Abstract
Ionotropic glutamate receptors (iGluRs) are enticing targets for pharmaceutical research; however, the search for selective ligands is a laborious experimental process. Here we introduce a purely computational procedure as an approach to evaluate ligand–iGluR pharmacology. The ligands are docked into the closed ligand-binding domain and during the molecular dynamics (MD) simulation the bi-lobed interface either opens (partial agonist/antagonist) or stays closed (agonist) according to the properties of the ligand. The procedure is tested with closely related set of analogs of the marine toxin dysiherbaine bound to GluK1 kainate receptor. The modeling is set against the abundant binding data and electrophysiological analyses to test reproducibility and predictive value of the procedure. The MD simulations produce detailed binding modes for analogs, which in turn are used to define structure–activity relationships. The simulations suggest correctly that majority of the analogs induce full domain closure (agonists) but also distinguish exceptions generated by partial agonists and antagonists. Moreover, we report ligand-induced opening of the GluK1 ligand-binding domain in free MD simulations. The strong correlation between in silico analysis and the experimental data imply that MD simulations can be utilized as a predictive tool for iGluR pharmacology and functional classification of ligands.
Keywords: Molecular dynamics, Agonism, Partial agonism, Antagonism, Kainate receptor, Ionotropic glutamate receptor
1. Introduction
Ionotropic glutamate receptors (iGluRs) form (S)-glutamate-gated ion channels that mediate neuronal excitation through synapses of the central nervous system. The iGluR family is divided into NMDA (N-methyl-d-aspartate), kainate (2S,3S,4S)-3-(carboxymethyl)-4-prop-1-en-2-ylpyrrolidine-2-carboxylic acid), and AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid) receptors based on their sequence identity and pharmacology (Hollmann and Heinemann, 1994). Kainate receptors (KARs) are assembled as a dimer of dimers to form membrane-bound tetrameric ion channels (Safferling et al., 2001), either as homomeric assemblies of GluK1 (GluR5; GLUK5), GluK2 (GluR6; GLUK6), and GluK3 (GluR7; GLUK7) subunits or in heteromeric combinations with GluK4 (KA1; GLUK1) and GluK5 (KA2; GLUK2) subunits (Pinheiro and Mulle, 2006) (IUPHAR nomenclature used; Collingridge et al., 2009). KARs are linked to a number of neurological pathologies, including epilepsy (Barton et al., 2003), migraine (Filla et al., 2002; Sang et al., 2004), cerebral ischemia (O’Neill et al., 1998), pain (Dominguez et al., 2005; Gilron et al., 2000; Sang et al., 1998), and anxiety disorders (Alt et al., 2004). For determination of the exact functional and pathological roles of KARs comprised of distinct subunits, it will be necessary to develop subtype-specific antagonists. A number of such selective antagonists have been identified, but these largely are restricted to inhibition of GluK1 receptors (Weiss et al., 2006). Several non-competitive GluK1-specific antagonists have been described as well (Christensen et al., 2004; Valgeirsson et al., 2004).
A detailed molecular understanding of ligand–receptor interactions is required for the generation of KAR selective antagonists. The ligand-binding domains of the full-length iGluRs are formed from two discontinuous extracellular segments, S1 and S2, of the subunit proteins. Agonists such as (S)-glutamate bind to the ligand-binding domain or restructured ligand-binding core (LBC) between the D1 and D2 lobes (or domains) formed from the S1 and S2 primary segments (Stern-Bach et al.,1994) and induce rotation (or closure) of the LBC. The extent of relative closure (or opening) of the cleft between D1 and D2, which are linked by a flexible hinge area formed primarily by antiparallel β-strands, is variable and dependent upon the molecular interactions between the ligand and the LBC. Screw-axis bending resembling a Venus flytrap mechanism, generally referred to as “domain closure”, has been correlated with receptor activation (Armstrong and Gouaux, 2000; Armstrong et al., 2003) and agonist efficacy (Jin et al., 2003), supporting the hypothesis that gating transitions are downstream of lobe closure and may be sensitive to the magnitude of induced rotation, possibly through linker domains that connect the LBC with transmembrane domains that form the channel pore. However, exceptions to the correlation between LBC closure and agonist efficacy have been noted for iGluRs (Inanobe et al., 2005; Frydenvang et al., 2009; Fay et al., 2009). With respect to KARs, these include a fully closed GluK1–LBC crystal structure with the bound weak partial agonist MSVIII-19 (Frydenvang et al., 2009) and inferences derived from a docking study by Fay et al. (2009), which concluded that some partial agonists can bind into closed receptor conformation. Competitive antagonists inhibit closure of the bi-lobed LBC and thereby prevent opening of the ion channel.
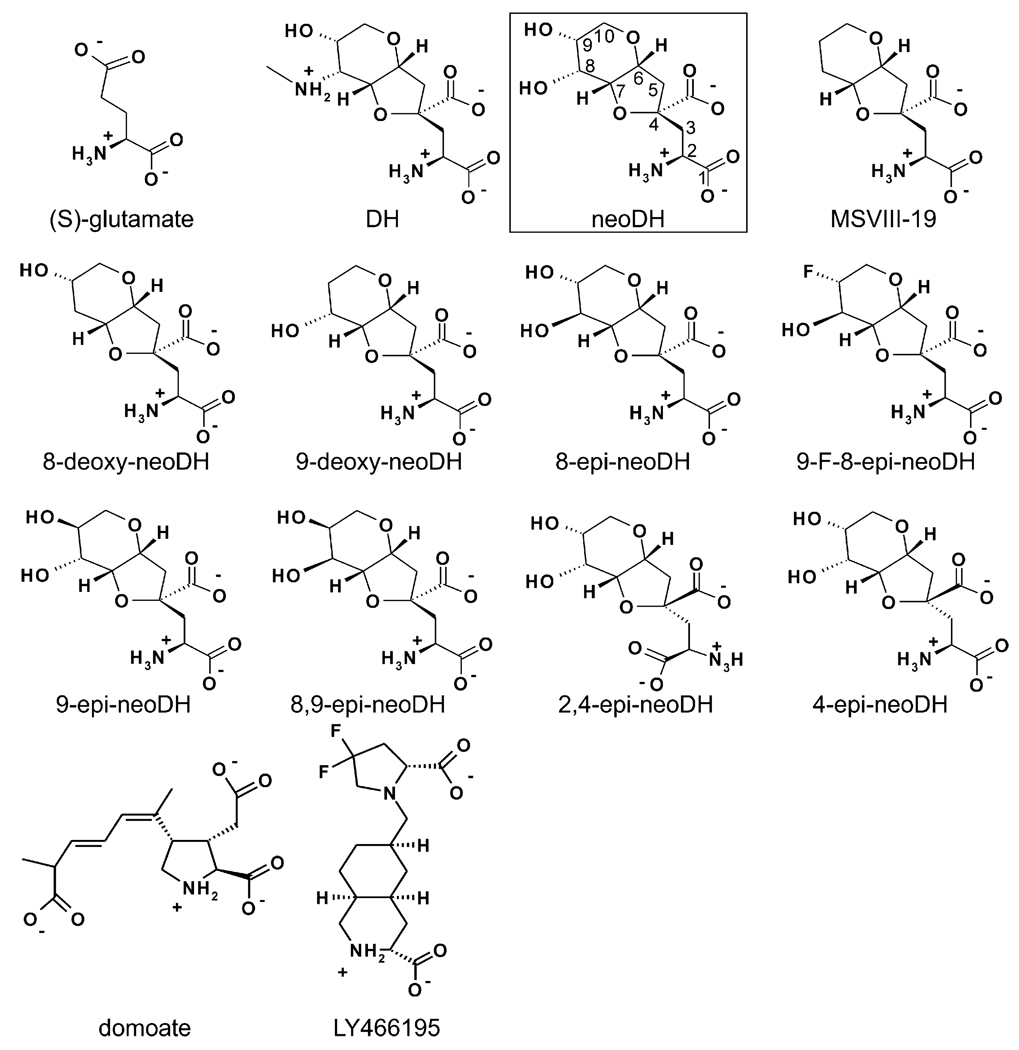
To clarify molecular interactions within iGluR binding domains, we previously derived models that rationalized agonist binding selectivity for AMPA and kainate receptor subunits (Pentikäinen et al., 2003), used molecular dynamics (MD) simulations to probe antagonist binding to GluA2- (GluR2; GLUA2) and GluK1–LBCs (Pentikäinen et al., 2006), and studied the binding of dysiherbaine (DH) and its analogs to KARs (Frydenvang et al., 2009; Lash et al., 2008; Sanders et al., 2005, 2006). The highly potent KAR agonist ligands, DH and neodysiherbaine (neoDH) were originally isolated from Micronesian marine sponge Lendenfeldia chodrodes (Sakai et al., 2001), and these molecules and their synthetic analogs have been of particular use for structure-function studies in KARs because they exhibit a wide range of pharmacological activities. DH and its derivatives contain the conserved amino acid backbone of (S)-glutamate fused into a hydrophobic hydrofuropyran ring system (Fig. 1). The two natural toxins differ only at the C8 position of the ring system: DH contains a methylamine substituent and neoDH a hydroxyl group. Most of the DH analogs bind exclusively to GluK1 or have higher affinity for this subunit relative to GluK2 or other KAR subunits (Lash et al., 2008). While the majority of the DH analogs were categorized as agonists with a range of affinities, 2,4-epi-neoDH appeared to act as an antagonist (Lash et al., 2008) and 8,9-dideoxy-neoDH (or MSVIII-19) (Sanders et al., 2005) as a very weak partial agonist or functional antagonist with minimal agonist efficacy (Frydenvang et al., 2009), demonstrating that relatively small differences in ligand structure could profoundly impact pharmacological activity.
Fig. 1.
The 2D structures of all simulated ligands. The carbon atom numbering used for DH analogs is shown for boxed neoDH.
Here, we determine if binding modes derived from computationally demanding MD simulations are predictive for the pharmacological properties of DH analogs on GluK1. The ligands (Fig. 1) were docked flexibly into the ligand-binding site of the closed, rotated GluK1–D1D2 structure, and then ligand–LBC movements and interactions were simulated with MD. We demonstrate for the first time that partial agonists and antagonists mechanistically induce opening of the closed structural model of GluK1–LBC in a free MD simulation. The dissimilar binding characteristics of each DH analog are used to identify particular molecular interactions required for activation or desensitization of GluK1 receptors. Subtle rearrangements of the LBC that underlie these physiological processes are relayed by a meshwork of interconnected water molecules in response to (S)-glutamate binding (Armstrong and Gouaux, 2000), but this highly organized system behaves differently with the larger DH analogs that contain rigid ring systems (Fig. 1). In most cases, our results conform to a simple mechanistic model in which the pharmacological behavior can be directly predicted by comparing the opening of the receptor cleft with published crystal structures (e.g. Naur et al., 2005; Hald et al., 2007; Mayer et al., 2006), while the magnitude of opening can vary. The behavior of MSVIII-19 in simulations constitutes an exception to this model and suggests that the mechanism of action must be somewhat different for this ligand, as was noted in our previous crystallographic study (Frydenvang et al., 2009). For this we provide a mechanistic hypothesis. These analyses will be of use in future efforts to design selective pharmacological agents for iGluRs.
