Abstract
SIMtoEXP is a software package designed to facilitate the comparison of biomembrane simulations with experimental X-ray and neutron scattering data. It has the following features: (1) Accepts number density profiles from simulations in a standard but flexible format. (2) Calculates the electron density ε(z) and neutron scattering length density ν(z) profiles along the z direction (i.e., normal to the membrane) and their respective Fourier transforms (i.e., Fε[qz] and Fν[qz]). The resultant four functions are then displayed graphically. (3) Accepts experimental Fε(qz) and Fν(qz) data for graphical comparison with simulations. (4) Allows for lipids and other large molecules to be parsed into component groups by the user and calculates the component volumes following Petrache et al. (Biophys J 72:2237–2242, 1997). The software then calculates and displays the contributions of each component group as volume probability profiles, ρ(z), as well as the contributions of each component to ε(z) and ν(z).
Keywords: MD simulation, X-ray scattering, Neutron scattering, Computer software
Introduction
It is well recognized that atomic-level simulations can provide quantitative detail that surpasses any known experimental data. It is also recognized that simulations may produce invalid results due to incorrect force fields or insufficient equilibration times. Based on this, it is therefore imperative to compare simulation with experiment. The NMR SCD order parameter has long been used as one such appropriate test, although it focuses only on the lipid’s hydrocarbon chains. Moreover, experimental NMR SCD data are subject to interpretive ambiguity when perdeuterated chains are employed, even for saturated chains; and the problem is more severe for unsaturated hydrocarbon chains for which SCD data are usually lacking. More recently, however, simulations have been tested using X-ray scattering data (Sachs et al. 2003; Tristram-Nagle and Nagle 2004; Klauda et al. 2006; Pandit et al. 2008) and both X-ray and neutron scattering data (Benz et al. 2005; Kučerka et al. 2008a). Because X-ray and neutron scattering data yield complementary information along the z direction (i.e., normal to the bilayer plane), the SIMtoEXP software is designed to facilitate comparisons with both kinds of data. It may be noted that there are also scattering data that provide information in other spatial directions and which may be used to further test simulations. However, those data are not addressed by the SIMtoEXP software.
Two quantities intrinsic to the SIMtoEXP software are the commonly used electron density (ED) profile, ε(z), and the neutron scattering length density (NSLD) profile, ν(z). Experimental studies often report ε(z) and ν(z), and simulators often compare their results to these one-dimensional profiles. However, ε(z) and ν(z) are not direct experimental data but are constructed using a number of assumptions. A better method is to compare simulations to the primary experimental scattering data. In the case of X-rays and neutrons, these are the form factors Fε(qz) and Fν(qz), the Fourier transforms (FTs) of ε(z) and ν(z), respectively. The FT of simulated ε(z) and ν(z) is very accurate and does not involve any assumptions, in contrast to what is required to produce ε(z) and ν(z) from experiments. However, physical insight is often more easily obtained in real space, so SIMtoEXP allows the user to compare simulated and experimentally derived ε(z) and ν(z) profiles in real z space, in addition to the more direct comparison in reciprocal qz space.
Other physical quantities that SIMtoEXP calculates from simulations are component volumes, Vi, for the various parts of lipids and other molecules. Component volumes are very useful quantities when one uses models to obtain ε(z) and ν(z) from experimental Fε(qz) and Fν(qz) data. This utility is an example of the synergy between simulation and experiment. Although the volumetric feature in SIMtoEXP was developed over a decade ago (Petrache et al. 1997), it has not been widely used by simulators.
Our interaction with simulators over the years has suggested that there is considerable value in having a uniform file format and well-tested software for comparison between simulation and experiment. We have also tried to make SIMtoEXP as flexible as possible so as to be able to accommodate the preferences of different researchers working on a variety of systems, including complex lipid mixtures and peptides.
