Abstract
Homeodomain-interacting protein kinases including HIPK1, HIPK2 and HIPK3 are serine/threonine kinases that form a family of highly conserved kinases. HIPKs are involved in diverse cellular functions including regulation cell death, survival, proliferation and differentiation. Here we report the characterization of a human HIPK4 that we identified in a proteomic screen during our efforts to unravel novel markers linked to cell death and survival. Human HIPK4 protein is composed of 616 residues with predicted molecular mass of 69.425 kDa and harbors a serine/threonine protein kinase catalytic domain at its N-terminal end. In the in vitro kinase assay, HIPK4 exhibits kinase activity and mutation of the conserved lysine 40 or aspartic acid 136 residue in its catalytic domain inactivates its kinase function. Human HIPK4 harbors multiple putative serine/threonine- and tyrosine-specific phsophorylation sites and also contains four high probability sumoylation sites, findings that suggest its function to be modulated by post-translational modifications. HIPK4 has been so named in the database because of its sequence homology to HIPK1, 2 and 3 predominantly within its catalytic domain. However, HIPK4 is smaller in size than the known HIPKs and has additional distinct features suggesting it to be a unique member of the HIPK family. Further functional characterization of HIPK4 is needed and will prove valuable to ascertain whether it performs distinct functions or share overlapping functions with other HIPKs.
Keywords: Kinase, HIPK family, HIPK4, Phosphorylation, Sumoylation, Site-directed mutagenesis
Introduction
Homeodomain-interacting protein kinases including HIPK1, HIPK2 and HIPK3 (collectively designated as HIPKs) are serine/threonine kinases that were originally identified in a yeast two-hybrid screen as homeoprotein-interacting kinases (1 and reviewed in reference 2). The HIPKs were found to predominantly exhibit nuclear localization and enhance the repressor activities of NK homeoproteins (1). Since their original discovery, a considerable progress has been made to better understand their function. Several lines of evidence now indicate that the HIPKs are involved in a number of important cellular processes and mediate their effects by phosphorylation of several important proteins. For example, HIKP1, 2 and 3 have been reported to phosphorylate the homeodoamin protein NKx2.1 (1). HIPK2 phosphorylates tumor suppressor p53 on serine 46 (3, 4) and HIPK3 has been reported to phosphorylate FADD, the death domain-containing adaptor protein that is critical in mediating death signals originating from the extrinsic pathway of apoptosis (5).
Clearly, HIPKs form a family of highly conserved kinases that are involved in diverse cellular functions including regulation cell death, survival, proliferation and differentiation (2). In addition, they have been implicated in modulating the cellular response to DNA damage i.e. genotoxic stress (2). In our ongoing efforts to identify novel markers of cell death and survival, we had identified several novel human genes/proteins (6–10) including HIPK4. Here we report the characterization of this novel kinase.
Material and Methods
Materials
DMEM, Ham’s F-12 and RPMI-1640 cell culture media, Penicillin/Streptomycin and L-Glutamine were purchased from Mediatech, Inc (Manassas, VA, USA) and the fetal bovine serum was bought from Gemini Bioproducts (Calabasas, CA, USA). PrEBM and MEBM and their growth supplements were purchased from Lonza (Walkersville, MD, USA).
Cell lines and cell culture
Prostate cancer cell lines PC-3, DU145 and LNCap as well as breast cancer cell MCF-7 and HEK 293 cells used in this study have been described in our previous publications (6–12). DU145, MCF-7 and HEK 293 Cells were normally kept in DMEM (Mediatech, VA, USA) supplemented with 10% fetal bovine serum, 100 U/ml Penicillin, 100 ug/ml Streptomycin and 2 mM L-Glutamine. PC-3 and LNCap cells were kept in Ham’s F-12 and RPMI-1640 with supplements, respectively. PrEC, the normal prostate cells, were from Cambrex (Walkersville, MD, USA). PrEC (normal prostate) and MCF10A (normal breast) cells were normally kept in PrEBM and MEBM with the respective supplements.
Western blotting
Western blot analyses were performed by standard procedures as described previously (13, 14). HIPK4 signals were detected by using commercially available polyclonal anti-HIPK4 antibody (Abgent, San Diego, CA, USA) and also the one generated in our laboratory. β-actin signals were detected via a monoclonal antibody purchased from Sigma Chemicals (St. Louis, MO, USA).