2. Methods
2.1. Starting structures for molecular dynamics simulations
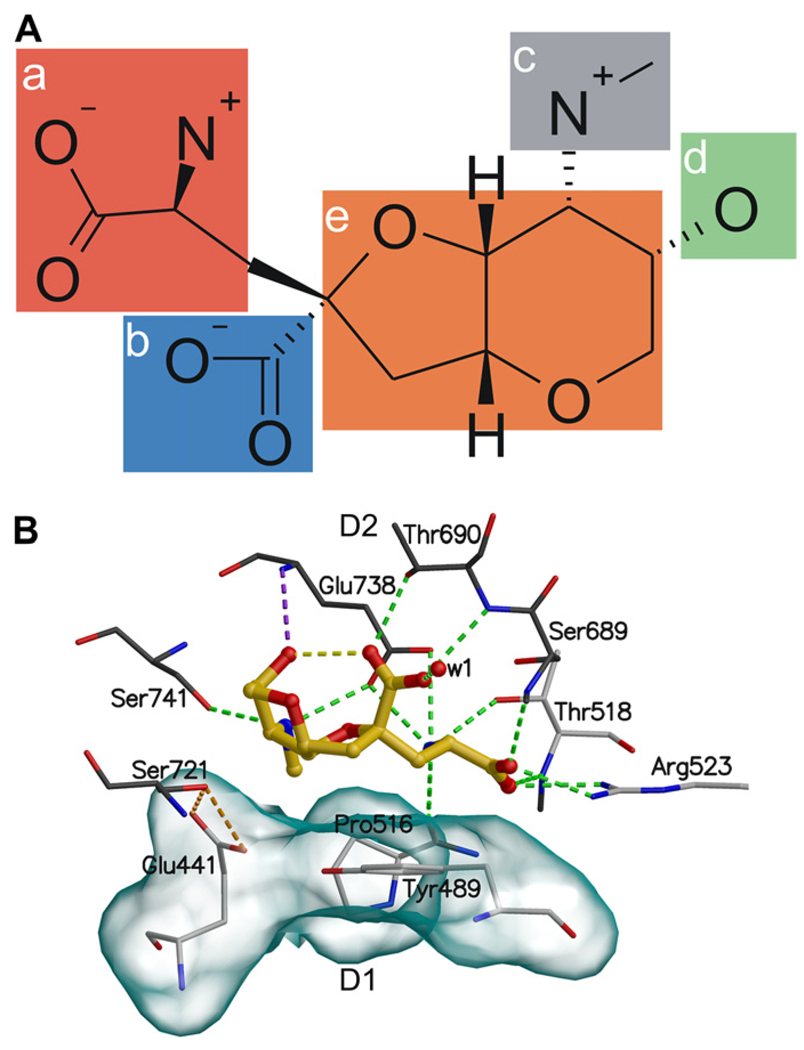
The structures of ligands (Fig. 1) sketched with SYBYL7.3 (Tripos, Inc., St Louis, MO, USA) were geometry-optimized quantum mechanically with GAUSSIAN03 (Gaussian Inc., Wallingford, CT, USA) at the HF/6–31+G* level with the continuum water (PCM) model. The 3D structure of GluK1–LBC bound to (S)-glutamate (PDB-code: 1YCJ; Naur et al., 2005) was acquired from the PDB (Berman et al., 2000) (http://www.pdb.org/). The dimer structures of GluK1–LBC were customized using BODIL modeling environment (Lehtonen et al., 2004). The use of dimer structures doubles the yield of a single MD simulation and reduces unnatural solitary movements of the LBCs, thus increasing the amount of data and possibly dependability of the results. The optimized molecules were docked flexibly into both ligand-binding sites with GOLD3.1.1 (Jones et al., 1995, 1997). The search area was predefined to a 15 Å radius sphere centered at the OOH/O.3-atom of Tyr489, which has a central location in the ligand-binding site. As a rule, the docked ligand poses used in MD simulations had most of the canonical binding interactions with GluK1 (Fig. 2). To assure that the initial docking of the DH analogs led to reasonable predictions for ligand-binding conformations, some of the ligands were intentionally inserted into the GluK1–LBC in configurations that lacked all of the interactions formed by (S)-glutamate with the LBC (Fig. 2A: groups a and b). During the equilibration stage of the simulations, however, these ligands rotated to establish interactions observed between the (S)-glutamate congener in DH analogs and (S)-glutamate itself (Naur et al., 2005) (Fig. 2). Accordingly, data from these aberrant initializations are not differentiated in the study.
Fig. 2.
The binding mode of DH. In (A) the 2D structure of DH with five key binding groups (a–e) are highlighted and in (B) the binding interactions of DH with GluK1–LBC are shown in detail. (a) The side chain guanidinium group of Arg523 and Ser689N, and Thr518N hydrogen bond to the α-carboxylate group of DH and the side chain carboxylate group of Glu738, the side chain hydroxyl of Thr518, and Pro516O hydrogen bond to the α-amine group of DH. (b) Thr690N and the side chain hydroxyl of Thr690 hydrogen bond to the γ-carboxylate group of DH. (c) The C8 aminomethyl group hydrogen bonds to the side chain hydroxyl of Ser741 and the side chain carboxylate group of Glu738, and (d) the C9 hydroxyl of the ring system hydrogen bond to Glu738N. (e) The tetrahydrofuropyran ring (referred as the ring system) of DH hydrophobically packs against the side chains of the binding pocket residues Tyr489, Glu441, and Pro516 at the D1 face of the binding pocket. The hydrogen bonds are shown as green dotted lines, the yellow line represent intra-ligand hydrogen bonds, the orange lines highlight the hydrogen bonds between the side chains of Glu441 and Ser721, and the purple line connect Glu738N to the C9 hydroxyl. The solvent accessible surface (transparent surface) visualizes the hydrophobic face. Amino acids at the D1 face are shown with white carbon atoms and at the D2 face with black carbon atoms. The oxygen atom of the water molecule is presented as red sphere, and the ligand skeleton is shown as yellow ball-and-stick representation.
Protonation of histidines was selected on the basis of possible hydrogen bonds with nearby residues and water molecules present in the initial crystal structure. The artificial Gly545-Thr546-linker in GluK1–LBC, which was used for the purposes of connecting S1 and S2 domains in crystallization, was removed from both chains. The movements of C-termini of S1 and N-termini of S2 during the MD simulation of GluK1 with bound (1) neoDH, (2) 8,9-epi-neoDH, and (3) LY466195 are shown in Supplementary Fig. S1. After the docking of ligands, and prior to the MD simulation, water molecules in the crystal structures that were situated too close (1.4 Å radius) to the ligand were removed using BODIL. TLEAP in ANTECHAMBER-1.27 (Wang et al., 2006) was used to: (a) set the force field parameters for the protein (parm99) and ligands (gaff), (b) add hydrogen atoms, (c) neutralize the system with chloride ions, and (d) to solvate the system with a rectangular box of transferable intermolecular potential three-point water molecules (TIP3P) 13 Å in every dimension.
The constructed LBC–ligand complexes were used as starting structures for the MD simulations. The electrostatic potentials of the ligands were computed with GAUSSIAN03 (HF/6–31+G*) for the optimized ligands. The RESP methodology (Bayly et al., 1993; Cieplak et al., 1995; Cornell et al., 1993) was used to create the atom-centered point charges from the electrostatic potentials, where charges of chemically comparable atoms in the ligands were simultaneously set to identical values (Supplementary Fig. S2).
2.2. Molecular dynamics simulations
The two-step energy minimization and the three-step MD were run with NAMD 2.6 (Phillips et al., 2005). Firstly, the water molecules, counter-ions, and amino acid side chains were minimized with the conjugate gradient algorithm (15,000 steps) as rest of the complex was constrained by restraining Cα-atoms into their initial positions with the harmonic force of 5 kcal mol−1 Å−2. Secondly, the whole complex was minimized without constraints (15,000 steps) to assure complete equilibration of the system. The initial MD simulations, in which the Cα-atoms were restrained, as in the energy minimization, were performed in constant volume (30,000 steps) and then in constant pressure (30,000 steps) as well. The production simulation was performed without constraints for 14 ns for all ligands with the GluK1–LBC. In general each ligand–dimer complex was simulated no more than once and the ligands that induced opening of the GluK1–LBC were simulated twice, however, the behavior was reproduced in corresponding simulations, and thus data is shown only for single dimer simulations.
The simulated complex was held at constant temperature of 300 K with Langevin dynamics for all non-hydrogen atoms, using a Langevin damping coefficient of 5 ps−1. A constant pressure of 1 atm was upheld by a Nosé-Hoover Langevin piston (Feller et al., 1995) with an oscillation timescale of 200 fs and a damping timescale of 100 fs. An integration time step of 2 fs was used under a multiple time stepping scheme (Schlick et al., 1999). The bonded and short-range interactions were calculated every time step and long-range electrostatic interactions every third step. A cutoff value of 12 Å was used for the van der Waals and short-range electrostatic interactions. A switching function was enforced for the van der Waals forces to smoothen the cutoff. The simulations were conducted under the periodic boundary conditions with the full-system, and the long-range electrostatics were counted with the particle-mesh Ewald method (Darden et al., 1993; Essmann et al., 1995; Sagui and Darden,1999; Toukmaji et al., 2000). The bonds involving hydrogen atoms were restrained by the SHAKE algorithm (Ryckaert et al., 1977).
2.3. Trajectory analysis
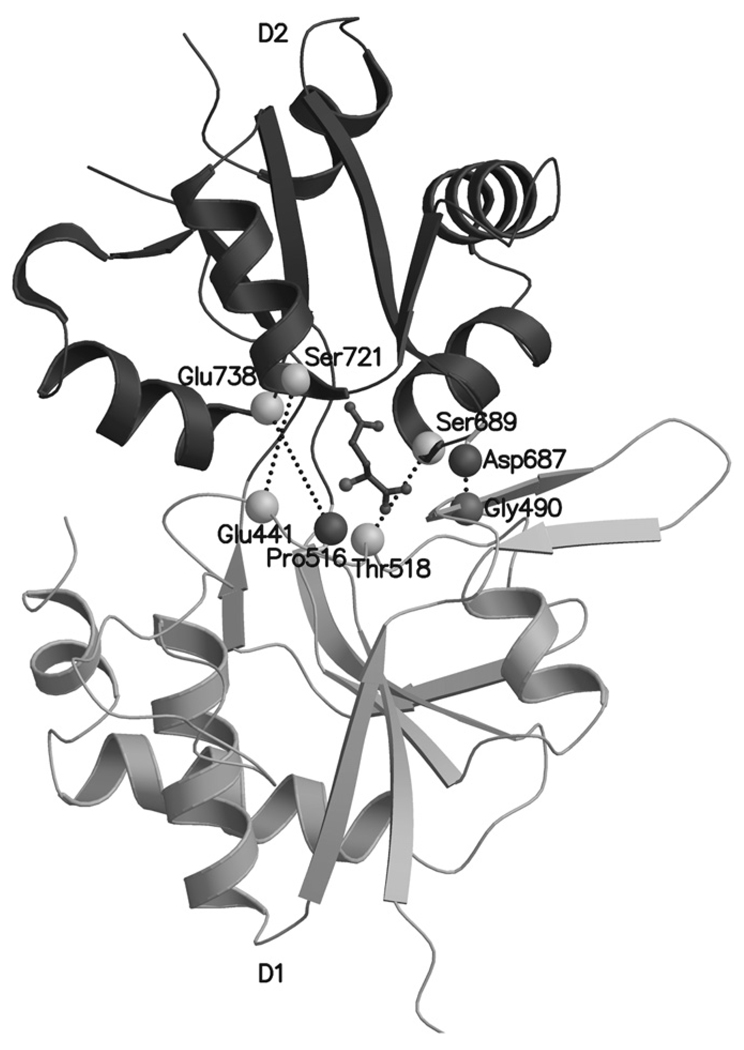
Snapshots at 360 ps intervals were extracted from the MD trajectories with PTRAJ 6.5 (standalone version: www.chpc.utah.edu/~cheatham/software.html, accessed 1.5.2007). Complete trajectories were analyzed in detail, but for Figs. 4,6, and 7 snapshots were chosen at around 14 ns that reflected well the binding processes. Various atom distances were extracted with PTRAJ 6.5 from key amino acid residues in the ligand-binding pocket (Fig. 3) to study the receptor cleft opening in detail. These particular atom pairs were selected from the main chain of the key amino acid residues to avoid misleading measurement of flexible rotamer pose changes of the side chains.