Description of SIMtoEXP
Simulation Input
SIMtoEXP reads an ASCII data file (sim is the default extension) that contains the z distributions of each atom, or assembly of united atoms, in the solvent, lipids and any other molecules that are present. The file’s first row assigns the column names, starting with “z” for the z location and continuing with the names of the various atoms. Usually, an atom will have the same name when located at the same position on any molecule in the simulation (an exception might be to distinguish atoms in the two monolayers, although that would not affect the computation of ε[z] and ν[z]). The only strict SIMtoEXP requirement for the computation of ε(z) and ν(z) is that the name of each atom begins with its (case-insensitive) commonly used abbreviation, i.e., C, N, O, P, H and D for deuterium. For united atoms, the following abbreviations are used: M (CH2), T (CH3), W (H2O) and V (D2O). If simulations involve other atoms or groups of atoms, suitable first-letter identifiers may be developed.
Beyond the first letter, additional atom identifiers in the *.sim file can be assigned by the user. Figure 1 shows our current preference for the names of all of the atoms in the commonly studied DOPC lipid. The first number is the “distance” from the lipid backbone to a given atom with different hydrogens denoted by the letters a, b and c. The hydrocarbon chains (i.e., c1 and c2) and the headgroup (h) identifiers then follow. For example, H16bc2 signifies the “second” hydrogen atom (b) that is bonded to the sixteenth carbon in the sn-2 chain (c2). Although one might question the utility of being able to distinguish H18bc1 from H18cc1, NMR (Klauda et al. 2008) indicates that the four D2_c_ atoms are different. As such, our preference is to include more, rather than less, information in the *.sim file.
Fig. 1.
Schematic showing one possible naming convention for atoms in DOPC. Other naming conventions can be used in SIMtoEXP input simulation files, subject to the restriction that the first letter is used as the atom type identifier
The first column of the simulation input file contains the z location of atoms, in discrete bins. The bin size should be chosen to be commensurate with the statistics of the simulation; a well-equilibrated simulation with many statistically independent frames (i.e., snapshots) should contain sufficient statistics for a bin size of ~0.2 Å, resulting in smooth, one-dimensional profiles. Subsequent columns, separated by spaces or tabs, give the number density (ND) distribution functions nα(z) for each atom of type α. The number of occurrences of type α atoms located within a z bin is counted for each frame and divided by the bin volume, resulting in nα(z) as the average over all frames. Bin volume is defined as the bin height multiplied by the area of the simulation box for a given frame, which will generally be different for different frames in constant pressure simulations. For nonperiodic boundary conditions, the analysis box should be drawn within any intruding boundaries, which means that it should be well inside the simulation box to avoid any edge effects while comprising all atoms of the bilayer.
When averaging over many frames in a long simulation with periodic boundary conditions in the z direction, the center of the bilayer may move in the z direction, artificially broadening the real space profiles. It is then very important to recenter the bilayer within each frame before averaging over all frames—this can be done with simple algorithms. Also, when the simulated system has a large area and the bilayer bending modulus is small, undulation fluctuations can lead to broadening of the real space profiles, even within each frame. This correction is considerably less trivial to perform than the recentering correction and is currently under development (Braun et al. unpublished data).
Display of Simulation Data
Upon reading the *.sim file, SIMtoEXP calculates total ε(z) and ν(z), based on the atom type identifiers defined above. Each ND nα(z) is first multiplied by the appropriate number of electrons or neutron scattering length (Sears 1992), and then a summation over all atoms α is carried out. It is worth noting that the software currently uses atomic number to assign the number of electrons and does not account for the partial charges. However, the user can easily change this in the scattering length table by defining a new atom type with different scattering properties. Nevertheless, this option is not really necessary. While partial charges are important in running an MD simulation, the ED profile is hardly affected by approximating them with atomic number charges because the net local charge is the same with partial charges as for atomic number charges. For example, the charge on the phosphate PO4 is the same even though the partial charges on the phosphorus and the oxygens differ from the atomic number charges. Moreover, such a difference affects Ångström-level resolution in the ED profile, which only affects data at q values higher than those observable experimentally.