Plasmids
To generate mammalian expression vector carrying human HIPK4 (pSR-Nstag-HIPK4 and pSR-HIPK4-Cstag), the following PCR primers were used for amplification of the HIPK4 open reading frames from clone MGC-26964 (accession number BC034501, purchased from ATCC):
pSR-Nstag-HIPK4
sense: 5′-GGGGGTACCATGTCCACCATCCAGTCG GAG-3′
antisense: 5′-ATAAGAATGCGGCCGCTCAGTGGTG CCCGGTGAC-3′
pSR-HIPK4-Cstag
sense: 5′ TGCTCTAGAATGTCCACCATCCAGTCG GAG-3′
antisense: 5′-TCCCCGCGGGTGGTGCCCGGTGACA TG-3′
The amplified products were respectively digested with kpnI-NotI and XbaI-SacII and then subcloned into the mammalian expression vector pSR. The resulting vectors were sequenced for confirmation of correct sequence.
pSR-HIPK4-Cstag kinase-dead mutant vectors
To generate kinase-dead mutants, site-directed mutagenesis was performed using QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). The following primers were used for respective kinase-dead mutants K40A, D136N and double mutant:
sense: 5′-GAGATGGTGGCCATTGCAATCCTCAAG AATGACG-3′
antisense: 5′-CGTCATTCTTGAGGATTGCAATGGC CACCATCTC-3′
sense: 5′-CTGGCTATCATCCACGCTAATCTCAAGC CTGAGAACATC-3′
antisense: 5′-GATGTTCTCAGGCTTGAGATTAGCG TGGATGATAGCCAG-3′.
S-tag protein-pull down and in vitro kinase assays
The S-tag protein pull-down and in vitro kinase assays were modified from the procedures described previously (12). Briefly, 293T cells were transiently transfected with pSR-HIPK4-Cstag or mutant vectors according to manufacturer’s suggested procedure. Cells were harvested and lysed in lysis buffer [50 mM Tris-HCl(pH 7.5), 120 mM NaCl, 0.1% NP-40, 5 mM EDTA, 2 mM MgCl2, 10 mM KCl, 2 mM Na3VO4, 25 mM glycerophosphate, 10 mM NaF, 1 mM PMSF,2 μg/mL leupeptin, 1 μg/mL pepstatin A, 1 μg/mL chymostatin, 0.1 μg/mL okdaic acid and 10 μg/mL aprotinin]. Approximately 200 μg of cell lysates were further mixed with 50 μl of S-protein conjugated agarose beads for 4 hours, and then the beads were washed with lysis buffer. The washed S-protein beads were mixed with 50 μl of kinase assay buffer [20 mM Tris-HCl (pH 7.5), 25 mM glycerophosphate, 2 mM DTT, 20 mM MgCl2, 40 μM ATP, 5 μCi γ-32P-ATP] and 2 μg substrate MBP for 30 min at 30°C. The samples were analyzed by SDS-PAGE and autoradiography.