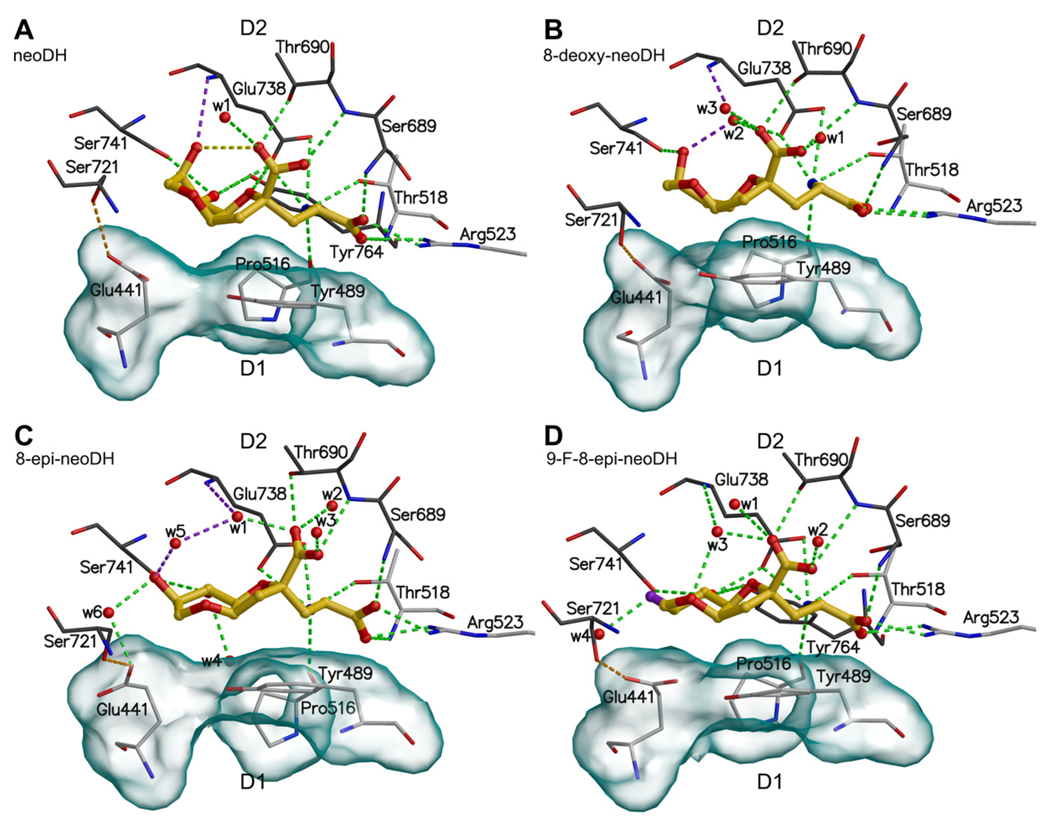
Fig. 4.
The binding modes of DH-based high affinity agonist ligands into GluK1–LBC: (A) neoDH, (B) 8-deoxy-neoDH, (C) 8-epi-neoDH, and (D) 9-F-8-epi-neoDH. For coloring and interpretation see Fig. 2B.
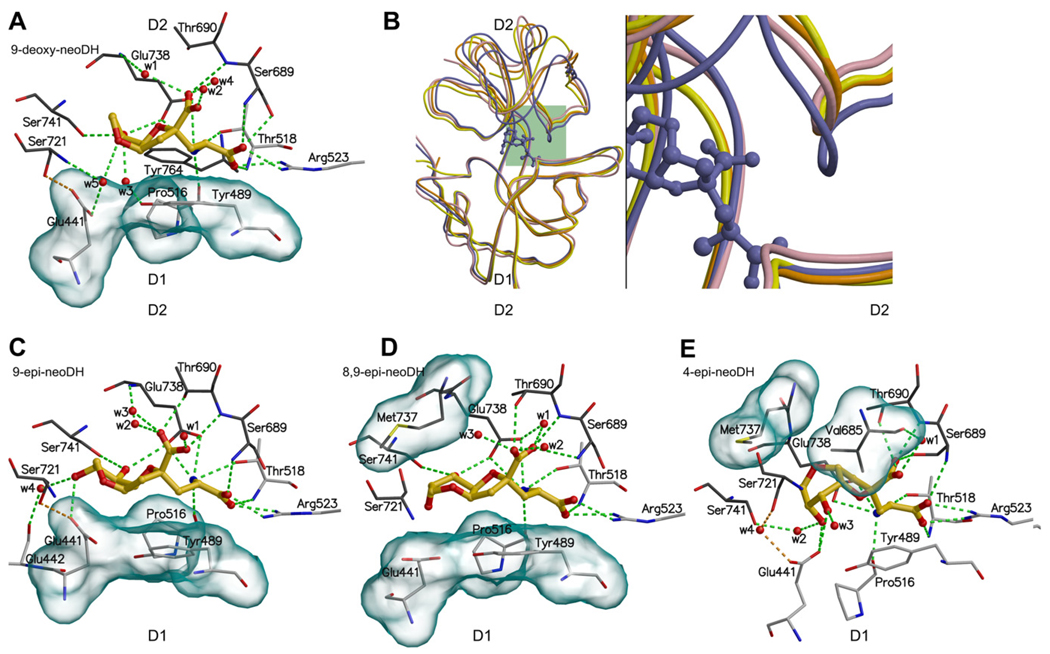
Fig. 6.
The binding modes of DH-based low affinity (or partial) agonist ligands for GluK1: (A) 9-deoxy-neoDH, (C) 9-epi-neoDH, (D) 8,9-epi-neoDH, and (E) 4-epi-neoDH. For coloring and interpretation (A, C–E) see Fig. 2B. In (B) is compared the crystal structures of two partial agonist–LBC complexes, GluA2–kainate (yellow) and GluK1–domoate (orange), agonist–LBC complex GluK1–DH (purple), and the MD simulated GluK1–9-deoxy-neoDH complex (pink). The structure comparison shows that 9-deoxy-neoDH is likely a partial agonist for GluK1, because its receptor-bound conformation resembles more the partial agonist structures than that of natural agonist DH (purple ball-and-stick).
Fig. 7.
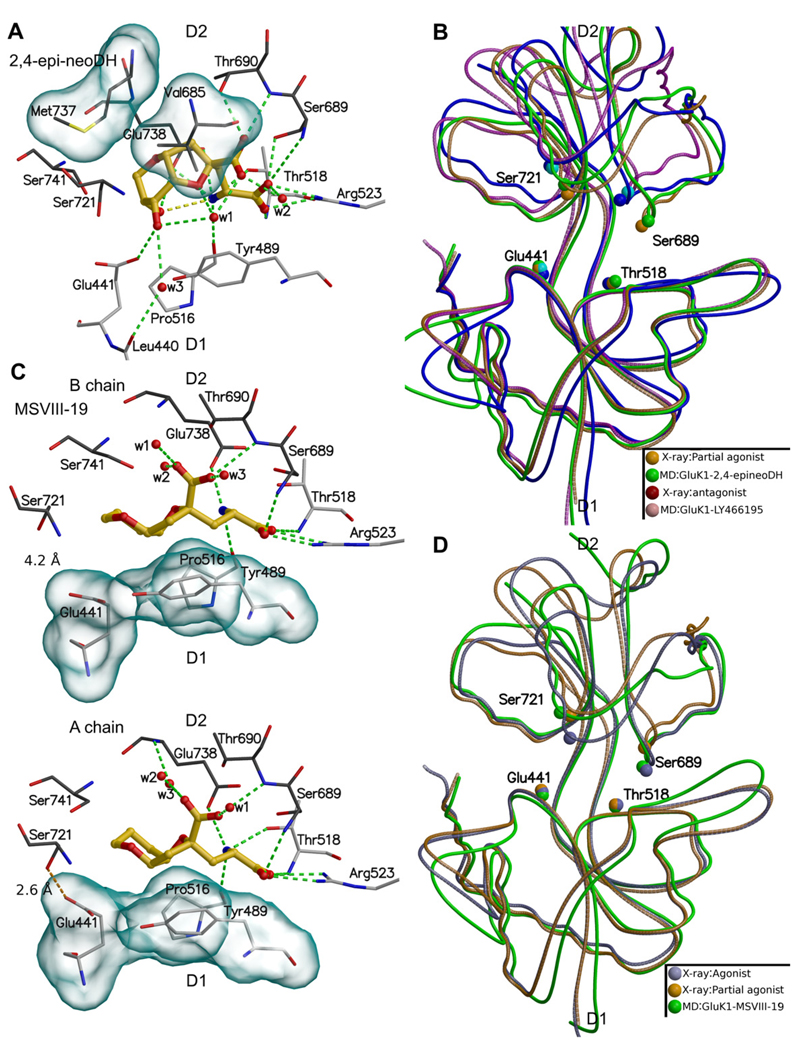
The binding modes of 2,4-epi-neoDH (A) and MSVIII-19 (C), and their effect on the GluK1–LBC opening (B) and (D), respectively. In (C), in the B chain (upper panel) the side chains of Glu441 and Ser721 are hydrogen bonded but in the A chain (lower panel) the bond does not exist. In (B), the crystal structure and MD simulation of GluK1–LY466195 complex is also shown. In (B) crystal structures of GluA2-kainate (partial agonist) and GluK1-LY466195 (antagonist) were used. In (D) crystal structures of GluK1-Glutamate (agonist) and GluA2-kainate (partial agonist) were used. For coloring and interpretation of panels (A) and (C), see Fig. 2B.
Fig. 3.
The atom pairs in the D1 and D2 lobes of the GluK1–LBC ligand-binding pocket selected for distance measurements from MD trajectories. The D1–D2 atom pairs indicated by dotted lines include: Glu441Cα-Ser721Cα, Thr518Cα-Ser689Cα, Pro516O-Glu738Cα, and Gly490O-Asp687N. The bound (S)-glutamate skeleton is shown as a black ball-and-stick representation. The D2 is shown with darker color than the D1.
The degree of the GluK1–LBC closure was measured by using the HINGEFIND algorithm (Wriggers and Schulten, 1997), which determines a flexible hinge between the domains of a protein. The closure of each ligand-bound GluK1–LBC was acquired by comparing the MD snapshots to a structure of GluK1–LBC co-crystallized with antagonist ligand UBP302 [(S)-1-(2-amino-2-carboxyethyl)-3-(2-carboxybenzyl)pyrimidine-2,4-dione] (PDB-code: 2F35; Mayer et al., 2006). As the angle comparison is not reasonable to be carried out for all the steps recorded into trajectory, the angles were measured from snapshot structures that were extracted every 0.36 ns, resulting in at least 40 study points for each complex. An increase in the angle value signifies the closure of the LBC and vice versa. The angle results are suggestive and supportive in nature, because the method is error-prone as the hinge has to be determined independently at every step.
