Summary of Recent Advances
Mycobacterium tuberculosis is the causative agent of the global tuberculosis epidemic. To combat this successful human pathogen we need a better understanding of the basic biology of mycobacterial pathogenesis. The use of mycobacterial model systems has the potential to greatly facilitate our understanding of how M. tuberculosis causes disease. Recently, studies using mycobacterial models, including M. bovis BCG, M. marinum and M. smegmatis have significantly contributed to understanding M. tuberculosis. Specifically, there have been advances in genetic manipulation of M. tuberculosis using inducible promoters and recombineering that alleviate technical limitations in working with mycobacteria. Model systems have helped elucidate how secretion systems function at both the molecular level and during virulence. Mycobacterial models have also led to interesting hypotheses about how M. tuberculosis mediates latent infection and host response. While there is utility in using model systems to understand tuberculosis, each of these models represent distinct mycobacterial species with unique environmental adaptations. Directly comparing findings in model mycobacteria to those in M. tuberculosis will illuminate the similarities and differences between these species and increase our understanding of why M. tuberculosis is such a potent human pathogen.
Introduction
The global tuberculosis (TB) epidemic annually accounts for more than 3 million deaths worldwide. Because of the capacity of Mycobacterium tuberculosis to cause latent disease, an estimated 1-2 billion people worldwide are infected with M. tuberculosis. Immunodeficiency caused by malnutrition, old age or HIV infection enhances development of active disease, either from a primary infection or by reactivation of a latent infection. The global TB epidemic is greatly exacerbated by insufficient public health measures to detect, prevent and treat TB, and the lengthy antibiotic course required to treat active TB results in non-adherence and development of bacterial resistance. The rising incidence of multi- and extremely-drug resistant (MDR and XDR) TB is worrisome, indicating that more effort should be directed towards understanding the basic biological pathways that underlie mycobacterial virulence. To this end, mycobacterial model systems have the potential to facilitate our understanding of M. tuberculosis pathogenesis. In this review, we highlight how models have been used over the past two years to study important aspects of M. tuberculosis biology, including virulence factor secretion, dormancy and host response.
Why are mycobacterial models useful?
The direct study of M. tuberculosis is vital to understanding its pathogenesis. However, use of this pathogen in the laboratory is labor-intensive for several reasons. First, M. tuberculosis is a Category three human pathogen, requiring dedicated biosafety level three laboratory and animal facilities, substantial training prior to handling, and carries with it a risk of accidental exposure [1]. Second, M. tuberculosis grows slowly, doubling every 22 hours in liquid culture. Thus, colony formation requires two to three weeks, making each experiment time consuming. While many laboratories successfully study M. tuberculosis, using mycobacterial model systems to gain insight into M. tuberculosis virulence mechanisms is becoming more common.
While multiple mycobacterial species have been harnessed in the past to understand M. tuberculosis virulence, three very different mycobacterial species, Mycobacterium bovis (BCG strain), Mycobacterium smegmatis and Mycobacterium marinum now predominate the recent literature as models for studying M. tuberculosis. M. bovis (BCG) is a member of the TB Complex that was attenuated by serial passage in the laboratory. While it grows slowly like M. tuberculosis, it is a Category 2 organism. M. smegmatis is a soil dwelling saprophytic mycobacterial species that is a distant relative of M. tuberculosis. This avirulent mycobacterial species is fast growing, with a doubling time of approximately four hours and colony generation in two to three days. M. smegmatis is a particularly convenient model due to its ease of genetic manipulation (due to much effort on the part of the mycobacterial community). M. marinum is an occasional human pathogen that causes a TB-like infection in ectotherms. It doubles every 10-12 hours, resulting in colony formation from one to two weeks, and is amenable to similar genetic manipulations as M. smegmatis. Importantly, M. marinum causes caseating granulomas in zebrafish, which resemble those formed by M. tuberculosis in humans (reviewed in [2]). Moreover, known virulence determinants are conserved between M. marinum and M. tuberculosis such that genes from M. tuberculosis can complement mutations in orthologous M. marinum genes [2].
Increasing the genetic tractability of M. tuberculosis using model mycobacteria
The use of M. smegmatis has greatly contributed to genetic tractability of pathogenic mycobacteria, beginning with the advent of plasmids for mycobacterial transformation in the late 80's and early 90's, and the isolation of transformation permissive M. smegmatis strains [3]. Recently, a technique known as “recombineering” was developed for both M. smegmatis and M. tuberculosis, through the exploitation of mycobacteriophage genes that promote recombination from PCR products [4,5]. Recombineering allows for direct manipulation of the mycobacterial chromosome, which formerly represented a major technical limitation [4,5].