Results and Discussion
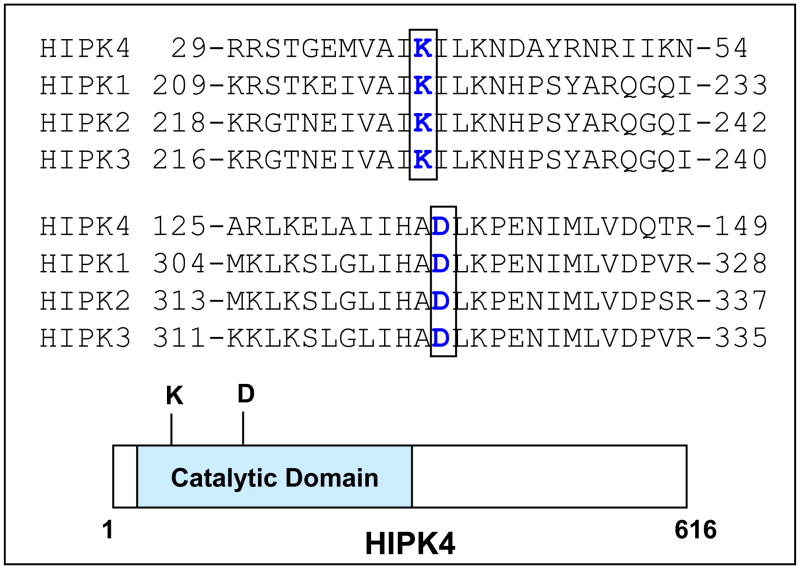
We identified human HIPK4 in a proteomic screen and found its sequence to be annotated in the GenBank as an uncharacterized hypothetical protein (accession # BC034501) so named because of its homology to HIPK1, 2 and 3. Figure 1 shows the amino acid sequence of HIPK4 and as is shown, HIPK4 protein is composed of 616 residues with predicted molecular mass of 69.425 kDa and pI of 6.18. Database searches revealed HIPK4 to harbor a “serine/threonine protein kinase catalytic domain” at its N-terminal end residing within residues 11 to 347. HIPK1, 2 and 3 also harbor the serine/threonine kinase catalytic domains at their N-termini (1–5) and their sequence comparisons with HIPK4 reveal that the residues within their kinase catalytic domains are conserved among all four kinases (Figures 2 and 3). Overall however, HIPK4 shows lesser sequence homology to HIPK1, 2 and 3 and has a unique sequence indicating it to be a novel serine/threonine kinase. Comparison of human HIPK4 sequence with comparable sequences from other species including mouse, rat, chimpanzee and monkey reveals high degree of sequence homology indicating this kinase to be highly conserved across species (Figure 4).
Figure 1. Amino acid sequence of human HIPK4.
Residues 11–347 corresponding to the catalytic domain are shown in blue. Conserved lysine (K) and aspartic acid (D) important for its catalytic activity are shown in red.
Figure 2.
Alignment of human HIPK4 amino acid sequence with sequences of human HIPK1, 2 and 3.
Figure 3. Amino acid sequence alignment of selected regions in the catalytic domains of HIPK4 and HIPK1, 2, 3.
Conserved critical residues lysine (K) and aspartic acid (D) are shown in blue color. Schematic illustration of HIPK4 depicts approximate location of K and D residues in the catalytic domain.
Figure 4. Alignment of human HIPK4 amino acid sequence with the corresponding sequences from various species.
Putative sumoylation sites are in blue color.
Further analysis of the HIPK amino acid sequence for post-translational modifications revealed human HIPK4 to harbor several putative phosphorylation sites including 18 serine, 5 threonine and 8 tyrosine phosphorylation sites. SUMOplot™ prediction was also performed and indicated that human HIPK4 contained several sumoylation sites. SUMO-1 is a small ubiquitin-related modifier (SUMO) that belongs to the ubiquitin and ubiquitin-like superfamily (15). The majority of sumoylation sites represent a four amino acid motif with a consensus sequence B-K-x-D/E (B: hydrophobic residue; K: SUMO conjugating lysine; x: any amino acid; D or E: acidic residue). Human HIPK4 harbors four such motifs (LKND; IKNE; LKPE and VKEP) with high probability of sumoylation occurring on lysine residues at positions 43, 53, 138 and 172 in these motifs. Interestingly, we note that all four motifs noted in human HIPK4 are conserved in other species including mouse, rat, chimpanzee and monkey (Figure 4).
