino acid residue is deleterious to the molecule’s function.
If ω < 1 at a specific site, then that site may have undergone purifying selection (i.e., a slow rate of change or no change over time). If ω = 1 at a given site, then the fixation of changes has occurred at the neutral rate of evolution, and, finally, if ω > 1 at any site, then that site may have been subject to adaptive selection. Calculating ω at each site may thus provide insights into the function of a particular amino acid within a protein. For example, Sawyer et al. (2005) determined that adaptive evolution took place in the primate TRIM5α, a gene encoding a protein responsible for limiting retroviral infection. Through their analyses, they identified a 13 residue long stretch in the protein that is necessary for resisting retroviruses. Similarly, measuring the rate of selection on the silk fibroins may reveal additional areas of conservation or areas undergoing positive selection.
Given the importance of silk to the ecology of Trichoptera and Lepidoptera, we measured the rate of molecular evolution at each codon site for L-fibroin and last 26 amino acids of the carboxyl-terminal region of H-fibroin (Fig. 3). The repetitive regions were not analyzed because of the sequence divergence across taxa and the high variability of repeat motifs even within an individual fibroin molecule. For the carboxyl-terminal region, there was no significant difference between log likelihood scores of the M7 (neutral) and M8 (selection) models, but there was a significant difference (P value > 0.0001) in log likelihood values of M1a (nearly neutral; −713.81) and M2a (selection; −736.55). Based on these comparisons we accepted M1a, the nearly neutral model.
Although model M1a (nearly neutral) implies that no sites have rapidly evolved under positive selection, the ω values for individual sites indicate that the carboxyl-terminal region has a history of purifying selection. All but two of the 26 sites had ω values <0.3, and most were below 0.1 (underlined positions, Fig. 3). These very low values signal a slow rate of evolution within the H-fibroin carboxyl-terminal region, consistent with mutations in this region being deleterious to silk solubility and fiber formation across Lepidoptera and Trichoptera.
We analyzed the rate of evolution at all 289 sites of the L-fibroin (Fig. 4). There was no significant difference between models M1a (−8393.39) versus M2a (−8393.39) or models M7 (−8354.73) versus M8 (−8354.20). Therefore, we chose M1a (nearly neutral model) as the best fit to the data. Most (80%) of the sites within L-fibroin had ω values <0.5, indicating that much of the molecule has undergone purifying selection (underlined positions, Fig. 4). For example, the conserved aromatic and cysteine sites (Fig. 4) had ω values of 0.15 or lower. These very low ratios can be contrasted with the 20% of amino acid sites with ω values greater than 0.5 that are interspersed throughout the protein. These more labile sites may have lower functional significance to the fibroin. The overall slow rate of change over 250 million years within the coding region of L-fibroin highlights the critical role of this molecule in silk protein transport and fiber formation, despite not constituting the bulk of the silk fiber (Yonemura et al. 2009).
Phylogenetic Signal
Amphiesmenoptera, the taxonomic grouping of Lepidoptera and Trichoptera, is a well established clade based on morphological and molecular data (Kristensen 1999; Whiting 2002). However, some higher-level groupings within Lepidoptera and Trichoptera have yet to be resolved. For example, phylogenetic relationships within the large lepidopteran subclades Ditrysia and Heteroneura are unclear (Wiegmann et al. 2000, 2002) and the position and monophyly of the trichopteran suborder Spicipalpia is uncertain (Morse 1997; Kjer et al. 2002). Given these issues, we test the phylogenetic utility of L-fibroin and the H-fibroin carboxyl-terminal region to resolve evolutionary relationships within Amphiesmenoptera.
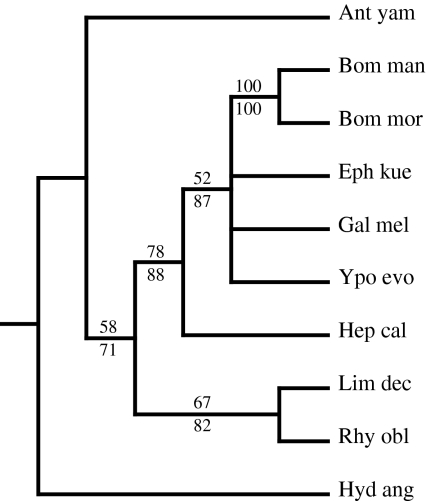
We utilized the 26 codon carboxyl-terminal alignment plus the stop codon to construct a gene tree of the H-fibroin carboxyl-terminal region. The reconstructed H-fibroin gene tree does not include a monophyletic Lepidoptera or Trichoptera (Fig. 5). This is due to the placement of A. yamamai outside of Lepidoptera because of its divergent carboxyl-terminal sequence, which most likely reflects its unusual silk filament composition of only H-fibroin dimers (see above). If the A. yamamai sequence is removed from the data matrix, Trichoptera and Lepidoptera are each recovered as monophyletic, although there is no resolution of individual clades within Lepidoptera. The carboxyl-terminal alignment of H-fibroin does not provide many phylogenetically informative characters due to its short length and sparse taxon sampling.
