Abstract
A robust, high throughput, two-tiered assay for screening small molecule inhibitors against botulinum neurotoxin serotype A was developed and employed to screen 16,544 compounds. Thirty-four compounds were identified as potent hits employing the first-tier assay. Subsequently, nine were confirmed as actives by our second-tier confirmatory assay. Of these, one displayed potent inhibitory efficacy, possessing an IC50 = 16 μM (± 1.6 μM) in our in vitro assay. This inhibitor (0831–1035) is highly water-soluble, and possesses an IC50 = 47 μM (± 7.0 μM) in our primary cell culture assay (with virtually no cytotoxicity up to 500 μM) suggesting that this inhibitor is a good candidate for further development as a therapeutic countermeasure to treat botulism resulting from botulinum neurotoxin serotype A intoxication. An enzyme kinetics study indicated that this inhibitor exhibits mixed non-competitive inhibition, with a KI = 9 μM.
Clostridial botulinum neurotoxins (BoNTs), secreted by strains of the genus Clostridium, are the most toxic biological substances presently known (Singh, 2000). Seven BoNT serotypes have been identified, and are designated A – G. The BoNT-mediated inhibition of acetylcholine release at neuromuscular junctions results in the life threatening flaccid paralysis associated with the disease botulism.
A combination of features including neuronal target sites, efficient cellular entry, and unique enzymatic activity contribute to the extreme toxicity of BoNTs (e.g., BoNT serotype A (BoNT/A) possesses a mouse lethal dose of about 0.3 ng/kg) (Montecucco and Schiavo, 1993). However, while all BoNT serotypes share similar function (i.e., the inhibition of neurotransmitter release) and epidemiology, only BoNT serotypes A, B, E and F are known to cause human botulism (Arnon et al., 2000). Of these, BoNT/A is the most potent and most common cause of human botulism. While naturally occurring botulism cases are rare, BoNTs have been weaponized, and due to their potencies and ease of production, represent serious biothreat agents (Arnon et al., 2000; Wein and Liu, 2005; Greenfield et al., 2002).
BoNTs are secreted as ~150 kDa single polypeptide chains that are activated by protease nicking to form di-chain molecules consisting of a 50 kDa light chain (LC) and a 100 kDa heavy chain (HC) linked via a disulfide bond (Montecucco and Schiavo, 1995; Li and Singh, 1999a). The BoNT LC is a zinc-endopeptidase that cleaves soluble NSF-attachment protein receptor (SNARE) proteins, which mediate synaptic vesicle docking and fusion in neurons, and therefore, BoNT blocks the release of acetylcholine (Montecucco and Schiavo, 1995; Li and Singh, 1999a; Poulain et al., 2008). BoNT serotypes A, E, and C cleave synaptosome associated protein of 25 kDa (SNAP-25), BoNT serotypes B, D, F, and G cleave vesicle associated membrane protein (VAMP, also referred to as synaptobrevin), and BoNT serotype C also cleaves syntaxin (Montecucco and Schiavo, 1995; Li and Singh, 1999a). It is this cleavage of SNARE proteins that inhibits exocytosis of the neurotransmitter. The BoNT HC plays an accessory role, binding to target neurons (via its C-terminus) and translocating the LC into the neuronal cytoplasm (via its N-terminus) (Simpson, 2004; Montecucco, 1986; Montecucco et al., 2004).
The current treatment for botulism involves the administration of antitoxin and respiratory supportive care. Available antitoxins include equine antitoxin consisting of neutralizing antibodies for BoNT serotypes A, B, and E (Cai and Singh, 2007); an investigational heptavalent equine antitoxin (to counter BoNT serotypes A, B, C, D, E, F, and G (Arnon et al., 2000); and BabyBIG®, which is derived from the blood of human donors vaccinated with a pentavalent (ABCDE) toxoid vaccine (Arnon et al., 2000). An important limitation of all above indicated antitoxin treatments is that they must be administered before toxin penetration into the neuronal cytosol; after such time they are no longer effective. Hence, the therapeutic window for administering antitoxins is very limited. Moreover, the flaccid muscle paralysis caused by BoNTs can last for several months (depending on the serotype, e.g., serotype A has the longest effect) (Greenfield et al., 2002; Rosenbloom et al., 2002; Poulain et al., 2008), with patients displaying paralysis of thoracic muscles requiring long-term respiratory care (Arnon et al., 2000; Greenfield et al., 2002; Rosenbloom et al., 2002). The estimated cost for treating a botulism patient with such intensive care could be as high as $350,000 (Wein and Liu, 2005). Hence, such treatments would place a large burden on hospitals, both financially and resource-wise, in the event of a bioterror attack employing BoNT(s). Moreover, while botulinum neurotoxin is also used as therapeutics for a range of neuromuscular disorders (Rossetto et al., 2001), with its increased usages, serious side effects (including fatal cases) have been reported, and FDA has put a black-box warnings on all botulinum neurotoxin-based therapeutics (http://www.fda.gov/downloads/Drugs/DrugSafety/UCM176360.pdf). Consequently, there is a pressing need for new, and more effective antidotes to treat botulism, for both prophylactic and post-exposure administration, and even for the antidotes against side effects of botulinum neurotoxin based therapeutics.
Inhibition of the endopeptidase activity of BoNTs with small, non-peptidic molecules is a valid strategy for developing new therapeutics to treat botulism, as such molecules possess the potential to penetrate neurons and counteract internalized BoNT activity (Cai and Singh, 2007). Compared to antibody based therapies, small molecule inhibitors are also more likely to be orally bioavailable, more stable when stored for long periods of time, and possess significantly better tissue and cell permeation (Cai and Singh, 2007).
Presently, a limited number of small molecule inhibitors of the BoNT/A LC with IC50 values in the low μM range have been identified (Dickerson and Janda, 2006; Boldt et al., 2006; Burnett et al., 2003; Burnett et al., 2005; Burnett et al., 2007a; Burnett et al., 2007b; Hermone et al., 2008). The three-dimensional analysis of these inhibitors has resulted in the development of a common pharmacophore for BoNT/A LC inhibition with demonstrated utility (Burnett et al., 2003; Burnett et al., 2005; Burnett et al., 2007a; Burnett et al., 2007b; Burnett et al., 2009; Hermone et al., 2008). However, there remains the need for the identification and/or development of small molecule BoNT/A LC inhibitors that are active with inhibitory constants in the nM range with no/low toxicity (Cai and Singh, 2007).