2.4. Figures
The Figs. 1 and 2A were prepared with ISISDRAW (Symyx Technologies, Inc., Santa Clara, CA, USA). The Figs. 2B,3,4,6, and 7 were constructed using BODIL (Lehtonen et al., 2004), MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merritt and Bacon, 1997).
3. Results
The following analyses describe the detailed binding of DH-based derivatives to the GluK1–LBC based on MD simulations and test the correlation between these binding modes, and in particular the receptor cleft opening, with the experimentally determined pharmacological properties (Lash et al., 2008) (Table 1). The results are divided according to the functional properties of ligands obtained by experimental measurements (Lash et al., 2008) (Table 1).
Table 1.
The key binding interactions of DH-based analogs in GluK1–LBC coupled to binding affinity, activation, and desensitization.
| Ligand | Ki (nM)a | Activationa | Desensitizationa | C8b | C9b | H1c | H2d | H3e |
|---|---|---|---|---|---|---|---|---|
| DH | 0.5 | ++ | ++ | |||||
| neoDH | 7.7 | ++ | ++ | Y | X | + | + | X |
| 8-deoxy-neoDH | 1.5 | ++ | ++ | − | X | + | + | X/wX |
| 8-epi-neoDH | 34 | ++ | ++ | R | X | + | + | wX |
| 9-F-8-epi-neoDH | 28 | ++ | ++ | R | F | + | + | wX |
| 9-deoxy-neoDH | 169 | + | + | Y | − | + | − | − |
| 9-epi-neoDH | 292 | + | + | Y | R | + | + | − |
| 8,9-epi-neoDH | 48,000 | − | − | R | R | − | + | − |
| 4-epi-neoDH | 559 | + | ++ | − | − | − | + | − |
| 2,4-epi-neoDH | 2400 | − | ++ | − | − | − | − | − |
| MSVIII-19 | 128 | − | + | − | − | + | +/− | − |
Experimental data from Lash et al. (2008), except for DH from Sakai et al. (2001). Activation refers to the strength of the receptor activation, and desensitization refers to the length of the receptor inaction after ligand binding.
The C8 and C9 groups of DH (X: C9 positioned hydroxyl with the same stereochemistry as DH, Y: hydroxyl with the same stereochemistry as C8 methylamine group of DH, R: hydroxyl with reversed stereochemistry from DH, F = hydroxyl substituted with fluorine) compared.
H1: Hydrogen-bonds connect the D1 and D2 lobes between Gly490O and Asp687N (from MD simulations).
H2: Hydrogen-bonds connect the D1 and D2 lobes between the side chains of Glu441 and Ser721 (from MD simulations).
H3: The Glu738N is hydrogen bonded to the ligand’s C9 hydroxyl (X: direct hydrogen bond, wX: connected via mediating water molecule or molecules) (from MD simulations).
3.1. Key interactions underlying DH binding and closure of the GluK1–LBC
In general, the binding interactions between the incorporated (S)-glutamate backbone within DH or its analogs and the GluK1–LBC (Sanders et al., 2005) (Fig. 2A: groups a and b) are conserved to a large extent with all bound DH-related structures. Accordingly, the apparent differences in ligand-binding affinities (Table 1), dissimilar KAR subunit specificity, and altered functionality (Lash et al., 2008) of the DH-based ligands result primarily from structural divergence within their ring systems (Fig. 2A: group e) and especially at their C8 (Fig. 2A: group c, Table 1) and C9 positions (Fig. 2A: group d, Table 1). The 2D structures of DH-related analogs and other MD simulated ligands are shown in Fig. 1.
3.2. The D1–D2 interface distances from MD simulations and crystal structures
The extent of GluK1 receptor cleft opening in MD simulations arising from binding of each DH-derived ligand was compared to the crystal structures of GluK1–LBC with an antagonist, ATPO (PDB-code: 1VSO; Hald et al., 2007), a partial agonist, domoate (PDB-code: 2PBW; Hald et al., 2007), and a full agonist, (S)-glutamate (PDB-code: 1YCJ; Naur et al., 2005) in order to categorize their pharmacological activity. Selected atom pairs were measured between the D1 and D2 lobes (Fig. 3) both from the reference structures and MD simulation trajectories (Table 2), providing suggestive guidelines for the three functional categories. The distance between Gly490N and Asp687O most prominently reflected the dissimilar extent in opening of the GluK1–D1D2 upon binding of ligands with distinct pharmacological properties: closure of the interface by a full agonist kept these atoms near enough to form a hydrogen bond (crystal structure: 3.0 Å vs. MD simulations: 3.0–3.6 Å), whereas a partial agonist (crystal structure: 6.7 Å vs. MD simulations: 3.0–4.7 Å), and an antagonist (crystal structure: 12.5 Å vs. MD simulations: 5.9–9.2 Å) induced progressively more distant apposition of the D1 and D2 lobes (Tables 1 and 2).
Table 2.
Distance measurements between the D1 and D2 lobes of GluK1–LBC.
| Ligand | Distance average (Å) from MD | Distance (Å) from X-ray |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Chain ID | Ae | Be | Ce | De | Ae | Be | Ce | De | |
| ATPOa | A | 11 | 10.5 | 12.5 | 12.5 | ||||
| (S)-glutamateb | A | 8.4 | 9 | 3 | 6.7 | 7.3 | 8.5 | 3 | 6.6 |
| B | 8.5 | 9 | 3.1 | 6.6 | 7.2 | 8.7 | 2.9 | 6.6 | |
| Domoatec | A | 9.5 | 9.3 | 3.9 | 6.8 | 6.9 | 9 | 6.7 | 9.2 |
| B | 9.6 | 9.8 | 3.6 | 7.5 | 6.9 | 9 | 6.7 | 9.2 | |
| MSVIII-19d | A | 9.1 | 9.1 | 3 | 6.7 | 7.6 | 8.9 | 3.3 | 6.6 |
| B | 8.4 | 9 | 3 | 6.7 | 7.5 | 8.7 | 2.9 | 6.6 | |
| apo | A | 7.7 | 9.6 | 3.3 | 7.1 | ||||
| B | 7.5 | 8.5 | 3.2 | 7 | |||||
| neoDH | A | 8.6 | 8.8 | 3.6 | 6.9 | ||||
| B | 8.2 | 8.9 | 3.2 | 6.7 | |||||
| 8-deoxy-neoDH | A | 8 | 9.1 | 3 | 6.6 | ||||
| B | 8 | 8.8 | 3.4 | 6.7 | |||||
| 8-epi-neoDH | A | 9.7 | 8.9 | 3 | 6.7 | ||||
| B | 8.5 | 9.1 | 3.1 | 7 | |||||
| 9-F-8-epi-neoDH | A | 9.3 | 8.9 | 3 | 6.6 | ||||
| B | 8.2 | 8.9 | 3.1 | 6.7 | |||||
| 9-deoxy-neoDH #1 | A | 8.5 | 8.7 | 4.2 | 7 | ||||
| B | 8.5 | 9.3 | 4.7 | 7 | |||||
| 9-deoxy-neoDH #2 | A | 8.6 | 8.9 | 3 | 6.6 | ||||
| B | 8.3 | 8.9 | 4.1 | 6.9 | |||||
| 9-epi-neoDH | A | 8.3 | 8.8 | 3.2 | 6.7 | ||||
| B | 9.5 | 9.1 | 3 | 6.7 | |||||
| 8,9-epi-neoDH | A | 9.3 | 9 | 3.1 | 6.9 | ||||
| B | 8.7 | 9.2 | 3.4 | 7 | |||||
| 4-epi-neoDH | A | 9 | 8.8 | 3.1 | 6.8 | ||||
| B | 8 | 8.8 | 3.1 | 6.8 | |||||
| 2,4-epi-neoDH | A | 9.6 | 8.7 | 5.9 | 7.5 | ||||
| B | 8.8 | 8.8 | 6.7 | 8.7 | |||||
| DNQX | A | 8.8 | 9.5 | 5.3 | 7.2 | ||||
| B | 11.8 | 11.2 | 9.2 | 11.2 | |||||
| LY466195 | A | 12 | 10.5 | 7.4 | 8.7 | ||||
| B | 10 | 9.2 | 5.5 | 6.6 | |||||
1VSO (Hald et al., 2007).
1YCJ (Naur et al., 2005).
2PBW (Hald et al., 2007).
GPB (Frydenvang et al., 2009).
A: Ser721Cα-Glu441Cα, B: Glu738Cα-Pro516O, C: Asp687N-Gly490O, D: Ser689Cα-Thr518Cα.
3.3. The (S)-glutamate backbone of DH derivatives is the basis for ligand-binding
As expected, the α groups (Fig. 2A: group a) and the γ-carboxylate group (Fig. 2A: group b) that comprise the (S)-glutamate backbone within DH-related ligands form similar interactions with KAR subunit LBCs as (S)-glutamate. These interactions are preserved for all DH-based high affinity ligands during the 14 ns MD simulations; minor exceptions were observed with weak affinity ligands and the antagonist ligand 2,4-epi-neoDH.
3.4. DH-based high affinity agonists
3.4.1. The DH-like stereochemistry at C9 position contributes to high affinity binding
DH analogs that act as high affinity agonists – neoDH, 8-deoxy-neoDH, 8-epi-neoDH, and 9-F-8-epi-neoDH – have (1) the same stereochemistry as DH at C2, C4, and C9 positions, and (2) possess a polar group at the C9 position (hydroxyl or fluorine moiety). They all bind with high affinity to GluK1 receptors (Table 1; Ki: 1.5–34 nM), and potently gate currents when applied to KARs in physiological experiments, while all those DH derivatives that have altered stereochemistry at C9 position show clearly weaker binding and reduced agonist effects (see Table 1) (Lash et al., 2008).
3.4.2. NeoDH
The C9 hydroxyl of quantum mechanically (QM) optimized neoDH donates an intramolecular hydrogen bond to the γ-carboxylate group, as is seen with DH in the crystal structure of GluK1–DH complex (Fig. 2B; PDB-code: 3GBA; Frydenvang et al., 2009). This intramolecular hydrogen bond persists throughout the 14 ns MD simulation of GluK1-LBC in complex with neoDH (Fig. 4A). Other interactions between neoDH and GluK1 also remain similar to that of DH; specifically, the C9 hydroxyl accepts a hydrogen bond from Glu738N (Fig. 4A, Table 1).
3.4.3. 8-deoxy-neoDH
The QM optimized 8-deoxy-neoDH lacks the intramolecular hydrogen bond between its C9 hydroxyl and γ-carboxylate group, distinguishing it from the parent molecule neoDH. The starting conformation acquired from docking studies was a critical factor in shaping the final receptor-bound conformation of 8-deoxy-neoDH; in fact, MD simulations produced two acceptable conformations: (a) one similar to that of DH and neoDH (data not shown), and (b) one in which the ligand is in extended conformation (with an axial C9 hydroxyl) and therefore does not form a direct hydrogen bond with Glu738N (Fig. 4B, Table 1). When C9 hydroxyl of 8-deoxy-neoDH is in the axial orientation, the ligand is close to the QM optimized conformation, the hydrogen bond network is maintained via water molecules, and the receptor cleft is firmly closed (Fig. 4B). Because both of these simulations are stable, the MD simulations alone cannot discriminate which of these binding conformations is biologically more relevant.