Fig. 5.
50% majority rule parsimony gene tree based on the carboxyl-terminal region of heavy chain fibroin. >50% bootstrap support values shown above (parsimony) and below (maximum likelihood) nodes. Taxon abbreviations and Genbank accessions as in Fig. 3
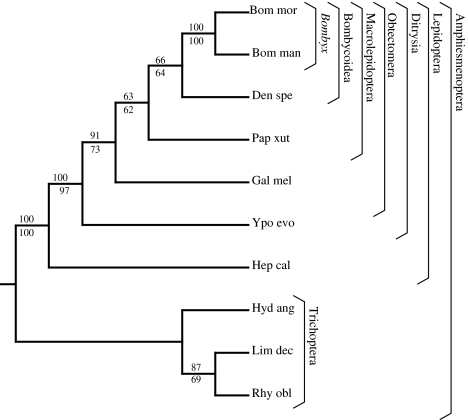
L-fibroin was more promising. The L-fibroin gene tree was constructed from the full-length coding region transcripts. Including gaps, the total alignment length was 867 nucleotides. The gene tree (Fig. 6) recovers monophyletic Trichoptera, monophyletic Lepidoptera, and is concordant with previous lepidopteran molecular phylogenies (Regier et al. 2008). Both parsimony and maximum likelihood trees agree at all nodes with moderate to high bootstrap support values for a number of clades within Amphiesmenoptera. Several clades are resolved within Lepidoptera, including Bombycoidea (66% parsimony bootstrap), Macrolepidoptera (63% parsimony bootstrap), Obtectomera (91% parsimony bootstrap), and Ditrysia (100% parsimony bootstrap). In Trichoptera, R. obliterata groups with L. decipiens, which is consistent with previous molecular studies (Kjer et al. 2002). Given the size and numerous introns of the L-fibroin gene, the best method to obtain sequence data would be from mRNA, using methodology such as in Regier et al. (2008). Ideally, mRNA should be isolated from last instar larvae to ensure an abundance of L-fibroin transcripts. Based on the resolved gene tree and bootstrap support values, L-fibroin is a promising molecular marker for reconstructing the higher-level relationships within Amphiesmenoptera.
Fig. 6.
Maximum likelihood and parsimony gene tree based on light chain fibroin. >50% bootstrap support values shown above (parsimony) and below (maximum likelihood) nodes. Brackets indicate recognized clades (Weller and Pashley 1995; Wiegmann et al. 2000, 2002). Taxon abbreviations and Genbank accessions as Fig. 4
In summary, we characterized the fibroins of the ghost moth, H. californicus. By involving H- and L-fibroin but apparently not P25, silk fiber formation in H. californicus is similar to that of Trichoptera (Yonemura et al. 2006, 2009), despite major differences in the spinning environment and ecological function of subterranean ghost moth silk versus aquatic caddisfly silk. The H. californicus L-fibroin had numerous attributes, such as an acidic pI and homologous placement of cysteine and aromatic residues that were maintained via purifying selection since the common ancestor of Lepidoptera and Trichoptera (Fig. 4). Similarly, the carboxyl-terminal region of the H. californicus H-fibroin contained several conserved amino acids, such as a cysteine located 22 residues upstream of the protein end (Fig. 3). The slow pace of substitutions suggests that mutations within L-fibroin and the H-fibroin carboxyl-terminal region are deleterious to silk fiber formation. In contrast, the repetitive region of H-fibroin is divergent across taxa and prone to rearrangements. Despite its more rapid rate of evolution, the repetitive region has constraints on its sequence. Like the silk proteins of other insects and spiders, the H. californicus H-fibroin is internally repetitive (tandem arrayed ensemble repeats) and dominated by glycine, alanine, and serine (Fig. 1; Gatesy et al. 2001).