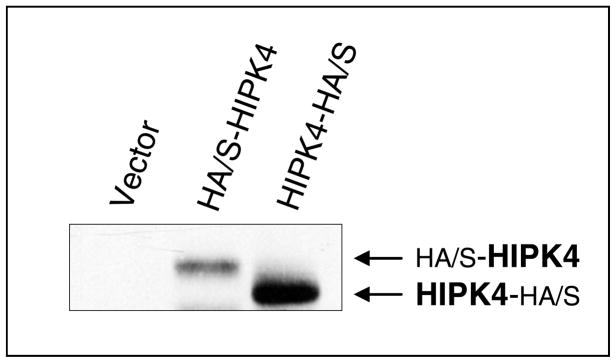
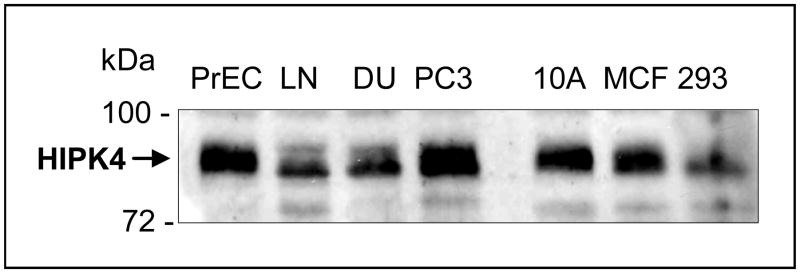
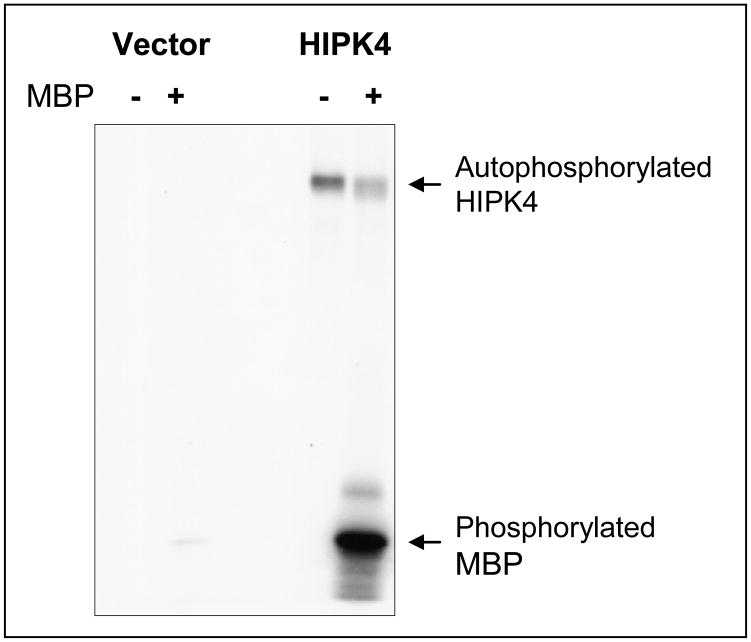
To test that human HIPK4 cDNA encodes bona fide protein, we performed Western blot analysis on the lysates of exogenous HIPK4-expressing HEK 293 cells and found that HIPK4 tagged with HA/S-tags at the Nor C-terminal end migrated in the expected size range (Figure 5). Figure 6 shows expression of endogenous HIKP4 in various human cell lines and as is shown endogenous HIPK4 migrates higher than the predicted molecular mass which could result due to post-translational modification. Next, we investigated whether the protein expressed by HIPKP4 cDNA had kinase activity and for that purpose performed in vitro kinase assay on lysates of HIPK4 expressing 293 kidney cells. As shown in Figure 7, the HIPK4 protein was capable of phosphorylating the universal kinase substrate, MBP (myelin basic protein) as well as exhibiting autophosphorylation in the in vitro kinase assay. HIPK1, 2 and 3 are all known to also exhibit autophosphorylation and have been reported to harbor important residues in their catalytic domains that are critical for their kinase activities. For example, the lysine (K) residue within their catalytic domains at positions 219, 228 and 226 for HIPK1, 2 and 3 respectively is highly conserved and mutation at this residue significantly affects their kinase activity (4, 5, 16, 17). We compared human HIPK4 sequence with those of other human HIPKs and noted that human HIPK4 also harbored the conserved lysine residue at position 40 within its catalytic domain (Figure 3). Furthermore, an aspartic acid (D) at position 315, 324 and 322 within the catalytic domains for HIPK1, 2 and 3 respectively is also highly conserved and critical for kinase activity (4, 5, 16, 17). HIPK4 also harbors the aspartic aid residue at position 136 within its catalytic domain (Figure 3).
Figure 5. Expression of exogenous human HIPK4 in HEK 293 cells.
Expression vectors carrying human HIPK4 cDNAs with HA/S dual tags at the N or C-terminal end were used for expression of HIPK4 in 293 human kidney cells. HIPK4 was detected by anti-HA tag antibody. HIPK4 with N-terminal tags migrates higher because of additional sequence added by the tags.
Figure 6. Expression of endogenous human HIPK4 in various human cells.