By employing a two tier assay (consisting of a high throughput primary screen and a secondary confirmatory assay), the ChemDiv 3 library (ChemDiv Inc., San Diego, CA), consisting of 16,544 compounds, was screened. Subsequently, we identified several small molecules that inhibit the endopeptidase activity of BoNT/A LC. Of these, one possesses an IC50 value = 16 μM (± 1.6 μM). This small molecule inhibitor forms the basis for the syntheses of more potent analogs as therapeutic candidates.
Materials and Methods
FRET Peptide
A fluorescence resonance energy transfer (FRET) peptide with 13 amino acids truncated from SNAP-25 was used to assay BoNT/A LC endopeptidase activity (Boldt, et al., 2006). In order to label the fluorophore (Fluorescein-5-isothiocyanate, FITC) more efficiently, we added β-alanine to the C-terminus. The sequence of the peptide is FITC-b(Ala)-Thr-(D-Arg)-Ile-Asp-Gln-Ala-Asn-Gln-Arg-Ala-Thr-Lys(DABCYL)-Norleucine-CONH2. The 4-dimethylaminoazobenzene-4′-carboxylic acid (DABCYL) component served as the FITC quencher. The peptide cleavage site is between its Gln and Arg residues, as highlighted in bold. The FRET peptide was synthesized by New England Peptide (Gardener, MA) and possessed a purity > than 95%.
BoNT/A and recombinant BoNT/A LC
Recombinant BoNT/A LC was produced and purified as described previously (Li and Singh, 1999b). The enzyme concentration was determined using 280 nm of 0.83 (mg/ml)−1 cm−1 (Li and Singh, 1999b). The BoNT/A LC (in 20% glycerol) was stored at −80°C, and endopeptidase activity was verified prior to screening. Native BoNT/A was produced and purified based on established methods in our laboratory (Cai et al., 1999), and purity and integrity were confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) before use.
Recombinant SNAP-25
SNAP-25 was expressed and purified according to Cai et al (Cai et al., 1999). Cells expressing GST/SNAP-25 fusion protein were collected by centrifugation and resuspended in 10 mM phosphate-buffered saline (PBS) (pH 7.4), containing 1 mM phenylmethylenesulfonyl fluoride (PMSF). Cells were lysed by sonication for 2 min, treated with 1% Triton X-100, and centrifuged to remove cell debris. The supernatant was applied to glutathione-agarose beads (Sigma Chemical Co., St. Louis, MO) and washed with PBS buffer to remove other cellular proteins, and GST/SNAP-25 fusion protein was eluted with 10 mM glutathione in 50 mM Tris-HCl buffer at pH 8.0. The fusion protein was precipitated with ammonium sulfate, redissolved in assay buffer (50 mM Tris-HCl, 10 mM sodium phosphate, 300 mM NaCl, 2 mM MgCl2, 0.3 mM CaCl2, 1 mM mercaptoethanol, and 0.1% NaN3 at pH 7.6), and dialyzed against the same buffer before being used for experiments.
Optimization of the Screening Assay
The FRET-based endopeptidase assay was optimized using a SpectraMax M5 fluorescence microplate reader (Molecular Devices). Peptide substrate was dissolved in distilled water, and the BoNT/A LC or Apo-BoNT/A LC were diluted with 20 mM HEPES, pH 8.0, containing 1.25 mM dithiothreitol (DTT) and 0.1% Tween 20 (assay buffer). Apo-BoNT/A LC was prepared by incubating the enzyme with 10 mM EDTA at 37°C for 30 min. The excitation wavelength used was 490 nm with an emission wavelength of 523 nm.
High throughput screening (HTS)-Assay
Peptide substrate stock solution was prepared using distilled water to obtain a 10 μM solution. BoNT/A LC was diluted with assay buffer to give a 100 nM stock solution. To carry out the screening assay, 30 μL of BoNT/A LC was transferred into 384-well black mictroplates (Corning, Corning, NY) using a liquid transfer robot (Matrix Technologies, Hudson, NH), following which, 100 nL of library compound (5 mg/ml in DMSO) was transferred into each well via a pin-transfer robot. The final compound concentration was 16.7 μg/ml in each well. Next, each plate was incubated at 37°C to allow the BoNT/A LC and library compounds to interact. After 30 min incubation, 30 μL peptide substrate was transferred to each well using the liquid transfer robotic. The final peptide substrate concentration was 5 μM. The final concentration of BoNT/A LC was 50 nM, and the final concentration of compound was 8.3 μg/ml. Each plate contained at least 16 wells for positive controls, and 16 wells for negative controls. The plates were incubated at 37°C for 1 h to allow the endopeptidase reaction to occur. The positive control was BoNT/A LC without library compound. The negative control was assay buffer without either BoNT/A LC or library compound. The plates were read using a Perkin-Elmer Envision fluorescence microplate reader with a plate stacker (PerkinElmer, Boston, MA). An FITC-485 (485 nm filter with 14 nm band width) was used as the excitation filter, and an FITC-535 (535 nm filter with 25 nm band width) was used as the emission filter. Each library plate was run in duplicate to increase the accuracy of the results.
Z′ Factor
Z′ factor has been used to accurately and rapidly evaluate HTS assays for the identification of active compounds (‘hits’) from large chemical libraries (Zhang et al., 1999). To evaluate our HTS assay, two full plates with only positive and negative controls were examined independently. The layout of the plate was 8×8×8, with 8 lanes of negative controls in the center of the plate, and 8 lanes of positive controls on each side of the plate. There were 256 positive controls and 128 negative controls in each plate.
Z′ factor is calculated using the following formula:
| (1) |
where, SD+ is the standard deviation of the positive controls, SD− is the standard deviation of negative controls; Ave+ is the positive control average, and Ave− is the negative control average.
Compound Library
The ChemDiv 3 library, which contained 16,554 compounds in forty seven 384-well plates, was obtained from ChemDiv Inc (San Diego, CA). The compounds were dissolved in DMSO to give a final concentration of 5 mg/ml. Individual compound ‘hits’ were ordered from ChemDiv Inc. for additional confirmation and investigational studies.