While they had many evolutionarily conserved characteristics, the ghost moth silk proteins exhibited key differences from other amphiesmenopteran silk proteins. Specifically, the H. californicus H-fibroin carboxyl-terminal region lacked the two cysteines present in most other lepidopteran H-fibroins at positions −1 and −4 (Fig. 3). These missing cysteines in conjunction with lack of evidence for P25, indicate that the ghost moth has an alternative method of fiber formation compared to more derived lepidopterans, as typified by Bombyx mori (Inoue et al. 2000). Thus, by filling in the substantial phylogenetic gap between caddisflies and more derived moths, H. californicus provides insight into the evolution of silk in Lepidoptera.
Acknowledgments
We thank J. Bastow, D. Gruner, J. Mclaughlin, M. Roberts, and D. Strong for providing larval H. californicus and Shun-ichi Sasanuma for the technical part of EST sequencing. N. Ayoub, M. McGowen, and J. Starrett improved the manuscript. Research was supported by NSF award DEB-0515868 to CYH and Czech grant ME32 to FS. EST sequencing was supported by a grant from Japan’s Ministry of Agriculture, Forestry and Fisheries.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Ashida J, Ohgo K, Komatsu K, Kubota A, Asakura T. Determination of the torsion angles of alanine and glycine residues of model compounds of spider silk (AGG)(10) using solid-state NMR methods. J Biomol NMR. 2003;25:91–103. doi: 10.1023/A:1022220428948. [DOI] [PubMed] [Google Scholar]
- Bini E, Knight DP, Kaplan DL. Mapping domain structures in silks from insects and spiders related to protein assembly. J Mol Biol. 2004;335:27–40. doi: 10.1016/j.jmb.2003.10.043. [DOI] [PubMed] [Google Scholar]
- Burley SK, Petsko GA. Aromatic-aromatic interaction a mechanism of protein-structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Craig CL. Evolution of arthropod silks. Annu Rev Entomol. 1997;42:231–267. doi: 10.1146/annurev.ento.42.1.231. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence-limits on phylogenies an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Friedlander TP, Regier JC, Mitter C, Wagner DL. A nuclear gene for higher level phylogenetics: phosphoenolpyruvate carboxykinase tracks Mesozoic-age divergences within Lepidoptera (Insecta) Mol Biol Evol. 1996;13:594–604. doi: 10.1093/oxfordjournals.molbev.a025619. [DOI] [PubMed] [Google Scholar]
- Garb JE, Hayashi CY. Modular evolution of egg case silk genes across orb-weaving spider superfamilies. Proc Natl Acad Sci USA. 2005;102:11379–11384. doi: 10.1073/pnas.0502473102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. New York: Humana Press; 2005. [Google Scholar]
- Gatesy J, Swanson WJ. Adaptive evolution and phylogenetic utility of Acr (acrosin), a rapidly evolving mammalian fertilization gene. J Mammal. 2007;88:32–42. doi: 10.1644/05-MAMM-S-345R1.1. [DOI] [Google Scholar]
- Gatesy J, Hayashi C, Motriuk D, Woods J, Lewis R. Extreme diversity, conservation, and convergence of spider silk fibroin sequences. Science. 2001;291:2603–2605. doi: 10.1126/science.1057561. [DOI] [PubMed] [Google Scholar]
- Gosline JM, Demont ME, Denny MW. The structure and properties of spider silk. Endeavour. 1986;10:37–43. doi: 10.1016/0160-9327(86)90049-9. [DOI] [Google Scholar]
- Green P (1994) Phrap. http://www.genome.washington.edu/UWGC/analysistools/phrap.htm
- Grimaldi DA, Engel MS. Evolution of the insects. Cambridge: Cambridge University Press; 2005. [Google Scholar]
- Guerette PA, Ginzinger DG, Weber BHF, Gosline JM. Silk properties determined by gland-specific expression of a spider fibroin gene family. Science. 1996;272:112–115. doi: 10.1126/science.272.5258.112. [DOI] [PubMed] [Google Scholar]
- Hayashi CY, Lewis RV. Spider flagelliform silk: lessons in protein design, gene structure, and molecular evolution. Bioessays. 2001;23:750–756. doi: 10.1002/bies.1105. [DOI] [PubMed] [Google Scholar]