3.4.4. 8-epi-neoDH and 9-F-8-epi-neoDH
The altered stereochemistry at the C8 position of 8-epi-neoDH and 9-F-8-epi-neoDH constrains their ring systems to flat conformations, preventing the formation of the intramolecular hydrogen bond (Fig. 4C,D, Table 1). In addition to this stereochemical restriction, substitution of C9 hydroxyl with a fluorine (in 9-F-8-epi-neoDH) prevents formation of an intramolecular hydrogen bond, because the electronegative fluorine (Fig. 2A: group d) and the negatively charged γ-carboxylate group (Fig. 2A: group b) repel each other. Despite this planar conformation, in which the fluorine cannot interact with Glu738N, a water molecule (Fig. 4D: w3) bridges the hydrogen bond from Glu738N to oxygen at the six-membered ring (Fig. 4D, Table 1), as is seen with the C9 hydroxyl of 8-deoxy-neoDH (see above; Fig. 4B: w2). The C9 hydroxyl of 8-epi-neoDH is able to make this connection to Glu738N through a water bridge (Fig. 4C: w1 and w5). In the extended binding conformation, the hydrophobic contact surface area between the ring system of 9-F-8-epi-neoDH and the hydrophobic residues in the D1 face is larger (Fig. 4D) than when the ligand is in a bent conformation (compare to neoDH in Fig. 4A).
3.4.5. Structure–activity relationship of high affinity agonist DH derivatives
Pharmacologically, the relatively small difference in the GluK1 binding affinities of DH (Table 1: Ki: 0.5 nM), neoDH (Table 1: Ki: 7.7 nM), and 8-deoxy-neoDH (Table 1: Ki: 1.5 nM) must arise from their differences at the C8 positions (Table 1). Although docking provides reasonable binding conformations for these ligands, the MD simulations can account for even the smallest variances. The binding affinity of DH, which is highest of all KAR ligands examined to date, is a consequence of its C8 aminomethyl group that donates hydrogen bonds to the side chains of Glu441 and Ser741 and packs its methyl group against the hydrophobic face of D1, especially Pro516 (Fig. 2B). The substitution of this aminomethyl with a hydroxyl group (to produce neoDH) alters both the hydrophobic packing against the D1 face and the strength of the hydrogen bonding. The aminomethyl group can form an ionic interaction with the side chain of Glu738 and a hydrogen bond with the side chain of Ser741, as was suggested by our previous modeling results (Sanders et al., 2005, 2006), whereas the interaction with a hydroxyl group in neoDH is clearly weaker. In addition, the C8 hydroxyl of neoDH (Table 1) induces the side chain of Ser741 into donating a hydrogen bond; this increases the intramolecular energy of GluK1, because the hydroxyl group of Ser741 prefers to donate a hydrogen bond to the surrounding amino acids directly or via a water molecule. MD simulations also suggest that the higher GluK1 binding affinity exhibited by 8-deoxy-neoDH relative to neoDH is attributable to more an extensive packing surface against the hydrophobic face of D1 (note the position of the pyran ring in Fig. 4A vs. B).
3.4.6. Stereochemistry at C8 position affects the hydrofurapyran ring conformation
Similar to 9-F-8-epi-neoDH, the hydroxyl at C8 position of 8-epi-neoDH (Table 1) favors the axial orientation over the equatorial. Accordingly, the hydrofurapyran ring system is flatter than neoDH, for example, suggesting that the stereochemistry at the C8 position impacts the overall structural characteristics of DH analogs (Table 1). The QM optimized geometries of 8-deoxy-neoDH and 8-epi-neoDH are very much alike; however, 8-deoxy-neoDH (Ki: 1.5 nM) binds with clearly higher affinity than 8-epi-neoDH (Ki: 34 nM), and 9-F-8-epi-neoDH (Ki: 28 nM) to GluK1. For this phenomenon there are two rational explanations: (1) the 8-deoxy-neoDH lacks the hydroxyl at C8 position, and, accordingly, the ring conformation adjusts more freely than in C8 epimer analogs, and (2) the packing of hydrophobic ring system against the hydrophobic D1 face formed by Glu441, Tyr489, and Pro516 (shown with transparent surfaces in Fig. 4), is more efficient without an obtrusive, polar C8 hydroxyl.
3.4.7. MD simulations predict high affinity agonism
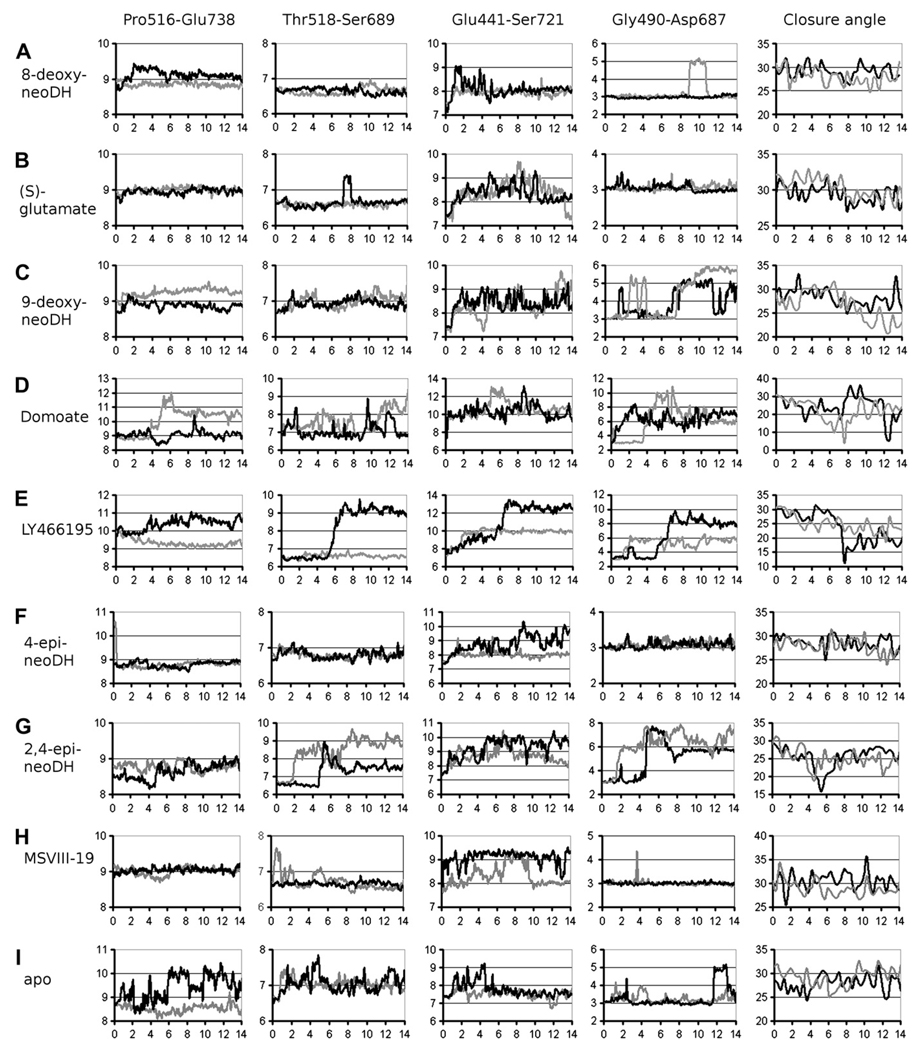
We tested if the stability of the ligand–receptor complex correlates with pharmacological activity using 14 ns MD simulations. The D1–D2 interface distances of GluK1–LBCs complexed with high affinity ligands were relatively invariant (for 8-deoxy-neoDH see Fig. 5A). The interactions linking the D1 and D2 lobes, associated interdomain distances derived from MD simulations, were similar to those in crystal structure of GluK1–LBC with bound (S)-glutamate (Naur et al., 2005) (Fig. 5B, Table 2). Specific examples of this close correlation include the two interdomain hydrogen bonds between atom pairs Glu441Cα-Ser721Cα and Gly490O-Asp687N (see Fig. 3 for reference, also Fig. 5A,B and Table 2) and the distances between the atom pairs Pro516O-Glu738Cα and Thr518Cα-Ser689Cα, which are located close to the hinge region in the ligand-binding cavity and are not hydrogen bonded (see Fig. 3 and Table 2 for reference, also Fig. 5A,B). Accordingly, these studies support the hypothesis that binding of DH-related high affinity agonists efficiently stabilizes the ligand-binding domain of GluK1 in a closed state conformation.
Fig. 5.
The distances (four leftmost panels) and domain closure angles (the rightmost panel) showing the D1–D2 separation and opening of the closed GluK1–LBC in complex with (A) 8-deoxy-neoDH, (B) (S)-glutamate, (C) 9-deoxy-neoDH, (D) domoate, (E) LY466195, (F) 4-epi-neoDH, (G) 2,4-epi-neoDH, (H) MSVIII-19, and (I) without a bound ligand. On the x axis is presented the timescale (ns) and on the y axis the distance (Å) as 100-moving average or receptor cleft closure angle degree. See Fig. 3 for details on the atoms used in the distance measurements. The measurements for A chain of the GluK1–LBC dimer are shown with darker color than for B chain.
3.5. DH analogs with weak agonist activity
3.5.1. The modified C9 position reduces the binding affinity for GluK1
In contrast to ligands that potently induce channel currents, those that gate only modest or marginally detectable currents have an alteration at their C9 position (Table 1; e.g., 9-deoxy-neoDH, 9-epi-neoDH, and 8,9-epi-neoDH). As a result, none of these ligands are able to form the intramolecular hydrogen bond with their γ-carboxylate group observed with high affinity agonists such as DH (Fig. 2B).
3.5.2. 9-deoxy-neoDH
To compare how molecules with lower apparent efficacy (at concentrations in the range of 10–50 µM) differ in their molecular interactions with the ligand-binding domain, we again carried out MD simulations, focusing in particular on 9-deoxy-neoDH (Fig. 6A). This analysis suggested that 9-deoxy-neoDH binds in a conformation highly similar to that of neoDH (Fig. 4A): (1) The C8 hydroxyl forms hydrogen bonds with the side chains of Ser741 and Glu738 (Table 1; Fig. 6A), and (2) the interdomain hydrogen bond is maintained between the side chains of Glu441 and Ser721 (Table 1). Because these stabilizing features are present in the simulation, the weaker binding affinity (9-deoxy-neoDH Ki: 169 nM vs. neoDH Ki: 7.7 nM) and very modest gating activity can be directly linked to the absence of C9 hydroxyl and its interaction with Glu738N. The MD simulation of GluK1–LBC with bound 9-deoxy-neoDH (Fig. 5C) showed a gradual opening of receptor cleft near the α groups (Fig. 2A: groups a), and consequently the distance between the D1 and D2 lobes increased (Fig. 5C, Table 2), in contrast to the marked stability of high affinity agonist complexes (e.g. with 8-deoxy-neoDH, Fig. 5A). An MD simulation initiated with a different 9-deoxy-neoDH pose that lacked the canonical interactions of (S)-glutamate resulted in an identical final conformation and degree of cleft opening, validating the fidelity of the simulation and independence from initial docking configuration (data not shown). The resulting receptor conformation closely resembled that of the iGluR–LBC crystal structures of two partial agonist ligand complexes: GluA2–kainate (Armstrong et al., 1998) and GluK1–domoate (Hald et al., 2007) (Fig. 6B). In addition, when compared to the full-length simulations of GluK1–LBC in complex with partial agonist domoate (Figs. 1 and 5D, Table 2) and the antagonist LY466195 (3S,4aR,6S,8aR)-6-[[(2S)-2-carboxy-4,4-difluoro-1-pyrrolidinyl]-methyl]decahydro-3-isoquinolinecarboxylic acid) (Weiss et al., 2006) (Figs. 1 and 5E, Table 2), it is evident that 9-deoxy-neoDH evokes similar trends of motion as domoate but of lower magnitude than LY466195. These MD simulations clarify the mechanistic basis for the pharmacological actions of 9-deoxy-neoDH and suggest that it likely represents a partial agonist for GluK1 (Lash et al., 2008).