Confirmatory Assay
An ELISA-based assay incorporating full length SNAP-25 as the substrate was used as the confirmatory assay to determine the endopeptidase activity of BoNT/A LC. Flat-bottom 384-well microtitter plates (Corning, Corning, NY) were coated with 10 μg/ml GST-SNAP-25 fusion protein. The plates were incubated at 4°C overnight. BoNT/A LC was diluted with assay buffer (using the same procedure outlined for the HTS-assay) to 100 nM to provide the working solution. BoNT/A LC working solution was mixed with the desired concentration of inhibitor candidates at 37°C for 30 min before use. To compare control samples versus samples containing inhibitors, the DMSO concentration was kept at 1% for all samples. Following appropriate washing and blocking steps (Sharma and Singh, 2004), 60 μl BoNT/A LC or BoNT/A LC + inhibitor were transferred to the plate. For negative controls, 60 μl of assay buffer, or 60 μl of Apo-BoNT/A LC were transferred to the plates. Science the inhibitor stock was dissolved in DMSO, the same amount of DMSO used in the BoNT/A LC working solution was also added to investigate the effect of DMSO on the endopeptidase activity in some controls. The cleavage reaction was carried out by incubating the plates at 37°C for 30 min (Sharma and Singh, 2004). After washing the plates, intact SNAP-25 was analyzed by adding 30 μL of rabbit antiC-terminal-SNAP-25 polyclonal antibody (3 ng/mL) (Stressgen Biotechnologies). Truncated SNAP-25 cleaved by the BoNT/A LC is not recognized by the anti-C-terminal-SNAP-25 antibody. Horseradish peroxidase (HRP) - labeled anti-rabbit antibody (1:10,000) was added to the plates as a secondary antibody, and the plates were incubated at 37°C for 30 min. A substrate solution containing 0.04% OPD (o-phenylenediamine dihydrochloride) and 0.012% hydrogen peroxide (in a citrate phosphate buffer at pH 5.0) was added to the wells, and the plates were incubated for 15 min at room temperature (25°C). The reaction was subsequently quenched with 50 μL of 2 M sulfuric acid, and the color was monitored by measuring the absorbance at 490 nm. The percent inhibition was calculated using the SNAP-25/DMSO control as the baseline (0% inhibition), and DMSO/BoNT/A LC as 0% inhibition.
Inhibition Concentration Evaluation
To further evaluate small molecule inhibition effects, concentration response curves for select compounds were performed using the peptide-based substrate (described in the HTS-assay section). The IC50 value was interpolated from the concentration response curve using a non-linear polynomial regression.
Enzyme Kinetics
Enzyme kinetics measurements were carried out using the peptide based substrate. The assay conditions were same as the HTS screening. Substrate concentrations from 5 to 25 μM were used (e.g., 5, 10, 20, and 25μM) for enzyme activity in the presence of 50 nM of BoNT/A LC. The reaction buffer was the same as used in the HTS-assay described above. The reactions were carried out at 37°C, with the monitoring of fluorescence in the first 10 min to calculate the initial reaction velocity. The fluorescence signals observed were within the linear range for the substrate concentrations chosen above. To evaluate inhibition kinetics, a given concentration of the inhibitor was pre-incubated with the BoNT/A LC at 37°C for 30 min before adding the substrate. The concentrations of inhibitor used were 19.5 and 39 μM. All the shown results are the average of triplicate measurements. The inhibition constant was calculated using a Lineweaver-Burk plot in the presence of the inhibitor, where the slope equals α*KM/Vmax, and the y-intercept equals α′/Vmax. The KI and KI′ were calculated from α and α′, respectively: α=1+[I]/KI; α′=1+[I]/KI′.
Data Analysis
Data was analyzed using the JMP 6.0 software (SAS Institute Inc., Cary, NC). To evaluate the inhibition effect observed during the ELISA-based assay, Tukey-Kramer pair analysis (α=0.05) was used.
Analysis of Inhibitor Specificity
To examine the specificity of compound 0831–1035, 78 μM of 0831–1035 was first incubated with either 100 nM or 1000 nM of thermolysin at 37°C for 30 min. The same volume of 10 μM of bovine serum albumin (BSA) was added to each thermolysin sample, and incubated at 37°C for 30 min. The reaction mixtures were then analyzed on SDS-PAGE (8 to 20% acrylamide gradient). The final concentration of BSA in the reaction mixture was 5 μM, the final concentration of compound 0831–1035 was 39 μM, and the final concentration of thermolysin was either 50 nM, or 500 nM.
Primary Rat Cerebellar Neuron-Based Assay
Neuronal granule cells from the pooled cerebellar tissue of 7–8 day old Sprague-Dawley rats were obtained using a previously described method (Skaper et al., 1979). Cells were suspended in DMEM/F-12 medium (Gibco-Invitrogen, Carlsbad, CA) supplemented with 50 U/mL penicillin, 50 μg/mL streptomycin, 10% FBS, N2 supplement, and 25 mM KCl. Cells were seeded onto poly(L-lysine)-coated 6-well plates at a density of 2.2×106 cells per well, and were maintained in a humidified 5% CO2 atmosphere at 37°C. After 24 hours, 10 μM cytosine-B-D-arabinoside (Sigma, St. Louis, MO ) was added to the culture to inhibit the replication of non-neuronal cells. The neurons were maintained by replacement every 7 days with freshly prepared medium.
Once the primary neuronal cell cultures were well established, which usually occurs 8–10 days after initial seeding, and remained viable for up to four weeks, varying concentrations of inhibitor were dissolved in DMSO and added to the culture to examine inhibitory effects on BoNT/A LC activity. Subsequently, the cultures were inoculated with 0.5 nM BoNT/A, and allowed to incubate at 37°C, 5% CO2, for 3 hours. The cells were washed with PBS and harvested into a pre-weighed eppendorf screw cap vial. The cells were then pelleted by centrifugation. After pelleting, the supernatant was carefully removed and the microcentrifuge tubes were re-weighed to determine the weight of the pellet. Four μl of M-PER, and 2 μl of SDS-PAGE sample buffer were added for each mg of cell pellet, and boiled for 10 minutes to inactivate the residual toxin. The samples were subsequently subjected to immunoblot analysis.
Immunoblot Analysis
Aliquots of protein were electrophoresed through 15% polyacrylamide gels. Proteins were transferred to polyvinylidene fluoride membranes (Millipore Corp., Bedford, MA) and processed for immunoblot analysis. Blotting was performed with 5% nonfat dry milk in TBST (10 mM Tris, 150 mM NaCl, 0.1% (v/v) Tween 20) for 1h at room temperature and blots were probed with rabbit anti-SNAP-25 antibody (1:2500, Sigma, St. Louis, MO) in 1% nonfat dry milk in TBST for 1 h at room temperature. Following washes with TBST, secondary antibody incubation was performed using HRP-conjugated goat anti-Rabbit IgG (1:10,000, Amersham Biosciences) in 1% nonfat dry milk in TBST. After 1 h at room temperature, blots were washed and developed using an ECL kit (GE Healthcare Bio-Sciences Corp.). Band densities for SNAP-25 and its cleaved product were normalized, and relative intensities were determined using scanning densitometry (Kodak Image station 200RT, Eastman Kodak Company, Rochester, NY).