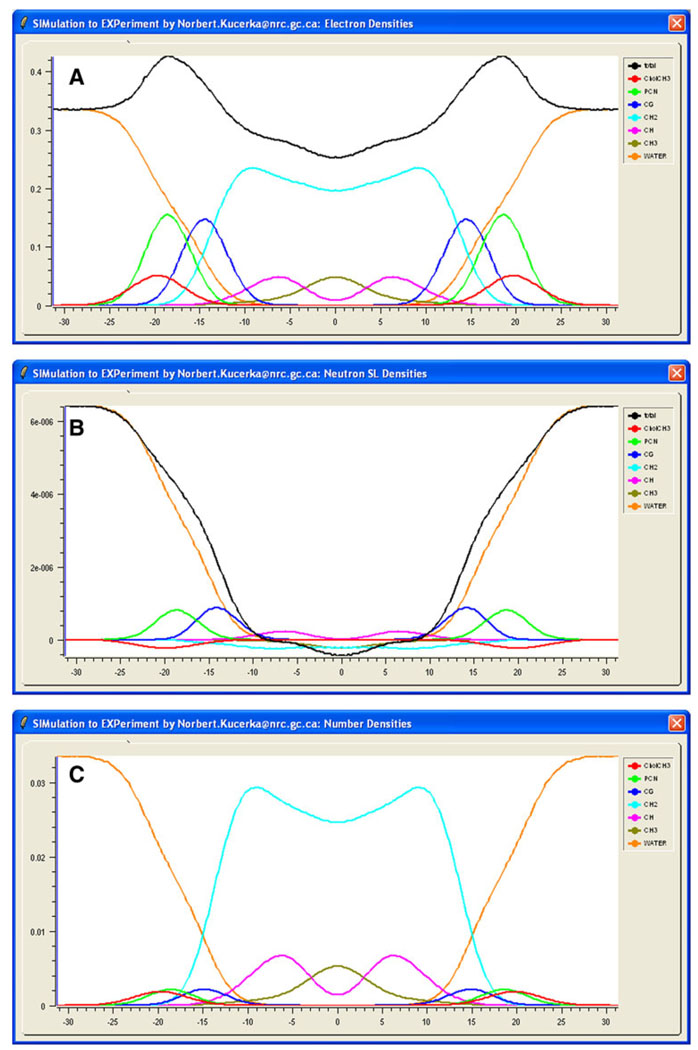
Total ε(z) is displayed graphically in the ED window (Fig. 2a), and ν(z) is displayed in the NSLD window (Fig. 2b) of the SIMtoEXP graphical user interface (GUI). In addition, it is useful to see which parts of the molecule contribute at different z levels, and this information can also be displayed in the ED and NSLD windows. Because there are usually more atom types in the *.sim file than one wishes to see displayed simultaneously, it is appropriate to define groups of atoms, which we call “molecular components.” A component file (cmp is the default extension) contains a list of user-defined names for the different components. For each component the user lists the names of the atoms defined in the simulation file that are to be included in the given component. For a proper evaluation of component volumes (see “Component Volume Determination”), each atom should be included in one and only one component of the space-filling component model, although for specific display purposes it may be convenient to relax this requirement. It is typical to parse the larger lipid molecules into several localized components (e.g., phosphates, glycerols, terminal methyls). An advantage of listing all atom types in the *.sim input file is that any variation in the parsing can easily be performed by small changes in the *.cmp component file rather than recompiling the entire simulation file.
Fig. 2.
(Color online) Electron densities (a), neutron scattering length densities (b) and number densities (c) determined from an MD simulation. SIMtoEXP displays graphically the distributions of all of the user-defined components (different color curves) as well as the total scattering density profiles (black curves in a and b)
Loading the *.cmp component file into SIMtoEXP adds the simulated scattering densities εi(z) and νi(z) for the user-defined components to the ED and NSLD windows, in addition to displaying the component number densities ni(z) (normalized by number of atoms per component) in the ND window (see Fig. 2). It is worth emphasizing that SIMtoEXP calculates component scattering densities by first multiplying the number density for each atom by the appropriate number of electrons or neutron scattering length and then summing over all component atoms. This is in contrast to summing atomic number densities and then multiplying them by the overall scattering length of each component. This way of calculating component distributions is especially important when a component includes different atoms with different scattering lengths that are distributed within that component asymmetrically (Kučerka et al. 2008a).