3.5.3. 9-epi-neoDH
8-epi-neoDH and 9-epi-neoDH complexed with GluK1–LBC produced similar conformations in MD simulations (Figs. 4C and 6C), underscoring the considerable plasticity in the binding modes of these DH-based ligands. Their C8 hydroxyls (Table 1) assumed the same position in 3D space and hydrogen bond to the side chain of Ser741 despite the reversal in orientation of the C9 hydroxyls. Thus, the 10-fold difference in binding affinity (8-epi-neoDH: Ki: 34 nM vs. 9-epi-neoDH: Ki: 292 nM) likely arises from the inability of the C9 hydroxyl of 9-epi-neoDH to hydrogen bond with Glu738N (Fig. 6C). The binding of both 8-epi-neoDH and 9-epi-neoDH permit hydrogen bonding between the side chains of Glu441 and Ser721, which in turn produce D1–D2 interface closure associated with agonism (Armstrong and Gouaux, 2000). In conclusion, despite the similar binding conformations, the opposite stereochemistry at C9 hydroxyls underlies the greater efficacy of 8-epi-neoDH for GluK1 relative to 9-epi-neoDH.
3.5.4. 8,9-epi-neoDH
Simultaneous epimerization of both C8 and C9 groups in neoDH nearly eliminates completely the binding for GluK1 (Sanders et al., 2005) (Table 1). MD simulations suggest that this loss of affinity observed with 8,9-epi-neoDH (Ki: 48 µM) occurs because the C9 hydroxyl fails to bond intramolecularly with γ-carboxylate group (compare to neoDH in Fig. 4A, for example) or with the receptor subunit (Fig. 6D). In addition, the conventional alignment of the ring system in 8,9-epi-neoDH against the hydrophobic face of D1 prevents strong bonding between the γ-carboxylate group and Thr690 (see DH in Fig. 2B). The C8 hydroxyl of 8,9-epi-neoDH hydrogen bonds simultaneously with the side chains of Ser741 and Glu738, and thus, the D1 face is linked to the D2 face only via the ligand rather than through direct interdomain interactions. This weakened linkage likely contributes modestly to the low apparent affinity of this molecule for GluK1. The distances between selected amino acid pairs (Fig. 3) for GluK1-8,9-epi-neoDH complex are shown in Supplementary Fig. S3.
3.5.5. 4-epi-neoDH
The final weak agonist we examined, 4-epi-neoDH (Fig. 6E, Table 1: Ki: 559 nM), has unfavorable ring system orientation (Fig. 2A: group e) relative to the D1 hydrophobic face. The D1 packing is compensated with less favorable positioning against the hydrophobic residues Val685 and Met737 on the D2 face (shown with transparent surfaces in Fig. 6E). Because of its altered stereochemistry at the C4 position, 4-epi-neoDH is unable to interact with the D2 face of the ligand-binding pocket directly via the C8 and C9 hydroxyls. However, the side chain of Ser741 is connected to the C9 hydroxyl of 4-epi-neoDH via a mediating water molecule (w2 in Fig. 6E). Note also that both the C8 and C9 hydroxyls of 4-epi-neoDH bind to Glu441, which stabilizes the ligand–receptor complex but prevents formation of a hydrogen bond between Glu441 and Ser721 across the D1–D2 interface. Thus, the weak binding affinity of 4-epi-neoDH and its instability inside the binding pocket (Fig. 5F) likely underlie the weak gating of ion channel currents followed by long-lasting desensitization (Lash et al., 2008). However, even though the D1–D2 linkage is disturbed between the side chains of Glu441 and Ser721, the domain–domain interaction exists between Gly490O and Asp687N throughout the whole 4-epi-neoDH-GluK1 simulation (Fig. 5F).
3.6. Two routes to receptor inactivation: 2,4-epi-neoDH and MSVIII-19
3.6.1. 2,4-epi-neoDH
2,4-epi-neoDH (Fig. 7A) adopts a similar binding conformation against the D2 face as 4-epi-neoDH (Fig. 6E); however, 2,4-epi-neoDH is an antagonist rather than a weak agonist like 4-epi-neoDH (Lash et al., 2008). In the MD simulation of GluK1–LBC in complex with 2,4-epi-neoDH, the stabilizing interactions between the D1 andD2lobes are lost, not only between the side chains of Glu441 and Ser721, as seen for 4-epi-neoDH, but also between Gly490O and Asp687N near the α groups of the ligand (Figs. 5G and 7A–B, Table 1). The latter divergence therefore must result from the modification of ligand stereochemistry at C2 position. Accordingly, the receptor cleft opening was slightly larger than was seen with the partial agonist domoate but nonetheless remained smaller than that induced by the antagonist LY466195 (Figs. 5 and 7B). The loss of these D1–D2 interactions with GluK1-2,4-epi–neoDH complex (Fig. 7A) is likely the mechanistic basis for its antagonist properties in pharmacological experiments (Lash et al., 2008). To ensure that the opening of the receptor cleft was reproducible, 2,4-epi-neoDH-GluK1 simulation was repeated with congruent results (data not shown).
As with 4-epi-neoDH (Fig. 5E), the C8 hydroxyl of 2,4-epi-neoDH hydrogen bonds with the side chain of Glu441 (Fig. 7A). When bound to GluK1–LBC, the C9 hydroxyl of 2,4-epi-neoDH makes an intramolecular hydrogen bond with the α-amine group (shown with yellow dotted line in Fig. 7A), further stabilizing its twisted ring system conformation. The lower binding affinity of 2,4-epi-neoDH (Table 1: Ki: 2.4 µM) in comparison to 4-epi-neoDH (Table 1: Ki: 556 nM), can be addressed by two observed differences: (1) the C9 hydroxyl of 2,4-epi-neoDH is unable to hydrogen bond to the side chain of Glu441, and (2) the loss of D1–D2 lobe interaction.
We tested if the simulation approach reproduces the open-cleft binding configuration of a known competitive antagonist, LY466195 (Fig. 1), which is structurally unrelated to the marine toxin DH. LY466195 is quite bulky ligand compared to DH-related derivatives, and accordingly, its binding conformation was far from optimal at the beginning of the MD simulation. However, during the MD simulation, the receptor cleft opened gradually, and the end-result closely resembled the crystallized complex of GluK1–LY466195 (Figs. 5E and 7B; PDB-code: 2QS4). This observation suggests that open-cleft conformations generated by rigorous MD simulations with DH-related antagonists such as 2,4-epi-neoDH, may reliably report their binding configuration.
3.6.2. MSVIII-19
MSVIII-19 (8,9-dideoxy-neoDH) is an unusual compound, in that it primarily acts as a functional KAR antagonist with very little apparent efficacy (Sanders et al., 2005), despite having been crystallized in a fully closed GluK1–LBC complex not substantially different that observed with (S)-glutamate (PDB-code: 1YCJ; Naur et al., 2005; Frydenvang et al., 2009). In MD simulations, the binding conformation of MSVIII-19 is based on the canonical interactions shared with (S)-glutamate and hydrophobic interactions with the D1 face of the GluK1–LBC, as the ligand does not have polar C8 and C9 groups (Table 1, Fig. 5H). These interactions are sufficient to produce relatively high binding affinity of MSVIII-19 for GluK1 (Table 1: Ki: 128 nM).
The binding conformation in our simulation was highly similar to that obtained in the recently published crystal structure of GluK1 with bound MSVIII-19 (Frydenvang et al., 2009). We proposed previously that this compound predominantly induces a desensitized state of the GluK1 receptor without prior activation of the receptor (Frydenvang et al., 2009), and that fully closed LBCs could therefore represent the structural correlate for at least two distinct functional states (activated and desensitized). The static crystal structures unfortunately did not yield insight into potential structural differences that might underlie the markedly divergent pharmacological properties of (S)-glutamate, DH, and MSVIII-19. In contrast, dynamic variations were observed in MD simulations. Some interdomain distances, such as the hydrogen bond between Gly490O and Asp687N, were maintained throughout the 14 ns MD simulations for both GluK1–MSVIII-19 and GluK1–(S)-glutamate complexes (Fig. 5). In contrast, the hydrogen bond between Glu441 and Ser721 is present throughout the simulation with (S)-glutamate but is rapidly lost with MSVIII-19 (Fig. 5H, Table 1). In chain B, the side chains of Glu441 and Ser721 remained hydrogen bonded for most of the simulation time (Fig. 7C upper panel), in a conformation similar to that of the crystal structure of GluK1–MSVIII-19 (Frydenvang et al., 2009). This hydrogen bond was absent from the receptor–ligand complex in the A chain (Fig. 7C lower panel), however, and this conformation better illustrates the instability present at the receptor cleft with bound MSVIII-19 (Fig. 5H). In addition, the positions of water molecules and amino acid side chains, e.g., the freely rotating side chain of Ser741, indicate instability near the C8 and C9 positions (Table 1). Whether this instability and the Glu441Cα-Ser721Cα distance fluctuation (especially notable in the A chain) is the reason for the unusual pharmacological activity of MSVIII-19 is yet to be determined. Interestingly, this conformational change closely resembles the conformational change seen in the crystal structures of partial agonist ligand kainate, both with GluA2 (PDB-code: 1FTK; Armstrong and Gouaux, 2000) and GluK2 (PDB-code: 1TT1; Mayer, 2005) (Fig. 7D). It is noteworthy that the GluK1 receptor can be inactivated by at least two different physical mechanisms: 2,4-epi-neoDH (and LY466195) force the GluK1–LBC into an open configuration (“conventional antagonism”), whereas MSVIII-19 induces closure of the LBC either by shifting the receptor directly into desensitized state without detectable channel gating or by failure to stabilize the receptor in a fully closed, activated conformation (Fig. 5G,H).
3.7. MD simulation of closed GluK1–LBC without a bound ligand
In these simulations, the DH-related ligands were inserted into a closed GluK1–LBC at the beginning. To test the influence of this initial protein conformation in the final binding modes, we measured the distance changes occurring in an MD simulation without a bound ligand (apo state). The distances measured in the GluK1-apo structure oscillated close to the initial closed conformation despite the absence of a ligand to stabilize the D1–D2 interface interactions (Fig. 5I). In the 4–14 ns time period the most noteworthy fluctuation with GluK1-apo was seen between the atom pairs of Pro516O-Glu738Cα and Thr518Cα-Ser689Cα (Fig. 5I), indicating that the “bottom” of the D1–D2 interface is unstable without a bound ligand. Opening of receptor cleft was not observed in the MD simulation, however, likely due the stabilizing hydrogen bonds between D1 and D2. This outcome is encouraging as it indicates that the force field based method produces stable results and the reported LBC opening in other MD simulations results from ligand binding.