Results and Discussion
Many structurally diverse, drug-like small molecule libraries are now available for screening against the BoNT/A LC. However, to examine such large compound libraries in an efficient manner, valid HTS-assay(s) are required. Specifically, the method must be robust, and result in a low number of false hits (either false positive or false negative). Here, we describe the development of a two-tier assay method to ensure the accurate measurement of BoNT/A LC activity in the presence of small molecules. The first assay tier utilizes a truncated sequence of SNAP-25 as the BoNT/A substrate, and employs fluorescence resonance energy transfer (FRET) as the signal readout (Boldt et al., 2006; Schmidt et al., 2001). The peptide sequence includes the cleavage site of the BoNT/A LC with a fluorophore linked to the N-terminus, and a quencher linked to the C-terminus. Upon BoNT/A LC substrate cleavage, the fluorescence signal increases due to the removal of the quencher. The second assay tier is a confirmatory method based on ELISA, which uses full length SNAP-25 as the substrate (Cai et al., 1999; Sharma and Singh, 2004).
HTS-Assay
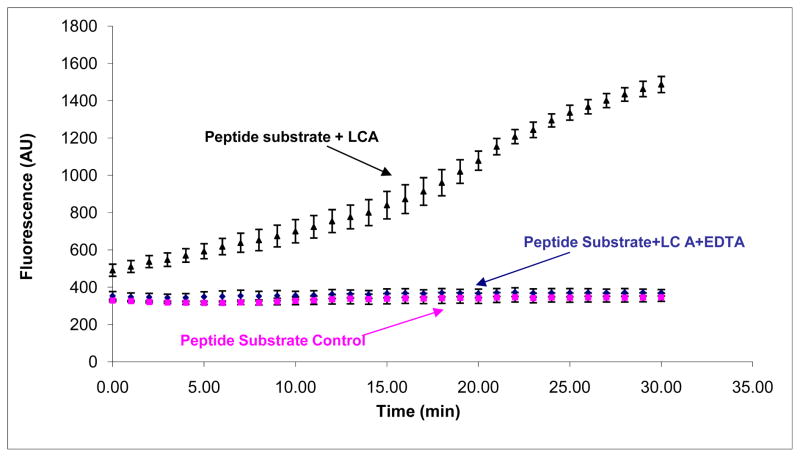
In this study, we employed a FRET peptide-based assay in which the fluorescence significantly increased upon peptide substrate cleavage (Fig. 1). For apo-BoNT/A LC, in which the endopeptidase activity is abolished due to the removal of zinc, failure to cleave the peptide substrate was evidenced by the low fluorescence signal of the peptide substrate (Fig. 1). The results shown in Figure 1 clearly indicate that the FRET peptide-based assay can differentiate between active and inactive BoNT/A LC, and can successfully be used to screen for BoNT/A LC inhibitors.
Figure 1.
High throughput inhibition assay using peptide based substrate. 5 μM peptide substrate, and 50 nM BoNT/A LC (black filled triangle) were used in the assay. For peptide substrate control (purple filled square), only peptide substrate was used. For EDTA inhibition (blue filled diamond), BoNT/A LC was incubated with 10 mM EDTA at 37°C for 30 min before adding substrate to the reaction wells. The assay was carried out at 37°C for 30 min. Fluorescence was read using a SpectraMax M5 fluorescence microplate reader (Molecular Devices). The excitation wavelength was 490 nm, and the emission wavelength was 523 nm, with an emission cutoff filter of 515 nm. The results shown are the average of five runs. The error bars represent plus/minus standard deviations from 5 runs.
To scale-up the HTS-assay, we further optimized the method by incorporating an Envision fluorescence microplate reader (Perkin-Elmer) with a plate stacker, which significantly increased the capacity of the assay to measure many plates automatically.
With regard to most HTS-assays, each compound is tested once, or at most in duplicate. Therefore, such assays require not only high throughput capability and robustness, but also adequate sensitivity, reproducibility, and accuracy (in order to discriminate bona fide ‘hits’ from among the large number of compounds being examined). Consequently, we implemented the commonly used Z′ factor as a statistical parameter to evaluate and validate our HTS-assay (Zhang et al., 1999). Z′ factor (see equation 1 in the Methods section) takes into account the data variability (standard deviation, or SD) between active groups (i.e., ‘hits’) and inactive groups, and the signal dynamic range (i.e., signals from actives and inactives); therefore, it takes into consideration all of the information necessary for HTS-assay data characterization and evaluation (Zhang et al., 1999). An ideal HTS-assay would possess a Z′ factor of 1, which would indicate that there is no variation (i.e., SD = 0), or that the data dynamic range is indefinite. In practical terms, for an assay with Z′ factor between 0.5 and 1, the separation band between positive and negative controls are large, and therefore, the assay is suitable for HTS. In contrast, for an HTS-assay with a Z′ factor between 0.5 and 0, the separation band is narrow, and can only provide dubious results. Furthermore, when the Z′ factor is less than 0, there is no separation band between positive and negative controls, and the data is therefore impossible to interpret with confidence.
To validate the HTS-assay, two full plates containing only positive and negative controls were examined. The Z′ factors from these two full plates are shown in Table 1. As observed in the Table, our FRET peptide-based assay provided Z′ factors greater than 0.8 for the two independent plates, thereby indicating its suitability for HTS.
Table 1.