Results for ε(z) and ν(z) obtained from modeling experimental data can be added to the ED and NSLD windows, facilitating comparison to simulated results. However, a more important comparison is the direct comparison to experimental Fε(qz) and Fν(qz) form factors, thereby avoiding any model assumptions. The simulated scattering form factors are readily calculated from the Fourier transformation of the spatial distribution of all the atoms α as
| (1) |
where nα(z) are the atomic number distributions obtained from simulation and ρs is the scattering density of solvent. In the case of neutron scattering, fα(qz) is the neutron scattering length density, which does not depend on the wave vector qz because nuclei are effectively point sources for neutrons with wavelengths corresponding to interatomic distances. In the case of X-ray scattering, fα(qz) is the atomic form factor which is given by the FT of the atomic ED (Benz et al. 2005); it gradually decays by ~5% as qz increases to 0.8 Å−1 for the X-ray scattering on principal atoms (Klauda et al. 2006). For computational convenience, the X-ray atomic form factors have been expressed by the analytical expression and the parameters aj, bj and c have been previously determined (Cromer and Mann 1968).
Integration limits in Eq. 1 (i.e., ±D/2) must be located in the pure solvent regions, where nα(z) = 0 for all the atoms except for those in the solvent; regions near the top or bottom of a simulation cell should be excluded, especially when they fluctuate under constant pressure, because unrealistic values would then be included. Note that the imaginary part of Eq. 1 becomes zero for centrosymmetric membranes. However, SIMtoEXP utilizes the complete complex form of the Fourier transformation (Kučerka et al. 2007). This is essential for treating asymmetric membranes, and it allows the user to observe the consequences of less than fully equilibrated simulations for symmetric membranes.
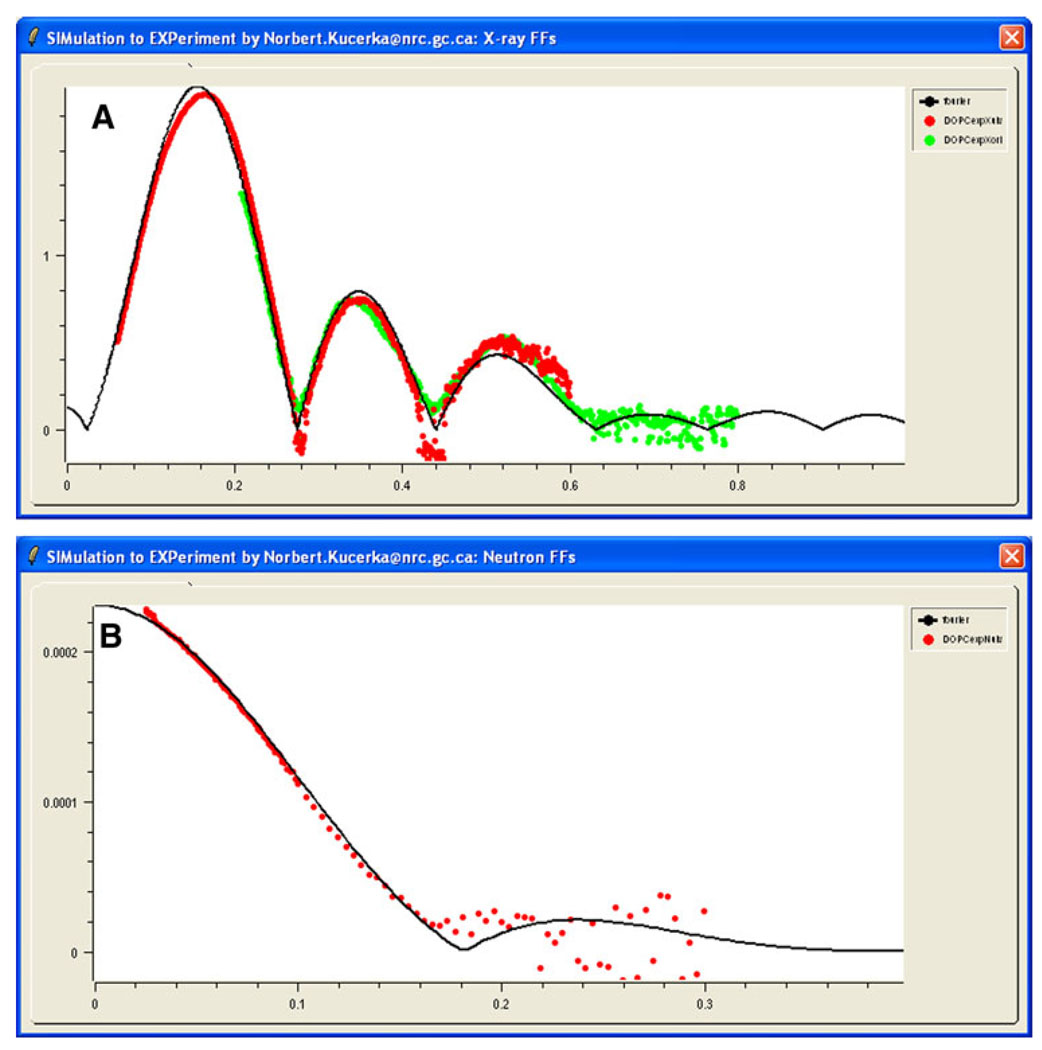
The simulated results for the Fourier-transformed form factors appear in the “X-ray FFs” (Fig. 3a) and “Neutron FFs” (Fig. 3b) windows on the GUI. Experimental data can then be imported into these windows.