Western blot analysis was performed using anti-HIPK4 antibody on the lysates prepared form the indicated cells. PrEC: Normal prostate epithelial cells, LN: LNCap prostate cancer cells, DU145: prostate cancer cells, PC3: PC-3 prostate cancer cells, 10A: MCF-10A normal breast cells, MCF: MCF-7 breast cancer cells, 293: HEK 293 cells.
Figure 7. Human HIPK4 exhibits kinase activity.
Human 293 kidney cells were transiently transfected with S-tagged HIPK4 expression vector or same vector without HIPK4 insert. Cell lysates were prepared for S-tag protein pull-down and the pull-down precipitates were subjected to in vitro kinase assay using [γ-32P]-ATP in the presence or absence of MBP as a substrate. Signals indicating phosphorylated proteins were detected by autoradiography. As is shown, MBP is phosphorylated only when HIPK4 is present. HIPK4 also exhibits autophosphorylation
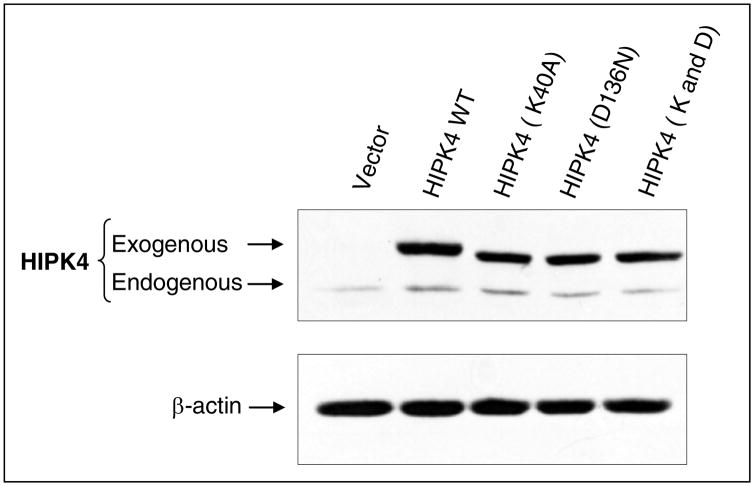
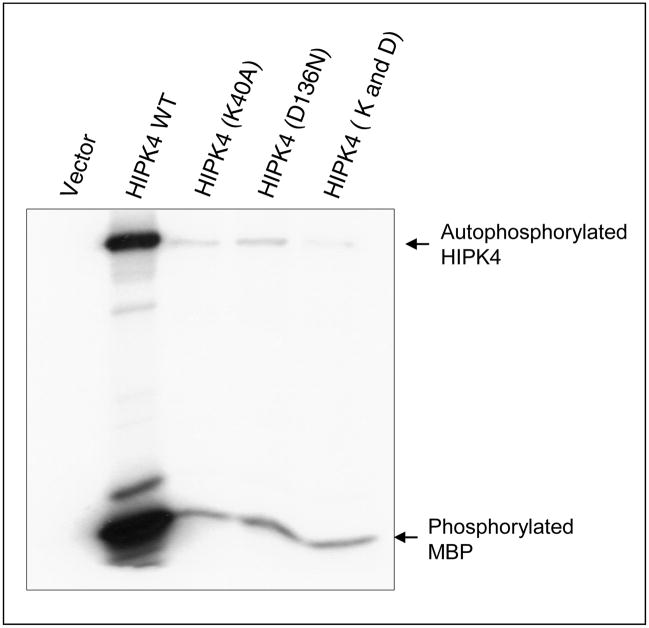
Next, we performed site-directed mutagenesis to mutate lysine 40 and replaced it with alanine. Similarly, aspartic acid 136 was mutated and replaced with asparagine. We also mutated both lysine 40 and aspartic acid 136 to generate double mutant. Independent expression vectors carrying wild type HIPK4 or these mutants were transiently transfected in 293 kidney cells and their expression was evaluated by Western blotting using anti-HIPK4 antibody (Abgent, San Diego, CA). As shown in Figure 8, wild type as well as mutant forms of HIPK4 are expressed and detected by the anti-HIPK4 antibody. Next, we sought to assess the kinase activity of these mutants and for that purpose these mutants were expressed in 293 kidney cells and their kinase activities were assessed by in vitro kinase assay. Our results indicated that mutations at these conserved residues significantly abolished HIPK4’s kinase activity (Figure 9). Together these results indicate HIPK4 to be a novel bona fide kinase.