Summary of the Zprime; factors from two independent experiments.
| Plate 1 | Plate 2 | ||
|---|---|---|---|
| Average Signal (AU) | Positive | 1357529 | 1469361 |
| Negative | 179600 | 196546 | |
| Standard Deviation of the Signal (AU) | Positive | 67563 | 63851 |
| Negative | 9736 | 7586 | |
| Z′ factor | 0.80 | 0.83 | |
To start, we needed to establish a consistent/standard method to qualify what would or would not be considered a hit from the HTS-assay. Typically, a hit rate of 0.3% or less in a compound library is thought to be acceptable (Devlin, 1997). Too high of a hit rate (>5%) usually indicates that the assay lacks robustness and/or is not specific enough for the target; too high of a hit rate also results in unmanageable secondary evaluation (Devlin, 1997). By analyzing the inhibition effect and the hit rate, we determined that 10 standard deviations (SD) below the average of the positive control signal was the cutoff point for the identification of hits, with potent hits defined as compounds that met this cutoff in duplicate (Figure S-1, in supplementary materials). The inhibition of peptide substrate cleavage by the hits was, in general, over 60% in the FRET-assay (Table S-1, in supplementary materials). Of the 16,544 examined compounds, 34 (a hit rate = 0.21%) were identified as hits by the FRET-assay.
Confirmatory Assay
The FRET peptide-based assay employed a truncated peptide as the substrate for the BoNT/A LC. However, this peptide may not be the optimized substrate for this enzyme, since its natural substrate is a large protein of 25 kDa (i.e., SNAP-25). In addition, the library compounds themselves may interfere with the fluorescence-based assay (e.g., by quenching the fluorescence signals). Therefore, the results from FRET peptide-based assay required secondary confirmation.
To this end, we developed an ELISA-based assay (employing full length SNAP-25 as the substrate) to confirm the protease activity of the BoNT/A LC in the presence of the small molecules (Sharma and Singh, 2004). While the ELISA-based assay is more time consuming than the FRET peptide-based assay, and involves multiple washing steps (making it unsuitable for large scale HTS), the sample throughput is nevertheless relatively high, and the sample and reagent consumption is relatively low; therefore, the ELISA-based assay is well suited for HTS hit confirmation. Moreover, as part of our strategy, Tukey-Kramer pair analysis (α=0.05) was used to determine which compounds analyzed via the ELISA-based assay were significantly different from positive controls (BoNT/A LC).
Out of the 34 initial hits, 9 were confirmed as inhibitors (Table S-1). These results strongly suggest that hits from the FRET peptide-based HTS-assays require secondary evaluation. However, it should be noted that the results also indicate that a substantial number of hits identified by the HTS-assay are bona fide inhibitors.
Of the 9 confirmed compounds, we initially chose our 2 most potent hits (0831–1035 and C151-0184 (Table 2) for further inhibition kinetics characterization and testing in neurons. However, due to C151-0184 insolubility, it was not amenable to either IC50 calculation or neuronal testing. In contrast, 0831–1035 was highly soluble and very amenable for further study. Hence, we proceeded with the characterization of this inhibitor - to preface its potential development as a therapeutic candidate.
Table 2.
The most potent hits from HTS
| Structures | Library | Vendor ID | Inhibition obtained from FRET-assay (%) | Inhibition obtained from ELISA-assay (%) |
|---|---|---|---|---|
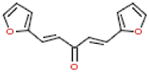 |
ChemDiv3 | 0831–1035 | 95 | 84 |
 |
ChemDiv3 | C151–0184 | 90 | 83 |
In Vitro Inhibition Concentration
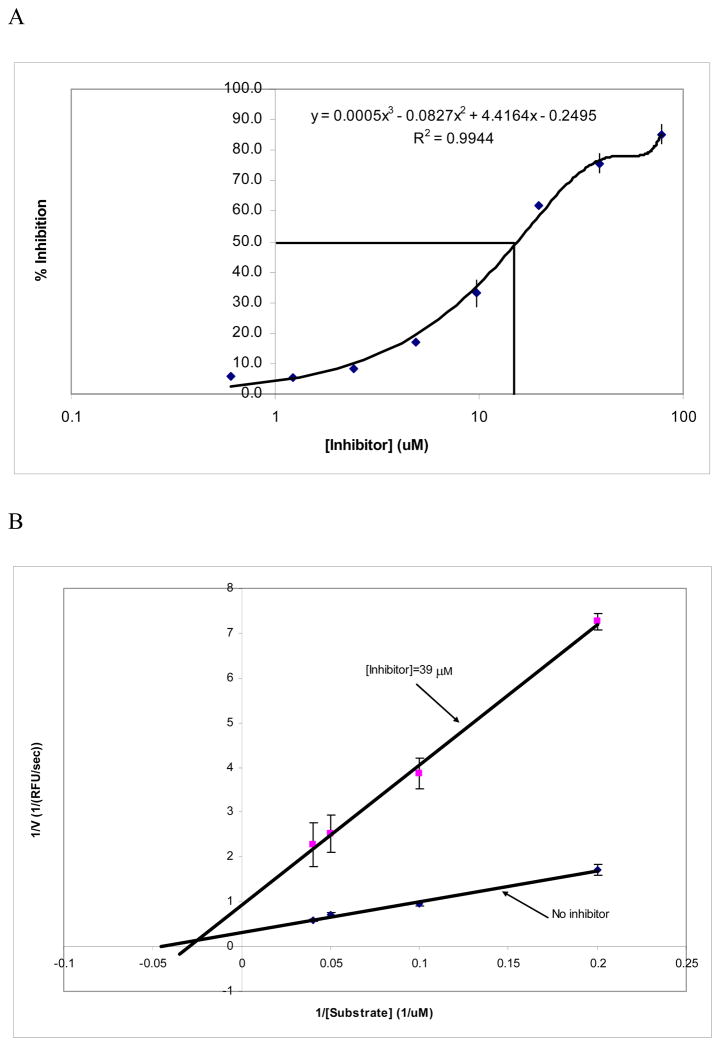
The chemical structures of inhibitors 0831–1035 and C151-0184 are very similar, and not surprisingly, both inhibit the BoNT/A LC with similar potencies (Table 2). But as indicated above, during serial dilutions, compound C151-0184 lacked the solubility necessary for further testing. Hence, only the IC50 for compound 0831–1035 was determined. As shown in Figure 2-A, the IC50 of 0831–1035 is 16 μM (± 1.6 μM) (with the BoNT/A LC concentration used in the experiment = 50 nM). This indicates that 50% BoNT/A LC inhibition is achieved when the inhibitor:BoNT/A LC molar ratio is 320:1.
Figure 2. Inhibition of the 0831–1035 on the endopeptidase activity of BoNT/A LC.
Panel A: the dose-response curve for 0831–1035 mediated BoNT/A LC endopepidase inhibition. The curve was fit using non-linear polynomial regression. Panel B: Lineweaver-Burk plot of BoNT/A LC inhibition with and without inhibitor 0831–1035 (using the13-mer peptide as the substrate (see the methods for details).