Fig. 3.
(Color online) X-ray (a) and neutron scattering form factors (b) calculated from MD simulation data (solid black line) and compared to experimental data (red and green points are the results of two sample preparations). Experimental points were scaled according to Eq. 2
Simultaneous Display of Experimental Data
Experimental scattering data consist of measured intensities that can be easily converted to form factors (Kučerka et al. 2005). While form factors calculated from simulations are obtained on an absolute scale, experimental data are usually obtained only on a relative scale. SIMtoEXP scales the experimental form factors, Fe(qz), to the simulated form factors, Fs(q). The scaling factor ke by which each independent set of Fe(q) is multiplied is obtained from (Kučerka et al. 2008b)
| (2) |
where the summation goes through all of the experimental data (i = 1 … Nq) and ΔFe(q) is the experimental uncertainty of each datum. The overall agreement/disagreement between the simulation and the experimental data can be visually observed from the plots as shown in Fig. 3, or it can be quantified via a reduced χ2, which is calculated by SIMtoEXP as
| (3) |
SIMtoEXP allows the user to open several different experimental data files. These include X-ray form factors for unilamellar vesicles (ULVs) and oriented samples (ORIs), and corresponding neutron form factors. Combining ULV with ORI data was motivated by our previous work (Kučerka et al. 2005). Both these data sets involve data that are essentially continuous in qz. However, the program also allows the user to import form factor data from Bragg peaks that provide form factors only at discrete values of qz. Since different sample preparations are measured on different relative scales, it is necessary to scale them separately. This is automatically executed by SIMtoEXP (Eq. 2), or it can be overridden manually through a user-selected scaling factor. In both cases, the reduced χ2 is obtained according to Eq. 3 so that it can be used to evaluate the comparison.