- Hayashi CY, Shipley NH, Lewis RV. Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins. Int J Biol Macromol. 1999;24:271–275. doi: 10.1016/S0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- Hwang J-S, Lee J-S, Goo T-W, Yun E-Y, Lee K-S, Kim Y-S, Jin B-R, Lee S-M, Kim K-Y, Kang S-W, Suh D-S. Cloning of the fibroin gene from the oak silkworm, Antheraea yamamai and its complete sequence. Biotechnol Lett. 2001;23:1321–1326. doi: 10.1023/A:1010542011150. [DOI] [Google Scholar]
- Inoue S, Tanaka K, Arisaka F, Kimura S, Ohtomo K, Mizuno S. Silk fibroin of Bombyx mori is secreted, assembling a high molecular mass elementary unit consisting of H-chain, L-chain, and P25, with a 6:6: 1 molar ratio. J Biol Chem. 2000;275:40517–40528. doi: 10.1074/jbc.M006897200. [DOI] [PubMed] [Google Scholar]
- Inoue S, Tanaka K, Tanaka H, Ohtomo K, Kanda T, Imamura M, Quan G-X, Kojima K, Yamashita T, Nakajima T, Taira H, Tamura T, Mizuno S. Assembly of the silk fibroin elementary unit in the endoplasmic reticulum and a role of L-chain for protection of α1, 2-mannose residues in N-linked oligosaccharide chains of fibrohexamerin/P25. Eur J Biochem. 2004;271:356–366. doi: 10.1046/j.1432-1033.2003.03934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittah S, Cohen S, Garty S, Cohn D, Gat U. An essential role for the C-terminal domain of a dragline spider silk protein in directing fiber formation. Biomacromolecules. 2006;7:1790–1795. doi: 10.1021/bm060120k. [DOI] [PubMed] [Google Scholar]
- Jin HJ, Kaplan DL. Mechanism of silk processing in insects and spiders. Nature. 2003;424:1057–1061. doi: 10.1038/nature01809. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Mori K, Suzuki S, Yamaguchi K, Mizuno S. Structure of the Bombyx mori fibroin light-chain-encoding gene upstream sequence elements common to the light and heavy-chain. Gene. 1992;110:151–158. doi: 10.1016/0378-1119(92)90642-3. [DOI] [PubMed] [Google Scholar]
- Kjer KM, Blahnik RJ, Holzenthal RW. Phylogeny of caddisflies (Insecta, Trichoptera) Zool Scr. 2002;31:83–91. doi: 10.1046/j.0300-3256.2001.00079.x. [DOI] [Google Scholar]
- Kristensen NP. Phylogeny of endopterygote insects, the most successful lineage of living organisms. Eur J Entomol. 1999;96:237–253. [Google Scholar]
- Mita K, Ichimura S, James TC. Highly repetitive structure and its organization of the silk fibroin gene. J Mol Evol. 1994;38:583–592. doi: 10.1007/BF00175878. [DOI] [PubMed] [Google Scholar]
- Mori K, Tanaka K, Kikuchi Y, Waga M, Waga S, Mizuno S. Production of a chimeric fibroin light-chain polypeptide in a fibroin secretion deficient naked pupa mutant of the silkworm Bombyx mori. J Mol Biol. 1995;251:217–228. doi: 10.1006/jmbi.1995.0429. [DOI] [PubMed] [Google Scholar]
- Morse JC. Phylogeny of Trichoptera. Annu Rev Entomol. 1997;42:427–450. doi: 10.1146/annurev.ento.42.1.427. [DOI] [PubMed] [Google Scholar]
- Nielsen ES, Robinson GS, Wagner DL. Ghost-moths of the world: a global inventory and bibliography of the Exoporia (Mnesarchaeoidea and Hepialoidea) (Lepidoptera) J Nat His. 2000;34:822–878. [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Regier JC, Cook CP, Mitter C, Hussey A. A phylogenetic study of the ‘bombycoid complex’ (Lepidoptera) using five protein-coding nuclear genes, with comments on the problem of macrolepidopteran phylogeny. Syst Entomol. 2008;33:175–189. doi: 10.1111/j.1365-3113.2007.00416.x. [DOI] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5 alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnal F, Žurovec M. Construction of silk fiber core in Lepidoptera. Biomacromolecules. 2004;5:666–674. doi: 10.1021/bm0344046. [DOI] [PubMed] [Google Scholar]
- Sezutsu H, Yukuhiro K. Dynamic rearrangement within the Antheraea pernyi silk fibroin gene is associated with four types of repetitive units. J Mol Evol. 2000;51:329–338. doi: 10.1007/s002390010095. [DOI] [PubMed] [Google Scholar]