Enzyme Kinetics
To evaluate the mechanism of 0831–1035 inhibition, enzyme kinetics studies were performed. From the Lineweaver-Burk plot (Figure 4), 0831–1035 demonstrated non-competitive inhibition. In both the absence and presence of the inhibitor (39 μM) the KM was 19.4 μM (±2.1 μM) and 28.0 μM (±2.8 μM), respectively, whereas the Vmax values were 3.1 (±0.4) and 0.9 (±0.3) RFU/s, respectively (Table 3). By examination of the Lineweaver-Burk plots (Figure 2-B), which clearly indicate different intercepts at the Y and X axes (yet not being parallel), it is clear that the inhibition provided by 0831–1035 is of the mixed non-competitive type. The KI of 0831–1035 is 9 μM (± 1 μM), and its KI′ 15 μM (± 2 μM). These data further confirm our conclusion that it is a mixed mode non-competitive inhibitor. However, the substantially lower KI of this inhibitor (versus its KI′ (and also the observed significant increase in the Km value in the presence of the inhibitor)) suggests that perhaps a substantial portion of the calculated BoNT/A LC inhibition mediated by 0831–1035 may still be of a competitive nature. The nature of non-competitive inhibition indicates that the inhibitor may possess a secondary enzyme binding site that interferes with both the enzyme active site and the enzyme-substrate complex. The co-crystal structure of SNAP-25 in complex with the BoNT/A LC suggests that there are two exosites (on the BoNT/A LC) with which the natural substrate binds (Breidenbach and Brunger, 2004). The α-exosite is located outside of the catalytic cleft of the enzyme, but is predicted to play a key role in whole protein substrate specificity (Breidenbach and Brunger, 2004); the β-exosite protrudes into the enzyme’s catalytic center, and also plays a key role in substrate recognition and enzymatic activity (Breidenbach and Brunger, 2004). However, since we used a peptide substrate that does not possess the capability to interfere with a potential inhibitor:BoNT/A LC interaction at the α-exosite, the observed reduction in the enzyme’s activity most likely results from interactions between 0831–1035 and both the BoNT/A LC’s β-exosite and its active site. Indeed, our kinetic evaluation of the binding mode of 0831–1035 appears to suggest that this inhibitor may: 1) bind to the β-exosite (and possibly the α-exosite) of the BoNT/A LC, and thereby obstruct substrate recognition in a location removed from the catalytic center and 2) bind within the BoNT/A LC catalytic cleft, thereby providing competitive inhibition.
Table 3.
BoNT/A LC Enzyme kinetics and inhibition kinetics by 0831–1035.
| Inhibitor Concentration (μM) | 0 | 19.5 | 39.0 |
|---|---|---|---|
| KM(μM) | 19.4 (2.1*) | 25.4 (2.4) | 28.0 (2.8) |
| Vmax(RFU/s) | 3.1 (0.4) | 1.2 (0.3) | 0.92 (0.3) |
Data in the parentheses represent the standard deviation.
Specificity Analysis
We next examined the specificity of 0831–1035. Since thermolysin shares the same zinc binding motif as the BoNT/A LC (Singh, 2000, 2006; Montecucco and Schiavo, 1993; Zuniga et al., 2008), this enzyme was used as a model to examine the potential promiscuity of 0831–1035. Results from this testing (data not shown) indicated that 0831–1035 does not inhibit the proteolytic activity of thermolysin on bovine serum albumin. Hence, the results strongly suggest that our inhibitor is specific for the BoNT/A LC, and lacks general protease promiscuity. This is further evidence that 0831–1035 is a good lead candidate for therapeutic development.
Primary Neuronal Cell Culture
To be effective as therapeutic agents to counter BoNT intoxication, small molecules must cross the neuronal cell membrane and inhibit the LC in the cytosol. Cellular assays can provide vital information in this respect. Moreover, while the crystal structure of the BoNT/A LC under in vitro conditions is well known, the nature of the BoNT/A LC in intoxicated neurons remains elusive, as it undergoes unfolding and refolding during its release from the endosome (Cai et al., 2006; Koriazova and Montal, 2003), possibly along with other enzymatic modifications and potential interactions with the cell membrane. Furthermore, in vivo assays are expensive, time-consuming, and provide only semi-quantitative results. Thus, cellular models based on neuronal cell cultures mimicking in vivo intoxication are good means for gauging the therapeutic potentials of BoNT LC small molecule inhibitors.
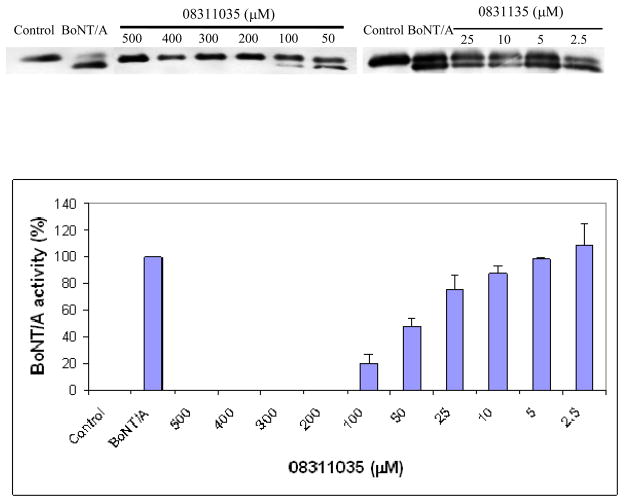
Examination of 0831–1035 in our neuronal assay indicated that this inhibitor provides complete protection of SNAP-25 (at 200 μM inhibitor concentration, and 0.5 nM concentration BoNT/A applied to the cell culture). Further investigation confirmed that this compound inhibited the toxin’s activity in neurons in a dose-dependent manner (Fig. 3), with an IC50 value of 47 μM (± 7.0 μM). The IC50 for the primary cell assay is in the same range as the in vitro assay (16 μM). This may be due to the different concentration of toxin used in the cell assay compared to the LC for the in vitro assay. The ratio of inhibitor to toxin for 50% inhibition was 94,000:1 in primary cell assay, whereas the ratio of inhibitor to LC for 50% inhibition in vitro is 320:1. The molar ratio of 50% inhibition provides better parameter for comparison between assays, since it normalizes the toxin used. The higher molar ratio in cellular assay reflects the inhibitor’s inefficiency of entering the cell, and presumably a different state of inhibition of the enzymatic activity intracellularly. Furthermore, it is not known what form of the toxin is available incellularly (LC only, or LC associated with HC). Nonetheless, the inhibition at the cellular level is the overall inhibition results, and is more relevant to in vivo conditions.