Component Volume Determination
SIMtoEXP incorporates a procedure for extracting component volumes, Vi, from simulations (Petrache et al. 1997). This method requires that the *.cmp file parses all atoms α, each into one and only one component i, and that the number of components (NC) be smaller than the number of bins (Nz). Component number distributions, ni(z), are obtained as summations over all of the atomic distributions, nα(z), making up a particular component divided by the number of atoms in the component i. The method assumes that the volume Vi of each component is independent of z and that the bin volume should be completely filled. In terms of probability this means that component probabilities defined as pi(z) = Vini(z) should add to 1 in each bin. In the usual case when there are many more z bins than components, there are more equations than unknowns, so the probabilities may not add precisely to 1, because of either statistical fluctuations or a breakdown in the assumption of a constant Vi(z) (Petrache et al. 1997). To determine Vi, one minimizes
| (4) |
by setting its partial derivatives with respect to each Vi parameter to zero. This results in the set of Nc linear equations
| (5) |
A simple linear equation solution by Gauss-Jordan elimination (Press et al. 2007) then provides component volumes and the root mean square deviation of total probability from unity, which is defined as
| (6) |
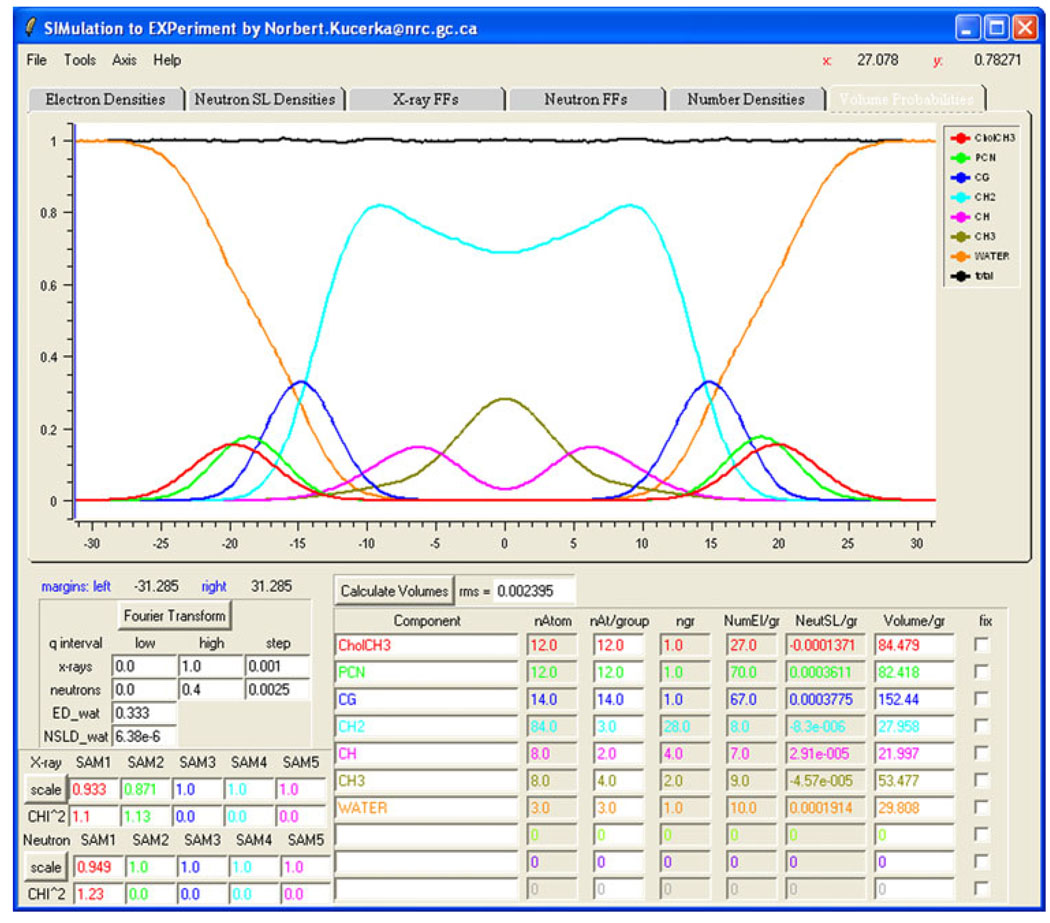
Figure 4 displays an example of such probability distributions, where the rms is approximately 0.2%.
Fig. 4.