- Strong DR, Kaya HK, Whipple AV, Child AL, Kraig S, Bondonno M, Dyer K, Maron JL. Entomopathogenic nematodes: natural enemies of root-feeding caterpillars on bush lupine. Oecologia. 1996;108:167–173. doi: 10.1007/BF00333228. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (* and other models) Sunderland: Sinauer Associates Inc., Publishers; 1998. [Google Scholar]
- Takei F, Kikuchi Y, Kikuchi A, Mizuno S, Shimura K. Further evidence for importance of the subunit combination of silk fibroin in its efficient secretion from the posterior silk gland-cells. J Cell Biol. 1987;105:175–180. doi: 10.1083/jcb.105.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Inoue H, Suzuki Y. The fibroin genes of Antheraea yamamai and Bombyx mori are different in their core regions but reveal a striking sequence similarity in their 5′ ends and 5′ flanking regions. Mol Gen Genet. 1987;207:189–195. doi: 10.1007/BF00331577. [DOI] [Google Scholar]
- Tanaka K, Mizuno S. Homologues of fibroin L-chain and P25 of Bombyx mori are present in Dendrolimus spectabilis and Papilio xuthus but not detectable in Antheraea yamamai. Insect Biochem Mol Biol. 2001;31:665–677. doi: 10.1016/S0965-1748(00)00173-9. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kajiyama N, Ishikura K, Waga S, Kikuchi A, Ohtomo K, Takagi T, Mizuno S. Determination of the site of disulfide linkage between heavy and light chains of the silk produced by Bombyx mori. Biochim Biophys Acta. 1999;1432:92–103. doi: 10.1016/s0167-4838(99)00088-6. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Mizuno S, Shimura K. Sequence polymorphisms around the 5′-end of the silkworm fibroin H-chain gene suggesting the occurrence of crossing-over between heteromorphic alleles. Gene. 1985;34:351–355. doi: 10.1016/0378-1119(85)90144-1. [DOI] [PubMed] [Google Scholar]
- Weller SJ, Pashley DP. In search of butterfly origins. Mol Phylogenet Evol. 1995;4:235–246. doi: 10.1006/mpev.1995.1022. [DOI] [PubMed] [Google Scholar]
- Whiting MF. Phylogeny of the holometabolous insect orders: molecular evidence. Zool Scr. 2002;31:3–15. doi: 10.1046/j.0300-3256.2001.00093.x. [DOI] [Google Scholar]
- Wiegmann BM, Mitter C, Regier JC, Friedlander TP, Wagner DM, Nielsen ES. Nuclear genes resolve mesozoic-aged divergences in the insect order Lepidoptera. Mol Phylogenet Evol. 2000;15:242–259. doi: 10.1006/mpev.1999.0746. [DOI] [PubMed] [Google Scholar]
- Wiegmann BM, Regier JC, Mitter C. Combined molecular and morphological evidence on the phylogeny of the earliest lepidopteran lineages. Zool Scr. 2002;31:67–81. doi: 10.1046/j.0300-3256.2001.00091.x. [DOI] [Google Scholar]
- Wong Po Foo C, Bini E, Hensman J, Knight DP, Lewis RV, Kaplan DL. Role of pH and charge on silk protein assembly in insects and spiders. Appl Phys A. 2006;A82:223–233. [Google Scholar]
- Yang Z. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski JP. Statistical methods for detecting molecular selection. Trends Ecol Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielson R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Yonemura N, Sehnal F. The design of silk fiber composition in moths has been conserved for more than 150 million years. J Mol Evol. 2006;63:42–53. doi: 10.1007/s00239-005-0119-y. [DOI] [PubMed] [Google Scholar]
- Yonemura N, Sehnal F, Mita K, Tamura T. Protein composition of silk filaments spun under water by caddisfly larvae. Biomacromolecules. 2006;7:3370–3378. doi: 10.1021/bm060663u. [DOI] [PubMed] [Google Scholar]
- Yonemura N, Mita K, Tamura T, Sehnal F. Conservation of silk genes in Trichoptera and Lepidoptera. J Mol Evol. 2009;68:641–653. doi: 10.1007/s00239-009-9234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žurovec M, Sehnal F. Unique molecular architecture of silk fibroin in the waxmoth, Galleria mellonella. J Biol Chem. 2002;277:22639–22647. doi: 10.1074/jbc.M201622200. [DOI] [PubMed] [Google Scholar]