Figure 8. Expression of wild type and kinase-dead mutants.
Human 293 kidney cells were transiently transfected with expression vectors carrying wild type or the indicated mutant HIPK4. Cell lysates were subjected to Western blot analysis using anti-HIPK4 antibody. The HIPK4 mutants migrate slightly faster than the wild type HIPK4. Similar feature has been reported for other HIPKs with mutations at the corresponding residues.
Figure 9. Mutation of lysine 40 or aspartic acid 136 or double mutations abolishe HIPK4 kinase activity.
Human 293 kidney cells were transiently transfected with expression vectors carrying wild type or mutant S-tagged HIPK4. Cells were also transfected with same vector without HIPK4 insert. Cell lysates were prepared for S-tag protein pull-down and the pull-down precipitates were subjected to in vitro kinase assay using [γ-32P]-ATP and MBP as a substrate. Signals indicating phosphorylated proteins were detected by autoradiography.
While we were characterizing human HIPK4, Arai et al., (18) reported the cloning and characterization of mouse Hipk4 cDNA. Mouse and human HIPK4 are highly homologous (Figure 4) and thus, functionally redundant. Arai et al., (18) found that mouse hipk4 functioned as a kinase both in vitro and in vivo (inside the cells). Furthermore, purified hipk4 also displayed kinase activity as well as autophosphorylation. Mouse hipk4 also harbors the conserved lysine at position 40 and mutation of lysine 40 also abolished kinase activity of the mouse hipk4 (18). Arai et al., (18) further reported that the mouse hipk4 was capable of phosphorylating human as well as mouse p53 on serine at position 9 and that led to p53 activation.
p53 is a transcription factor and a key regulator of apoptosis (19, 20). p53 is generally phosphorylated and activated by a variety of kinases and on various different residues including for example, serine and threonines at positions 6, 9, 15, 20, 33, 37, 46 and 392 (18, 21–23). It is believed that phosphorylation of p53 via multiple kinases (which do not always simultaneously phosphorylate p53) is warranted to fine tune p53 regulation and function (23). For example, in response to a signal, residue-specific phosphorylation on p53 determines which protein would selectively interact with p53 and that in turn enables p53 to selectively up or down-regulate some but not all of its downstream target genes. Residue-specific phosphorylation of p53 is also expected to fine tune its transcription-independent cytoplasmic function, as p53 resides in both nucleus and cytoplasm.
It is of note that HIPK2 also phsophorylates p53; in the case of human p53 such phophorylation occurs on serine 46 while for murine p53 it takes place on serine 58 (3, 4). HIPK2 is reported to be activated in response to severe genotoxic stress that induces apoptosis (2, 24). Thus, HIPK2 by phosphorylating p53 on serine 46 is believed to activate p53 to induce apoptosis in response to genotoxic stress (2–4, 24). It is believed that PML facilitates HIPK2-mediated phosphorylation on p53 and that HIPK2 also interacts with CBP which promotes acetylation on p53 residues lysine 373 and 382, an event critically linked to induction of apoptosis (24). Murine hipk4 phosphorylates p53 on serine 9 (18) and it is likely that human HIPK4 would also do the same. The functional significance of HIPK4-mediated p53 phosphorylation on serine 9 remains to be fully elucidated. Whether human HIPK4 is activated in response to genotoxic stress or other types of stresses also remains unclear and requires further investigation.
HIPK1, 2 and 3 were identified based on their interactions with homeodomain proteins and share higher degree of homology among them (1, 2). Of the three HIPKs, HIPK2 is the more extensively studied kinase and there is evidence to suggest that both HIPK1 and HIPK2 appear to have redundant function (25). HIPK4 has been so named because it shows some degree of sequence homology with HIPK1, 2 and 3 predominantly within its catalytic domain. HIPK 1, 2 and 3 are reported to be modified by sumoylation (2, 26–28). HIPK4 harbors four high probability sumoylation sites and one of the sites (LKPE) at position 137–140 is identical to that present in HIPK1, 2 and 3. It is therefore, possible that the function of HIPK4, just like that of the other HIPKs, is also regulated by sumoylation. However, unlike HIPK1, 2 and 3 that are composed of amino acids 1210, 1198 and 1215 respectively, HIPK4 is composed of 616 amino acids and thus, smaller in size. Furthermore, unlike HIPK1, 2 and 3 that are believed to predominantly reside in the nuclei (2), HIPK4 exhibits cytoplasmic localization (18).