(Color online) Snapshot of the SIMtoEXP GUI in which the volume probability distributions pi(z) = Vini(z) of the components (colors) and their sum (black) are displayed graphically. The lower part of the GUI has a column giving the names of the user-defined components, followed by characterization columns that include the calculated volume per group, given in angstroms cubed. The tabs at the top of the GUI allow the user to view graphs for electron density, neutron scattering length density, X-ray form factors, neutron form factors, number density and volume probability (the one shown here)
The example shown in Fig. 4 uses a recently developed space-filling model to define the various lipid components (Kučerka et al. 2008a). Three different components—CholCH3, PCN and CG—define the lipid headgroup; CH2, CH and CH3 define the hydrocarbon chains; and WATER represents water molecules. According to these definitions, SIMtoEXP automatically calculates the number of primitive groups in each component, as well as the corresponding number of electrons and neutron scattering lengths. The last column at the bottom of the GUI (see Fig. 4) displays the volumes calculated based on these user-defined components. Besides the SDP model used in this example, two other models are included in the current SIMtoEXP package, the HB model (Wiener et al. 1989) and the H2 model (Klauda et al. 2006). Of course, other models can easily be developed by the user.
Graphical User Interface
We believe that an attractive feature of SIMtoEXP is its user-friendly GUI, shown in Fig. 4. The most prominent portion of the software’s GUI is the graphical display, in which the various graphs can be displayed using the overlaying tabs. Successive tabs display the “Electron Densities” graph (see Fig. 2a), the “Neutron SL Densities” graph (Fig. 2b), the “X-ray FFs” graph (Fig. 3a), the “Neutron FFs” graph (Fig. 3b), the “Number Densities” graph (Fig. 2c) and the “Volume Probabilities” graph (Fig. 4). SIMtoEXP provides the user with basic manipulations (e.g., bring forward, hide, delete) to organize each graph and an option to export into postscript graphics. A zoom and an x–y coordinate readout allow for convenient quantitative comparisons.
Below the graphical display, the lower part of the GUI panel includes four buttons for performing the data manipulation described above (i.e., Fourier transform, scaling the X-ray and neutron data, and volume calculation). When each calculation is finished, the appropriate graphs are updated and quantitative results are displayed in the bottom part of the main panel. The bottom left location includes all of the user-entered information necessary for plotting the form factor results and scaling factors for the different experimental data files (Eqs. 2 and 3).
The bottom right part of the main panel shows the average rms of the volume calculation (Eq. 6) and a summary of the user-defined space-filling model for the molecular components. Most of the information here is automatically entered using *.cmp files, which can be chosen using the “File → OpenCMP” menu. Other “Open” commands in the File menu allow the user to load simulation data (OpenSIM), experimental data (OpenEXPx and OpenEXPn) consisting respectively of X-ray and neutron scattering form factors and ED or NSLD profiles (i.e., OpenEDP and OpenNSLD) obtained previously using SIMtoEXP or similar software. The latter two commands were designed to allow the user to perform a visual comparison between the various models in real space. Any of the inputted data can be exported after performing the desired operation and saved for future reference as an ASCII file.
Conclusions and Availability
Our aim has been to develop software that allows convenient comparison of simulation and experimental scattering data. Strong features of SIMtoEXP are its user-friendly graphical interface and the inclusion of all necessary procedures in one package. The program also features an integrated “Help” menu. The current functionalities of SIMtoEXP make it a suitable tool for the final stages of molecular dynamics simulations. The emphasis on inverse space allows for a direct comparison of simulation to experimental data, which can then be further scrutinized by comparing to the results of modeling experimental data in real space.
The current version of SIMtoEXP is written in C++ with GUI implemented in script language Tcl/Tk with BLT extension. The executable program has been compiled in the MS Windows operating system and is available in a bundle with all of the necessary libraries. Linux and Mac OS X executables are also available, although proper linking to the essential libraries (Tcl/Tk/BLT) is required of the user. The program bundle, a user guide that supplies additional details and tutorial commands, and sample input files are available upon request from any of the coauthors. Subsequent versions may be developed to accommodate user-suggested features that we have not anticipated.
Acknowledgements
We thank Scott Feller, Jonathan Sachs, Jason Perlmutter and Jeffery Klauda for providing us with simulation data, and Olle Edholm and Frank Heinrich for valuable feedback. N. K. acknowledges partial funding from the Advanced Foods and Materials Network (AFMNet), a Network of Centres of Excellence of Canada. J. F. N. was supported by the U.S. National Institutes of Health, Institute of General Medicine (grant GM44976).