Another important advance was the development of inducible mycobacterial promoters. By directly controling gene expression, researchers can study the role of essential genes in both viability and virulence, and can fine tune expression of genes during infection. The best developed inducible systems in mycobacteria are the Tet-ON and Tet-OFF systems, allowing for regulation of gene expression in the presence of anhydrotetracycline (atc) [6••], [7] and [8•]. In one example, the Tet-ON and Tet-OFF systems were used in M. smegmatis to make a conditional secA1 mutant [7] (Fig. 1). Moreover, the essential nature of the mycobacterial proteosome during growth of M. tuberculosis on agar, in liquid media, and in mice was characterized with this system [6••]. In another creative application of the Tet-inducible system, a plasmid was constructed that allows for tunable expression of multiple mycobacterial proteins in vivo [9•] (Figure 1). Both the Tet-ON, Tet-OFF and one-plasmid expression vectors were initially optimized in M. smegmatis and/or BCG and then applied to M. tuberculosis [7,9•].
Figure 1. Recent Advances in Mycobacterial Tool Development.
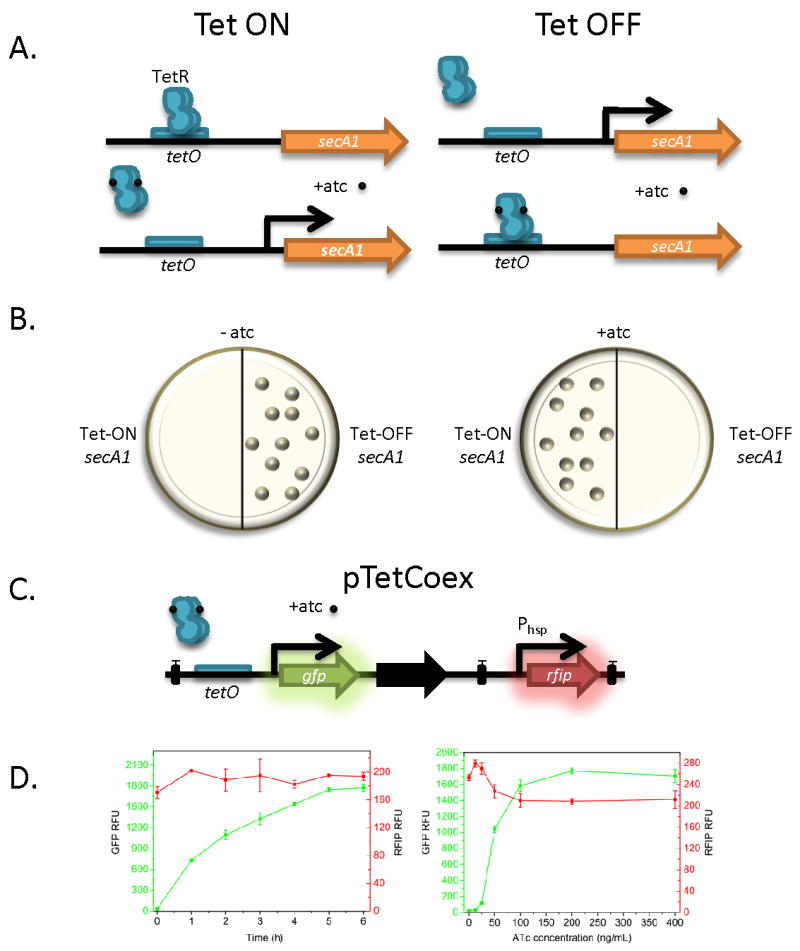
A. The Tet-ON and Tet-OFF inducible mycobacterial promoter systems, for example driving the essential secA1 gene. For the Tet-ON system, the TetR repressor occupies the tetO operator in the absence of anhydrotetracycline (atc) repressing transcription. This is relieved upon atc addition. Tet-OFF allows for transcription in the absence of atc, and transcriptional repression or silencing in the presence of atc. B. Mycobacterial strains lacking the essential secA1 gene. Strains with Tet-ON secA1 are only viable in the presence of atc, while strains with Tet-OFF secA1 are only viable in the absence of atc. C. The pTetCoex system. The inducible Tet-ON promoter drives expression of gfp, while the Phsp constitutive promoter drives expression of rfip, a newly discovered red fluorescent protein. Black boxes represent terminators, and the black arrow represents a stabilizing gene. D. Tunable expression of GFP and co-expression of RFIP using TetCoex. Left, increase of GFP expression in M. smegmatis over time in the presence of 200ng/ml atc relative to co-expression of RFIP (RFU, relative fluorescent units). Right, tunable expression of GFP in an atc dose-dependent manner, compared to constitutive expression of RFIP. From [9] with permission.
Virulence factor transport
One of the most active areas of mycobacterial research is the regulation, selection and transport of protein virulence factors that likely interact with the host cell during infection. Two systems being studied are the accessory Sec system and the ESX systems (known as Snm, Ess, and Type VII secretion systems).