Clearly, HIPK4 appears to be a unique member of the HIPK family and its further functional characterization will likely provide valuable information as to whether it has distinct functions and/or functions that overlap with those of other HIPKs. It is also possible that HIPK4 may belong to an independent family of kinases that are distantly related to HIPKs or that it belongs to a subfamily within the HIPK superfamily. Future in-depth studies are also expected to provide valuable information to clarify this issue.
Acknowledgments
This work was supported in part by NIH Grants ES016668, ES005633 (to MSS) and CA121850 (to YH).
Footnotes
Conflicts of Interest
No potential conflicts of interest to disclose.
References
- 1.Kim YH, Choi CY, Lee SJ, et al. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J Biol Chem. 1998;2(273):25875–79. doi: 10.1074/jbc.273.40.25875. [DOI] [PubMed] [Google Scholar]
- 2.Rinaldo C, Siepi F, Prodosmo A, et al. HIPKs: Jack of all trades in basic nuclear activities. Biochem Biophysica Acta. 2008:2124–29. doi: 10.1016/j.bbamcr.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 3.D’Orazi G, Cecchinelli B, Bruno T, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 2002;4:11–9. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- 4.Hofmann TG, Moller A, Sirma H, et al. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- 5.Rochat-Steiner V, Becker K, Micheau O, et al. FIST/HIPK3: a Fas/FADD-interacting serine/threonine kinase that induces FADD phosphorylation and inhibits fas-mediated Jun NH(2)-terminal kinase activation. J Exp Med. 2000;192:1165–74. doi: 10.1084/jem.192.8.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun H, Luo X, Montalbano J, et al. DOC45, a novel DNA damage-regulated nucleocytoplasmic ATPase that is overexpressed in multiple human malignancies. Mol Cancer Res. 2010;8:57–66. doi: 10.1158/1541-7786.MCR-09-0278. [DOI] [PubMed] [Google Scholar]
- 7.Montalbano J, Lui K, Sheikh MS, et al. Identification and characterization of RBEL1 subfamily of GTPases in the Ras superfamily involved in cell growth regulation. J Biol Chem. 2009;284:18129–42. doi: 10.1074/jbc.M109.009597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corcoran CA, He Q, Ponnusamy S, et al. Neutral sphingomyelinase-3 is a DNA damage and nongenotoxic stress-regulated gene that is deregulated in human malignancies. Mol Cancer Res. 2008;6:795–807. doi: 10.1158/1541-7786.MCR-07-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo X, He Q, Huang Y, Sheikh MS. Cloning and characterization of a p53 and DNA damage down-regulated gene PIQ that codes for a novel calmodulin-binding IQ motif and is upregulated in gastrointestinal cancers. Cancer Res. 2005;65:10725–33. doi: 10.1158/0008-5472.CAN-05-1132. [DOI] [PubMed] [Google Scholar]
- 10.Luo X, Huang Y, Sheikh MS. Cloning and characterization of a novel gene PDRG that is differentially regulated by p53 and ultraviolet radiation. Oncogene. 2003;22:7247–57. doi: 10.1038/sj.onc.1207010. [DOI] [PubMed] [Google Scholar]
- 11.He Q, Luo X, Jin W, Huang Y, et al. Celecoxib and a novel COX-2 inhibitor ON09310 upregulate death receptor 5 expression via GADD153/CHOP. Oncogene. 2008;27:2656–60. doi: 10.1038/sj.onc.1210894. [DOI] [PubMed] [Google Scholar]
- 12.Rong R, Jiang LY, Sheikh MS, Huang Y. Mitotic kinase Aurora-A phosphorylates RASSF1A and modulates RASSF1A-mediated microtubule interaction and M-phase cell cycle regulation. Oncogene. 2007;26:7700–8. doi: 10.1038/sj.onc.1210575. [DOI] [PubMed] [Google Scholar]