3.8. Determination of the degree of the GluK1–LBC closure
The general tendencies of angle measurements (Supplementary Table 1) performed with HINGEFIND algorithm (Wriggers and Schulten, 1997) support the results of the distance measurements (Table 2). The GluK1–LBC angles with bound agonists, e.g., 8-deoxy-neoDH and (S)-glutamate, are equivalent to those in the apo state (Fig. 5A,B,I, respectively). In the 4–14 ns time period, the DH analogs induced ~29° LBC closure, similar to (S)-glutamate and consistent with their agonist activity (Table 1). Closure angles of GluK1-LBC protomers within dimer complexes generally followed similar trends while exhibiting some variance (on average 1–2° apart) and non-concerted movements. This also is apparent in distance measurements (Fig. 5). It was clear that 2,4-epi-neoDH and 9-deoxy-neoDH presented the smallest closure angles of the DH analog set, thereby generating the greatest openings of their respective GluK1–LBCs. 9-deoxy-neoDH promoted gradual opening of the GluK1–LBC during the MD simulations and 2,4-epi-neoDH induced fast and extensive opening followed by a small degree of re-closure. As mentioned previously, and in contrast to typical antagonists such as LY466195 (Fig. 5E), the functional antagonist MSVIII-19 (Fig. 5H) induced domain closure to the same degree as efficacious agonists (Fig. 5A,B).
4. Discussion
In this study, we determined ligand-binding modes for wide selection of DH-derived ligands and correlated their relative receptor cleft closure with ligand structure–function relationships and pharmacological activity. The MD simulations generated reproducible results that were congruent with the experimental measurements; that is, receptor cleft opening occurred to differing degrees consistent with the known pharmacological properties of a divergent series of ligands. As our ligand set does not contain molecules that would exert counter-productive or extensive forces on the D1–D2 interface, we were able to use the closed LBC conformation in all simulations. We also considered use of an open receptor model, such as the crystal structure of GluK1–LBC with a bound antagonist (Hald et al., 2007; Mayer et al., 2006), because initiation of MD simulation from this conformation could be considered more biologically relevant. However, iGluR cleft closure in a classical MD simulation has been achieved only once (and in that case without a bound ligand) (Bjerrum and Biggin, 2008). Application of an umbrella sampling technique also induced closure of an agonist-bound complex (Lau and Roux, 2007). Despite numerous attempts, we did not observe full receptor closure from a completely open (antagonist-bound) conformation in a free MD simulation using explicit water molecules, with or without an agonist, suggesting that the routine use of open conformations for these types of simulations is impractical. Conversely, stabilizing interactions between the D1 and D2 lobes in a closed conformation could potentially prevent receptor cleft opening with bound ligands, as was seen in the MD simulation of a ligand-free, closed GluK1–LBC, but this turned out not to be the case. The crystal structure of the recently reported GluK1-partial agonist domoate complex (Hald et al., 2007) could also be used in the study of slightly bulkier antagonists that cannot be accommodated into a closed GluK1–LBC.
The experimental characterization of KAR-specific ligands into distinct pharmacological groups is generally complicated by rapid desensitization, which can reduce receptor activation or agonist efficacy. Cleft closure linked to receptor activity can be determined experimentally using fluorescence resonance energy transfer-based assay (Ramanoudjame et al., 2006), for example. MD simulations additionally provide an atom-level view of the ligand–receptor dynamics. For example, our simulations indicate that partial agonism by 9-deoxy-neoDH is not a function of steric occlusion of receptor cleft closure but rather is caused by subtle positioning or interactions within the binding pocket. Thus, it is plausible that during the binding process of 9-deoxy-neoDH the GluK1–LBC rapidly achieves full closure, however, this closed conformation cannot be stabilized (due to the electrostatic repulsions, for example), and instead the receptor cleft opens slightly to accommodate the ligand. The interpretation of such activity in physiological experiments can be difficult, and we suggest that our modeling protocol can provide extra insight to distinguish partial agonists from agonists and antagonists or at least provide initial hypotheses regarding the pharmacological properties of molecules with uncharacterized activities.
MSVIII-19 constituted a clear exception to the generally robust correlation between predicted ligand-binding domain closure and ligand activity both in our simulations as well as in the crystallization study (Frydenvang et al., 2009). Additionally, a recent docking simulation suggested that some partial agonists would induce full closure of the LBC on KARs (Fay et al., 2009). In contrast to the docking simulation (Fay et al., 2009) and crystallization (Frydenvang et al., 2009) studies, the LBC is considered as a flexible entity in solution in the MD simulations. Thus, we analyze continuous receptor–ligand complex movements instead of focusing on a limited number of static conformations, which likely represent a subset of possible ligand-binding modes. MD simulation of GluK1–MSVIII-19 suggests that the receptor cleft experiences differential opening along its face, with hydrogen bonds maintained between Gly490O and Asp687N but disrupted between the side chains of Glu441 and Ser741. The receptor cleft then remains open at the latter site for the majority of the simulation. This cleft opening was not observed in the crystal structure of GluK1–MSVIII-19 (Frydenvang et al., 2009), which could be explained by the tendency for crystallization to favor extreme conformations that do not encompass the multitude of movements possible in solution. This is a significant problem when considering the structure–activity relationship of MSVIII-19, because it cannot sterically hinder the receptor LBC closure through a similar mechanism as kainate or domoate, which have structural elements that occupy interdomain space. We propose, therefore, that partial agonism (or functional antagonism) could result from a slightly open conformation at one interdomain site (Glu441Cα-Ser721Cα) while remaining stably closed at another (Gly490O-Asp687N). In conclusion, the inability of MSVIII-19 to stabilize the essential interdomain interactions inside the ligand-binding pocket could be the cause behind the reported antagonist-like pharmacological activity.
There also is instability in the closed, ligand free apo-structure in the MD simulation manifested as minor movements close to the hinge area (based on distances of Pro516O-Glu738Cα and Thr518Cα-Ser689Cα) (Fig. 5I). This suggests that any lability in the cavity could be sufficient to keep the receptor inactive, even if the apo-iGluR was able to close in situ, and is thus in good agreement with the earlier MD simulations of GluA2 (Arinaminpathy et al., 2002).
5. Conclusions
The lengthy timescale (14 ns) and the number of our MD simulations ensure that we can predict reliably the different binding modes and explain the measured binding affinities and activities of the DH-based analogs interacting with GluK1 (Lash et al., 2008) (Table 1). Moreover, comparisons of subtle structural differences with the experimentally measured activities yield a simple structure–activity relationship. The DH analogs mediate the D1–D2 interactions via their C8 and C9 hydroxyls, the latter position being more important for high affinity agonism (Table 1), and if this linkage is non-existent or weak, no activation results or the channel gating is poor, respectively. Changes in several interdomain distances and D1-hinge-D2 angles in MD simulations correlate with the pharmacological activity of most of the DH-based analogs. Hydrogen bonds that connect the D1 and D2 lobes, between Gly490O and Asp687N and the side chains of Glu441 and Ser721 (Table 1), are disrupted or absent in the case of conventional competitive antagonism such as that observed with 2,4-epi-neoDH, which also rapidly opens the closed GluK1–LBC. The binding of the partial agonist 9-deoxy-neoDH disrupts only the Gly490O-Asp687N hydrogen bond, and therefore the receptor cleft opens gradually and only partially. Both the MD simulation and crystallization suggest that the functional antagonist MSVIII-19 adopts an agonist-like binding mode by inducing domain closure of GluK1–LBC. However, MSVIII-19 binding does not produce stable interdomain contacts required for either strong activation or long-lasting desensitization of GluK1, which likely contributes to its functional antagonism. Accordingly, it is possible that desensitization and/or partial agonism can be associated with unstable interdomain distance close to the Glu441Cα-Ser721Cα atom pair, as is the case with Gly490O-Asp687N on the opposite site of the binding pocket. This exhaustive analysis of the binding of DH analogs into the closed GluK1–LBC should prove useful for the development of novel ligands and future pharmacological research. Because of the high sequence similarity inside the iGluR family, the data collected on GluK1 pharmacology and dynamics can benefit the research involving other KARs and iGluRs as well.
Supplementary Material
Acknowledgements
The study was funded by the National Graduate School in Informational and Structural Biology (PAP), grant R01 NS44322 from the National Institutes of Health (GTS), and the Sigrid Jusélius Foundation (OTP). CSC, the Finnish IT Center for Science (Espoo, Finland), is acknowledged for the access to the computational resources (project: jyy2516).
Footnotes
Appendix. Supplementary information
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.neuropharm.2009.08.019.