Figure 3.
Dose-dependent inhibition of BoNT/A by 0831–1035 in a primary neuronal cell culture. Varying concentrations of 0831–1035 were added to the cell culture medium, followed by the addition of 0.5 nM BoNT/A. After 3 hr incubation, cell samples were subjected to immunoblot analysis. The degree of SNAP-25 cleavage was determined via scanning densitometry.
Importantly, we did not observe any significant morphological signs of neuronal damage during the experiment up to the application of 500 μM of 0831–1035, indicating that there is no cytotoxicity associated with this inhibitor within its range of efficacy in neurons. Thus, these results further indicate that 0831–1035 is an excellent lead candidate for further development as a drug to treat botulism resulting from BoNT/A intoxication.
Conclusions
We have reported a robust method for screening and confirming inhibitors of the endopeptidase activity of the BoNT/A LC. Using a two-tiered screening methodology, 16,554 compounds were evaluated, and of these, a lead for therapeutic development (inhibitor 0831–1035), possessing an IC50 value of 16 μM (± 1.6 μM) in vitro, was confirmed. Moreover, 0831–1035 possesses excellent solubility, and provides BoNT/A LC inhibition in a primary neuronal assay, with no cytotoxicity - even at a concentration as high as 500 μM. Furthermore, enzyme kinetics analysis indicated that 0813–1035 is a mixed mode non-competitive inhibitor, suggesting that it not only directly interferes with BoNT/A LC:substrate binding in the enzyme’s catalytic cleft, but also enzyme:substrate contacts located outside of the binding cleft. Based on the low toxicity and cellular activity of 0831–1035, this lead inhibitor will serve as the foundation for the generation of structure activity relationships to provide more potent derivatives, with the ultimate goal of developing a therapeutic countermeasure to treat BoNT/A LC mediated botulism.
Supplementary Material
Acknowledgments
This work was in part supported by National Institutes of Health through New England Center of Excellence for Biodefence (AI057159-01), and by NIH grant 5R21AI070787-02. We thank the National Small Molecule Screening and Medicinal Chemistry Core at the Harvard Medical School for aiding in the optimization of our HTS-assay, and for allowing us to use the HTS screening facility. The authors sincerely thank Ms. Koyel J. Ghosal for proof reading of the manuscript.
Abbreviations
- BoNT/A
botulinum neurotoxin serotype A
- LC
light chain
- HC
heavy chain
- FRET
fluorescence resonance energy transfer
- FITC
Fluorescein-5-isothiocyanate
- DABCYL
4-dimethylaminoazobenzene-4′-carboxylic acid
- HTS
High throughput screening
- HRP
Horseradish peroxidase
Footnotes
Conflict of Interest statement
The authors declare that there are no conflicts of interest.
This work was in part supported by the National Institutes of Health through the New England Center of Excellence for Biodefense (AI057159-01), and by NIH grant 1R21AI070787-02.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett DA, Scher MS, Eitzen E, Fine AD, Hauer J, Layton M, Lillibridge S, Osterholm MT, O’Toole T, Parker G, Perl TM, Russell PK, Swerdlow DL, Tonal K. Botulinum toxin as a biological weapon: medical and public health management. JAMA. 2000;285:1059–1070. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- Boldt GE, Eubanks LM, Janda KD. Identification of a botulinum neurotoxin A protease inhibitor displaying efficacy in a cellular model. Chem Commun (Camb) 2006;7:3063–3065. doi: 10.1039/b603099h. [DOI] [PubMed] [Google Scholar]
- Breidenbach MA, Brunger AT. Substrate recognition strategy for botulinum neurotoxin serotype A. Nature. 2004;432:925–929. doi: 10.1038/nature03123. [DOI] [PubMed] [Google Scholar]
- Burnett JC, Wang C, Nuss JE, Nguyen TL, Hermone AR, Schmidt JJ, Gussio R, Wipf P, Bavari S. Pharmacophore-guided lead optimization: the rational design of a non-zinc coordinating, sub-micromolar inhibitor of the botulinum neurotoxin serotype a metalloprotease. Bioorg Med Chem Lett. 2009;19:5811–5813. doi: 10.1016/j.bmcl.2009.01.111. [DOI] [PubMed] [Google Scholar]
- Burnett JC, Opsenica D, Sriraghavan K, Panchal RG, Ruthel G, Hermone AR, Nguyen TL, Kenny TA, Lane DJ, McGrath CF, Schmidt JJ, Vennerstrom JL, Gussio R, Solaja BA, Bavari S. A refined pharmacophore identifies potent 4-amino-7-chloroquinoline-based inhibitors of the botulinum neurotoxin serotype A metalloprotease. J Med Chem. 2007a;50:2127–2136. doi: 10.1021/jm061446e. [DOI] [PubMed] [Google Scholar]
- Burnett JC, Ruthel G, Stegmann CM, Panchal RG, Nguyen TL, Hermone AR, Stafford RG, Lane DJ, Kenny TA, McGrath CF, Wipf P, Stahl AM, Schmidt JJ, Gussio R, Brunger AT, Bavari S. Inhibition of metalloprotease botulinum serotype A from a pseudo-peptide binding mode to a small molecule that is active in primary neurons. J Biol Chem. 2007b;282:5004–5014. doi: 10.1074/jbc.M608166200. [DOI] [PubMed] [Google Scholar]
- Burnett JC, Schmidt JJ, McGrath CF, Nguyen TL, Hermone AR, Panchal RG, Vennerstrom JL, Kodukula K, Zaharevitz DW, Gussio R, Bavari S. Conformational sampling of the botulinum neurotoxin serotype A light chain: implications for inhibitor binding. Bioorg Med Chem. 2005;13:333–341. doi: 10.1016/j.bmc.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Burnett JC, Schmidt JJ, Stafford RG, Panchal RG, Nguyen TL, Hermone AR, Vennerstrom JL, McGrath CF, Lane DJ, Sausville EA, Zaharevitz DW, Gussio R, Bavari S. Novel small molecule inhibitors of botulinum neurotoxin A metalloprotease activity. Biochem Biophys Res Commun. 2003;310:84–93. doi: 10.1016/j.bbrc.2003.08.112. [DOI] [PubMed] [Google Scholar]