- 13.He Q, Shi J, Jones S, et al. Smac deficiency affects endoplasmic reticulum stress-induced apoptosis in human colon cancer cells. Mol Cell Pharmacol. 2009;1:23–28. doi: 10.4255/mcpharmacol.09.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J, He Q, An J, Sun H, Huang Y, Sheikh MS. Sulindac sulfide differentially induces apoptosis in Smac-proficient and -deficient human colon cancer cells. Mol Cell Pharmacol. 2009;1:92–97. doi: 10.4255/mcpharmacol.09.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melchior F. SUMO--nonclassical ubiquitin. Annu Rev Cell Dev Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 16.Ecsedy JA, Michaelson JS, Leder P. Homeodomain-interacting protein kinase 1 modulates Daxx localization, phosphorylation, and transcriptional activity. Mol Cell Biol. 2003;23:950–60. doi: 10.1128/MCB.23.3.950-960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Zhang R, Luo D, et al. Tumor necrosis factor alpha-induced desumoylation and cytoplasmic translocation of homeodomain-interacting protein kinase 1 are critical for apoptosis signal-regulating kinase 1-JNK/p38 activation. J Biol Chem. 2005;280:15061–70. doi: 10.1074/jbc.M414262200. [DOI] [PubMed] [Google Scholar]
- 18.Arai S, Matsushita A, Du K, Yagi K, Okazaki Y, Kurokawa R. Novel homeodomain-interacting protein kinase family member, HIPK4, phosphorylates human p53 at serine 9. FEBS Lett. 2007;581:5649–57. doi: 10.1016/j.febslet.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 19.Sheikh MS, Fornace AJ., Jr Role of p53 family members in apoptosis. J Cell Physiol. 2000;182:171–81. doi: 10.1002/(SICI)1097-4652(200002)182:2<171::AID-JCP5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 20.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 21.Higashimoto Y, Saito S, Tong XH, et al. Human p53 is phosphorylated on serines 6 and 9 in response to DNA damage-inducing agents. J Biol Chem. 2000;275:23199–203. doi: 10.1074/jbc.M002674200. [DOI] [PubMed] [Google Scholar]
- 22.Saito S, Goodarzi AA, Higashimoto Y, et al. ATM mediates phosphorylation at multiple p53 sites, including Ser(46), in response to ionizing radiation. J Biol Chem. 2002;277:12491–4. doi: 10.1074/jbc.C200093200. [DOI] [PubMed] [Google Scholar]
- 23.Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–50. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 24.Sombroek D, Hofmann TG. How cells switch HIPK2 on and off. Cell Death Differ. 2009;16:187–94. doi: 10.1038/cdd.2008.154. [DOI] [PubMed] [Google Scholar]
- 25.Isono K, Nemoto K, Li Y, et al. Overlapping roles for homeodomain-interacting protein kinases hipk1 and hipk2 in the mediation of cell growth in response to morphogenetic and genotoxic signals. Mol Cell Biol. 2006;26:2758–71. doi: 10.1128/MCB.26.7.2758-2771.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofmann TG, Jaffray E, Stollberg N, Hay RT, Will H. Regulation of homeodomain-interacting protein kinase 2 (HIPK2) effector function through dynamic small ubiquitin-related modifier-1 (SUMO-1) modification. J Biol Chem. 2005;280:29224–32. doi: 10.1074/jbc.M503921200. [DOI] [PubMed] [Google Scholar]
- 27.Gresko E, Moller A, Roscic A, Schmitz ML. Covalent modification of human homeodomain interacting protein kinase 2 by SUMO-1 at lysine 25 affects its stability. Biochem Biophys Res Commun. 2005;329:1293–9. doi: 10.1016/j.bbrc.2005.02.113. [DOI] [PubMed] [Google Scholar]
- 28.Engelhardt OG, Boutell C, Orr A, Ullrich E, Haller O, Everett RD. The homeodomain-interacting kinase PKM (HIPK-2) modifies ND10 through both its kinase domain and a SUMO-1 interaction motif and alters the posttranslational modification of PML. Exp Cell Res. 2003;283:36–50. doi: 10.1016/s0014-4827(02)00025-3. [DOI] [PubMed] [Google Scholar]