Contributor Information
Norbert Kučerka, Canadian Neutron Beam Centre, National Research Council, Chalk River, ON K0J 1J0, Canada; Department of Physical Chemistry of Drugs, Faculty of Pharmacy, Comenius University, 832 32 Bratislava, Slovakia.
John Katsaras, Canadian Neutron Beam Centre, National Research Council, Chalk River, ON K0J 1J0, Canada; Guelph-Waterloo Physics Institute and Biophysics Interdepartmental Group, University of Guelph, Guelph, ON N1G 2W1, Canada; Department of Physics, Brock University, 500 Glenridge Avenue, St, Catharines, ON L2S 3A1, Canada.
John F. Nagle, Department of Physics, Carnegie Mellon University, Pittsburgh, PA 15213, USA, nagle@cmu.edu Department of Biological Sciences, Carnegie Mellon University, Pittsburgh, PA 15213, USA.
References
- Benz RW, Castro-Roman F, Tobias DJ, White SH. Experimental validation of molecular dynamics simulations of lipid bilayers: a new approach. Biophys J. 2005;88:805–817. doi: 10.1529/biophysj.104.046821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromer D, Mann J. X-ray scattering factors computed from numerical Hartee-Fock wave functions. Acta Crystallogr A. 1968;24:321–324. [Google Scholar]
- Klauda JB, Kučerka N, Brooks BR, Pastor RW, Nagle JF. Simulation-based methods for interpreting X-ray data from lipid bilayers. Biophys J. 2006;90:2796–2807. doi: 10.1529/biophysj.105.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda JB, Venable RM, Pastor RW, MacKerell AD. Considerations for lipid force field development: computational modeling of membrane bilayers. In: Feller SE, editor. Current topics in membranes. vol 60. San Diego: Elsevier; 2008. pp. 1–48. [Google Scholar]
- Kučerka N, Liu Y, Chu N, Petrache HI, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys J. 2005;88:2626–2637. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kučerka N, Pencer J, Sachs JN, Nagle JF, Katsaras J. Curvature effect on the structure of phospholipid bilayers. Langmuir. 2007;23:1292–1299. doi: 10.1021/la062455t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kučerka N, Nagle JF, Sachs JN, Feller SE, Pencer J, Jackson A, Katsaras J. Lipid bilayer structure determined by the simultaneous analysis of neutron and X-ray scattering data. Biophys J. 2008a;95:2356–2367. doi: 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kučerka N, Perlmutter JD, Pan J, Tristram-Nagle S, Katsaras J, Sachs JN. The effect of cholesterol on short- and long-chain monounsaturated lipid bilayers as determined by molecular dynamics simulations and X-ray scattering. Biophys J. 2008b;95:2792–2805. doi: 10.1529/biophysj.107.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit SA, Chiu SW, Jakobsson E, Grama A, Scott HL. Cholesterol packing around lipids with saturated and unsaturated chains: a simulation study. Langmuir. 2008;24:6858–6865. doi: 10.1021/la8004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrache HI, Feller SE, Nagle JF. Determination of component volumes of lipid bilayers from simulations. Biophys J. 1997;72:2237–2242. doi: 10.1016/S0006-3495(97)78867-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical recipes: the art of scientific computing. New York: Cambridge University Press; 2007. [Google Scholar]
- Sachs JN, Petrache HI, Woolf TB. Interpretation of small angle X-ray measurements guided by molecular dynamics simulations of lipid bilayers. Chem Phys Lipids. 2003;126:211–223. doi: 10.1016/j.chemphyslip.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Sears VF. Neutron scattering lengths and cross sections. Neutron News. 1992;3:26–37. [Google Scholar]
- Tristram-Nagle S, Nagle JF. Lipid bilayers: thermodynamics, structure, fluctuations and interactions. Chem Phys Lipids. 2004;127:3–14. doi: 10.1016/j.chemphyslip.2003.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener MC, Suter RM, Nagle JF. Structure of the fully hydrated gel phase of dipalmitoylphosphatidylcholine. Biophys J. 1989;55:315–325. doi: 10.1016/S0006-3495(89)82807-3. [DOI] [PMC free article] [PubMed] [Google Scholar]