- Cai S, Kukreja R, Shoesmith S, Chang TW, Singh BR. Botulinum neurotoxin light chain refolds at endosomal pH for its translocation. Protein J. 2006;25:455–462. doi: 10.1007/s10930-006-9028-1. [DOI] [PubMed] [Google Scholar]
- Cai S, Sarkar HK, Singh BR. Enhancement of the endopeptidase activity of botulinum neurotoxin by its associated proteins and dithiothreitol. Biochemistry. 1999;38:6903–6910. doi: 10.1021/bi990086c. [DOI] [PubMed] [Google Scholar]
- Cai S, Singh BR. Strategies to Design Inhibitors of Clostridium Botulinum Neurotoxins. Infectious Disorders-Drug Targets. 2007;7:47–57. doi: 10.2174/187152607780090667. [DOI] [PubMed] [Google Scholar]
- Devlin JP, editor. High Throughput Screening. CRC Press; 1997. [Google Scholar]
- Dickerson TJ, Janda KD. The use of small molecules to investigate molecular mechanisms and therapeutic targets for treatment of botulinum neurotoxin A intoxication. ACS Chem Biol. 2006;1:359–369. doi: 10.1021/cb600179d. [DOI] [PubMed] [Google Scholar]
- Greenfield RA, Brown BR, Hutchins JB, Iandolo JJ, Jackson R, Slater LN, Bronze MS. Microbiological, biological, and chemical weapons of warfare and terrorism. Am J Med Sci. 2002;323:326–340. doi: 10.1097/00000441-200206000-00005. [DOI] [PubMed] [Google Scholar]
- Hermone AR, Burnett JC, Nuss JE, Tressler LE, Nguyen TL, Solaja BA, Vennerstrom JL, Schmidt JJ, Wipf P, Bavari S, Gussio R. Three-dimensional database mining identifies a unique chemotype that unites structurally diverse botulinum neurotoxin serotype A inhibitors in a three-zone pharmacophore. ChemMedChem. 2008;3:1905–1912. doi: 10.1002/cmdc.200800241. [DOI] [PubMed] [Google Scholar]
- Koriazova LK, Montal M. Translocation of botulinum neurotoxin light chain protease through the heavy chain channel. Nat Struct Biol. 2003;10:13–18. doi: 10.1038/nsb879. [DOI] [PubMed] [Google Scholar]
- Li L, Singh BR. Structure-function relationship of clostridial neurotoxins. J Toxicol -Toxin Reviews. 1999a;18:95–112. [Google Scholar]
- Li L, Singh BR. High-level expression, purification, and characterization of recombinant type A botulinum neurotoxin light chain. Protein Expr Purif. 1999b;17:339–344. doi: 10.1006/prep.1999.1138. [DOI] [PubMed] [Google Scholar]
- Montecucco C. How do tetanus and botulinum toxins bind to neuronal membranes? Trends Biochem Sci. 1986;11:314–317. [Google Scholar]
- Montecucco C, Schiavo G. Tetanus and botulism neurotoxins: a new group of zinc proteases. Trends Biochem Sci. 1993;18:324–327. doi: 10.1016/0968-0004(93)90065-u. [DOI] [PubMed] [Google Scholar]
- Montecucco C, Schiavo G. Structure and function of tetanus and botulinum neurotoxins. Quarterly Rev Biophys. 1995;28:423–472. doi: 10.1017/s0033583500003292. [DOI] [PubMed] [Google Scholar]
- Montecucco C, Rossetto O, Schiavo G. Presynaptic receptor arrays for clostridial neurotoxins. Trends Microbiol. 2004;12:442–446. doi: 10.1016/j.tim.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Poulain B, Popoff MR, Molgo J. How do the Botulinum Neurotoxins block neurotransmitter release: from botulism to the molecular mechanism of action. The Botulinum J. 2008;1:14–87. [Google Scholar]
- Rosenbloom M, Leikin JB, Vogel SN, Chaudry ZA. Biological and chemical agents: a brief synopsis. Am J Therapeutics. 2002;9:5–14. doi: 10.1097/00045391-200201000-00003. [DOI] [PubMed] [Google Scholar]
- Rossetto O, Seveso M, Caccin P, Schiavo G, Montecucco C. Tetanus and botulinum neurotoxins: turning bad guys into good by research. Toxicon. 2001;39:27–41. doi: 10.1016/s0041-0101(00)00163-x. [DOI] [PubMed] [Google Scholar]
- Schmidt JJ, Stafford RG, Millard CB. High-throughput assays for botulinum neurotoxin proteolytic activity: serotypes A, B, D, and F. Anal Biochem. 2001;296:130–137. doi: 10.1006/abio.2001.5236. [DOI] [PubMed] [Google Scholar]
- Sharma SK, Singh BR. Enhancement of the endopeptidase activity of purified botulinum neurotoxins A and E by an isolated component of the native neurotoxin associated proteins. Biochemistry. 2004;43:4791–4798. doi: 10.1021/bi0355544. [DOI] [PubMed] [Google Scholar]
- Simpson LL. Identification of the major steps in botulinum toxin action. Annu Rev Pharmacol Toxicol. 2004;44:167–193. doi: 10.1146/annurev.pharmtox.44.101802.121554. [DOI] [PubMed] [Google Scholar]
- Singh BR. Intimate details of the most poisonous poison. Nat Struct Biol. 2000;7:617–619. doi: 10.1038/77900. [DOI] [PubMed] [Google Scholar]
- Singh BR. Botulinum neurotoxin structure, engineering, and novel cellular trafficking and targeting. Neurotox Res. 2006;9:73–92. doi: 10.1007/BF03033925. [DOI] [PubMed] [Google Scholar]
- Skaper SD, Adler R, Varon S. A procedure for purifying neuron-like cells in cultures from central nervous tissue with a defined medium. Develop NeuroSci. 1979;2:233–237. doi: 10.1159/000112485. [DOI] [PubMed] [Google Scholar]
- Wein LM, Liu Y. Analyzing a bioterror attack on the food supply: the case of botulinum toxin in milk. Proc Natl Acad Sci USA. 2005;102:9984–9989. doi: 10.1073/pnas.0408526102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zuniga JE, Schmidt JJ, Fenn T, Burnett JC, Araç D, Gussio R, Stafford RG, Badie SS, Bavari S, Brunger AT. A potent peptidomimetic inhibitor of botulinum neurotoxin serotype A has a very different conformation than SNAP-25 substrate. Structure. 2008;16:1588–1597. doi: 10.1016/j.str.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.