References
- Alt A, Weiss B, Ogden AM, Knauss JL, Oler J, Ho K, Large TH, Bleakman D. Pharmacological characterization of glutamatergic agonists and antagonists at recombinant human homomeric and heteromeric kainate receptors in vitro. Neuropharmacology. 2004;46:793–806. doi: 10.1016/j.neuropharm.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Arinaminpathy Y, Sansom MSP, Biggin PC. Molecular dynamics simulations of the ligand-binding domain of the ionotropic glutamate receptor glur2. Biophys. J. 2002;82:676–683. doi: 10.1016/S0006-3495(02)75430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an ampa-sensitive glutamate receptor: crystal structures of the glur2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Mayer M, Gouaux E. Tuning activation of the AMPA-sensitive GluR2 ion channel by genetic adjustment of agonist-induced conformational changes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5736–5741. doi: 10.1073/pnas.1037393100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton ME, Peters SC, Shannon HE. Comparison of the effect of glutamate receptor modulators in the 6 hz and maximal electroshock seizure models. Epilepsy Res. 2003;56:17–26. doi: 10.1016/j.eplepsyres.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Bayly C, Cieplak P, Cornell W, Kollman P. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerrum EJ, Biggin PC. Rigid body essential x-ray crystallography: distinguishing the bend and twist of glutamate receptor ligand binding domains. Proteins. 2008;72:434–446. doi: 10.1002/prot.21941. [DOI] [PubMed] [Google Scholar]
- Christensen JK, Varming T, Ahring PK, Jørgensen TD, NielsenØ E. In vitro characterization of 5-carboxyl-2,4-di-benzamidobenzoic acid (ns3763), a noncompetitive antagonist of gluk5 receptors. J. Pharmacol. Exp. Ther. 2004;309:1003–1010. doi: 10.1124/jpet.103.062794. [DOI] [PubMed] [Google Scholar]
- Cieplak P, Cornell W, Bayly C, Kollman P. Application of the multimolecule and multiconformational resp methodology to biopolymers-charge derivation for dna, rna, and proteins. J. Comput. Chem. 1995;16:1357–1377. [Google Scholar]
- Collingridge GL, Olsen RW, Peters J, Spedding M. A nomenclature for ligand-gated ion channels. Neuropharmacology. 2009;56:2–5. doi: 10.1016/j.neuropharm.2008.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell W, Cieplak P, Bayly C, Kollman P. Application of the RESP charges to calculate conformational energies, hydrogen bond energies, and free energies of solvation. J. Am. Chem. Soc. 1993;115:9620–9631. [Google Scholar]
- Darden T, York Y, Pedersen L. Particle mesh ewald: an w log(n) method for ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- Dominguez E, Iyengar S, Shannon HE, Bleakman D, Alt A, Arnold BM, Bell MG, Bleisch TJ, Buckmaster JL, Castano AM, Del Prado M, Escribano A, Filla SA, Ho KH, Hudziak KJ, Jones CK, Martinez-Perez JA, Mateo A, Mathes BM, Mattiuz EL, Ogden AML, Simmons RMA, Stack DR, Stratford RE, Winter MA, Wu Z, Ornstein PL. Two pro-drugs of potent and selective glur5 kainate receptor antagonists actives in three animal models of pain. J. Med. Chem. 2005;48:4200–4203. doi: 10.1021/jm0491952. [DOI] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML. The origin of the hydration interaction of lipid bilayers from md simulation of dipalmitoylphosphatidylcholine membranes in gel and liquid crystalline phases. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- Fay AL, Corbeil C, Brown P, Moitessier N, Bowie D. Functional characterisation and in silico docking of full and partial gluk2 kainate receptor agonists. Mol. Pharmacol. 2009 doi: 10.1124/mol.108.054254. doi:10.1124/mol.108.054254. [DOI] [PubMed] [Google Scholar]
- Feller S, Zhang Y, Pastor R, Brooks B. Constant pressure molecular dynamics simulation: the langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- Filla SA, Winter MA, Johnson KW, Bleakman D, Bell MG, Bleisch TJ, Castaño AM, Clemens-Smith A, del Prado M, Dieckman DK, Dominguez E, Escribano A, Ho KH, Hudziak KJ, Katofiasc MA, Martinez-Perez JA, Mateo A, Mathes BM, Mattiuz EL, Ogden AML, Phebus LA, Stack DR, Stratford RE, Ornstein PL. Ethyl (3s,4ar,6s,8ar)-6-(4-ethoxycar-bonylimidazol-1-ylmethyl)decahydroiso-quinoline-3-carboxylic ester: a prodrug of a glur5 kainate receptor antagonist active in two animal models of acute migraine. J. Med. Chem. 2002;45:4383–4386. doi: 10.1021/jm025548q. [DOI] [PubMed] [Google Scholar]
- Frydenvang K, Lash LL, Naur P, Postila PA, Pickering DS, Smith CM, Gajhede M, Sasaki M, Sakai R, Pentikäinen OT, Swanson GT, Kastrup JS. Full domain closure of the ligand-binding core of the ionotropic glutamate receptor iglur5 induced by the high-affinity agonist dysiherbaine and the functional antagonist msviii-19. J. Biol. Chem. 2009;284:14219–14229. doi: 10.1074/jbc.M808547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilron I, Max MB, Lee G, Booher SL, Sang CN, Chappell AS, Dionne RA. Effects of the 2-amino-3-hydroxy-5-methyl-4-isoxazole-proprionic acid/kainate antagonist ly293558 on spontaneous and evoked postoperative pain. Clin. Pharmacol. Ther. 2000;68:320–327. doi: 10.1067/mcp.2000.108677. [DOI] [PubMed] [Google Scholar]
- Hald H, Naur P, Pickering DS, Sprogøe D, Madsen U, Timmermann DB, Ahring PK, Liljefors T, Schousboe A, Egebjerg J, Gajhede M, Kastrup JS. Partial agonism and antagonism of the ionotropic glutamate receptor iglur5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid. J. Biol. Chem. 2007;282:25726–25736. doi: 10.1074/jbc.M700137200. [DOI] [PubMed] [Google Scholar]
- Hollmann M, Heinemann S. Cloned glutamate receptors. Annu. Rev. Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- Inanobe A, Furukawa H, Gouaux E. Mechanism of partial agonist action at the nr1 subunit of nmda receptors. Neuron. 2005;47:71–84. doi: 10.1016/j.neuron.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat. Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 1995;245:43–53. doi: 10.1016/s0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Kraulis P. Molscript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystal. 1991;24:946–950. [Google Scholar]
- Lash LL, Sanders JM, Akiyama N, Shoji M, Postila P, Pentikäinen OT, Sasaki M, Sakai R, Swanson GT. Novel analogs and stereoisomers of the marine toxin neodysiherbaine with specificity for kainate receptors. J. Pharmacol. Exp. Ther. 2008;324:484–496. doi: 10.1124/jpet.107.129890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AY, Roux B. The free energy landscapes governing conformational changes in a glutamate receptor ligand-binding domain. Structure. 2007;15:1203–1214. doi: 10.1016/j.str.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen JV, Still D, Rantanen V, Ekholm J, Björklund D, Iftikhar Z, Huhtala M, Repo S, Jussila A, Jaakkola J, Pentikäinen O, Nyrönen T, Salminen T, Gyllenberg M, Johnson MS. Bodil: a molecular modeling environment for structure-function analysis and drug design. J. Comput. Aided Mol. Des. 2004;18:401–419. doi: 10.1007/s10822-004-3752-4. [DOI] [PubMed] [Google Scholar]
- Mayer ML. Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Ghosal A, Dolman NP, Jane DE. Crystal structures of the kainate receptor glur5 ligand binding core dimer with novel glur5-selective antagonists. J. Neurosci. 2006;26:2852–2861. doi: 10.1523/JNEUROSCI.0123-06.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ. Raster3d: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Naur P, Vestergaard B, Skov LK, Egebjerg J, Gajhede M, Kastrup JS. Crystal structure of the kainate receptor glur5 ligand-binding core in complex with (s)-glutamate. FEBS Lett. 2005;579:1154–1160. doi: 10.1016/j.febslet.2005.01.012. [DOI] [PubMed] [Google Scholar]
- O’Neill MJ, Bond A, Ornstein PL, Ward MA, Hicks CA, Hoo K, Bleakman D, Lodge D. Decahydroisoquinolines: novel competitive ampa/kainate antagonists with neuroprotective effects in global cerebral ischaemia. Neuropharmacology. 1998;37:1211–1222. doi: 10.1016/s0028-3908(98)00134-8. [DOI] [PubMed] [Google Scholar]
- Pentikäinen OT, Settimo L, Keinänen K, Johnson MS. Selective agonist binding of (s)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid (ampa) and 2s-(2alpha,3beta,4beta)-2-carboxy-4-(1-methylethenyl)-3-pyrrolidineacetic acid (kainate) receptors: a molecular modeling study. Biochem. Pharmacol. 2003;66:2413–2425. doi: 10.1016/j.bcp.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Pentikäinen U, Settimo L, Johnson MS, Pentikäinen OT. Subtype selectivity and flexibility of ionotropic glutamate receptors upon antagonist ligand binding. Org. Biomol. Chem. 2006;4:1058–1070. doi: 10.1039/b515111b. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with namd. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro P, Mulle C. Kainate receptors. Cell Tissue Res. 2006;326:457–482. doi: 10.1007/s00441-006-0265-6. [DOI] [PubMed] [Google Scholar]
- Ramanoudjame G, Du M, Mankiewicz KA, Jayaraman V. Allosteric mechanism in ampa receptors: a fret-based investigation of conformational changes. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10473–10478. doi: 10.1073/pnas.0603225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert J, Ciccotti G, Berendsen H. Numerical integration of the cartesian equations of proteins and nucleic acids. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- Safferling M, Tichelaar W, Kümmerle G, Jouppila A, Kuusinen A, Keinänen K, Madden DR. First images of a glutamate receptor ion channel: oligomeric state and molecular dimensions of glurb homomers. Biochemistry. 2001;40:13948–13953. doi: 10.1021/bi011143g. [DOI] [PubMed] [Google Scholar]
- Sagui C, Darden TA. Molecular dynamics simulations of biomolecules: longrange electrostatic effects. Annu. Rev. Biophys. Biomol. Struct. 1999;28:155–179. doi: 10.1146/annurev.biophys.28.1.155. [DOI] [PubMed] [Google Scholar]
- Sakai R, Swanson GT, Shimamoto K, Green T, Contractor A, Ghetti A, Tamura-Horikawa Y, Oiwa C, Kamiya H. Pharmacological properties of the potent epileptogenic amino acid dysiherbaine, a novel glutamate receptor agonist isolated from the marine sponge dysidea herbacea. J. Pharmacol. Exp. Ther. 2001;296:650–658. [PubMed] [Google Scholar]
- Sanders JM, Ito K, Settimo L, Pentikäinen OT, Shoji M, Sasaki M, Johnson MS, Sakai R, Swanson GT. Divergent pharmacological activity of novel marine-derived excitatory amino acids on glutamate receptors. J. Pharmacol. Exp. Ther. 2005;314:1068–1078. doi: 10.1124/jpet.105.086389. [DOI] [PubMed] [Google Scholar]
- Sanders JM, Pentikäinen OT, Settimo L, Pentikäinen U, Shoji M, Sasaki M, Sakai R, Johnson MS, Swanson GT. Determination of binding site residues responsible for the subunit selectivity of novel marine-derived compounds on kainate receptors. Mol. Pharmacol. 2006;69:1849–1860. doi: 10.1124/mol.106.022772. [DOI] [PubMed] [Google Scholar]
- Sang CN, Hostetter MP, Gracely RH, Chappell AS, Schoepp DD, Lee G, Whitcup S, Caruso R, Max MB. Ampa/kainate antagonist ly293558 reduces capsaicin-evoked hyperalgesia but not pain in normal skin in humans. Anesthesiology. 1998;89:1060–1067. doi: 10.1097/00000542-199811000-00005. [DOI] [PubMed] [Google Scholar]
- Sang CN, Ramadan NM, Wallihan RG, Chappell AS, Freitag FG, Smith TR, Silberstein SD, Johnson KW, Phebus LA, Bleakman D, Ornstein PL, Arnold B, Tepper SJ, Vandenhende F. Ly293558, a novel ampa/glur5 antagonist, is efficacious and well-tolerated in acute migraine. Cephalalgia. 2004;24:596–602. doi: 10.1111/j.1468-2982.2004.00723.x. [DOI] [PubMed] [Google Scholar]
- Schlick T, Skeel RD, Brunger AT, Kalé LV, Board JAJ, Hermans J, Schulteny K. Algorithmic challenges in computational molecular biophysics. J. Comput. Phys. 1999;151:9–48. [Google Scholar]
- Stern-Bach Y, Bettler B, Hartley M, Sheppard PO, O’Hara PJ, Heinemann SF. Agonist selectivity of glutamate receptors is specified by two domains structurally related to bacterial amino acid-binding proteins. Neuron. 1994;13:1345–1357. doi: 10.1016/0896-6273(94)90420-0. [DOI] [PubMed] [Google Scholar]
- Toukmaji A, Sagui C, Board J, Darden T. Efficient particle-mesh ewald based approach to fixed and induced dipolar interactions. J. Chem. Phys. 2000;113:10913–10927. [Google Scholar]
- Valgeirsson J, Nielsen EO, Peters D, Mathiesen C, Kristensen AS, Madsen U. Bioisosteric modifications of 2-arylureidobenzoic acids: selective noncompetitive antagonists for the homomeric kainate receptor subtype glur5. J. Med. Chem. 2004;47:6948–6957. doi: 10.1021/jm030638w. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Weiss B, Alt A, Ogden AM, Gates M, Dieckman DK, Clemens-Smith A, Ho KH, Jarvie K, Rizkalla G, Wright RA, Calligaro DO, Schoepp D, Mattiuz EL, Stratford RE, Johnson B, Salhoff C, Katofiasc M, Phebus LA, Schenck K, Cohen M, Filla SA, Ornstein PL, Johnson KW, Bleakman D. Pharmacological characterization of the competitive gluk5 receptor antagonist decahydroisoquinoline ly466195 in vitro and in vivo. J. Pharmacol. Exp. Ther. 2006;318:772–781. doi: 10.1124/jpet.106.101428. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Schulten K. Protein domain movements: detection of rigid domains and visualization of hinges in comparisons of atomic coordinates. Proteins. 1997;29:1–14. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.