The general secretion pathway (or Sec) functions to translocate proteins from the bacterial cytosol to extracytosolic compartments. In many Gram-positive bacteria and mycobacteria, there exists a second, non-essential copy of the SecA ATPase, called SecA2. SecA2 and the accessory Sec system mediate secretion of proteins that are usually glycosylated, thereby precluding secretion by the Sec pathway. (reviewed in [10•]). Thus far, a handful of SecA2-dependent substrates have been identified in both M. tuberculosis and M. smegmatis [10•,11] and the molecular workings of the SecA2 ATPase have been elucidated in M. smegmatis [12,13]. Interestingly, the role of SecA2 in secreting virulence factors has recently been exploited to develop a new HIV/MTB vaccine platform [14•]. Deletion of the M. tuberculosis secA2 gene in combination with lysA deletion and expression of HIV envelope protein induced a strong CD8+ T cell response in mice that was greater than that elicited by a BCG strain expressing the same HIV envelope protein [14•].
The ESX secretion systems (ESAT-6 secretion systems) are alternative secretion machines that are duplicated multiple times in the chromosome of both pathogenic and non-pathogenic mycobacterial species [15]. The most well studied system is ESX-1, which is required for mycobacterial virulence and is absent from the genome of the BCG strain(reviewed in [16•]). Both M. smegmatis and M. marinum have been used to elucidate the molecular mechanisms of ESX-1 secretion, and to decipher its role in mycobacterial virulence (Recent examples include [17-20]). In the past year, studies using M. marinum to understand ESX-1 have identified two novel substrates (Mh3864 and EspF) [18,21••]. How are substrates secreted? Champion et al. showed that multiple ATPases function to select and transport ESX-1 substrates [18]. Additionally, Carlsson et al. showed that the ESX-1 system localizes at the poles of the bacterial cell, and provided the first visualization of the ESX-1 system in vivo [21••]. Finally, studies in M. smegmatis have suggested that ESX-1 regulates DNA conjugation in non-pathogenic mycobacteria [17,22].
Two additional ESX systems (ESX-3 and ESX-5) have been studied, primarily in non-tuberculous mycobacteria. First, the presumed ESX-3 genes were found to be regulated by iron in both M. tuberculosis and M.smegmatis, and by zinc in M. tuberculosis [23,24]. Using both M. smegmatis and BCG, Siegrist et al. demonstrated that ESX-3 is required for growth during iron deprivation, for iron acquisition, and for survival in the macrophage [25••]. Moreover they provide the first demonstration that ESX-3 functions as a secretion machine, reporting the secretion of the ESAT-6 ortholog, EsxH [25••]. Further, the Tet-inducible system was used to create a conditional mutant of the ESX-3 system, showing that it is essential for growth in M. tuberculosis [26]. ESX-5 has solely been studied in M. marinum, and was the focus of a recent review [27•]. ESX-5 functions to secrete a novel class of mycobacterial proteins, the PE/PPE and PGRS proteins, and appears to modulate the response of macrophages to infection [28•,29•].
Dormancy
Since most people exposed to M. tuberculosis develop latent (dormant) infection, understanding the molecular basis of this stage is critical. Recent experiments using model organisms have elucidated many important features of dormancy, but not all characteristics are shared. Nonetheless, model organisms are well-suited to in vitro dormancy systems as many known triggers of dormancy, including nutrient deprivation, hypoxia, nitric oxide, carbon monoxide and acid pH lead to similar responses in M. tuberculosis and non-tuberculous mycobacteria.
Three of these signals, hypoxia (Reviewed in [30]), NO [31,32] and CO [32,33], operate via the highly conserved two-component system, DosS-DosR (DevS-DevR) [34•]. The DosR transcriptional regulator and most of the dormancy regulon genes are shared in M. tuberculosis and BCG, M. marinum and M. smegmatis [34•]. Thus, further characterization of the dormancy response in model organisms is likely to yield important and comparable results to studies in M. tuberculosis, possibly accelerating development of drugs to inhibit or modulate the dormancy response [35].
Recently, both M. marinum and BCG grown into stationary phase for 6 months were found to sporulate at a low rate [36••]. Interestingly, when M. smegmatis was cultured in nitrogen-limited conditions, “spore-like” forms were observed that exhibited antibiotic resistance, diminished metabolic activity and diminished viability by CFU [37••]. These surprising observations remain untested in M. tuberculosis, and the majority of predicted sporulation genes are not required for virulence in macrophages [38]. However, since many sporulation genes are conserved, it seems plausible that sporulation may be a general pathway for mycobacterial dormancy.
Latent cells need to exit dormancy, and recent work in both M. smegmatis and M. tuberculosis has identified the lytic transglycosylases known as resuscitation promoting factors (rpfs) [39] as vital for revival and virulence [40-42]. Rpfs also interact with the RipA endopeptidase, an essential gene in M. tuberculosis [43]. Using the Tet-inducible system described above, Hett et al. depleted RipA in M. smegmatis, and showed that RipA cleaves peptidoglycan and is essential for cell division [44•]. Interestingly, mutation of a M. marinum homologue of ripA resulted in a filamentation phenotype, and in attenuated growth in macrophages [45]. Thus, Rpf/RipA mediated reemergence from dormancy appears to be a conserved mechanism in mycobacteria.
Host response
A vital component of M. tuberculosis pathogenesis is its ability to parasitize macrophages. Accordingly, much effort has been devoted to understanding this process, including critical experiments with non-tuberculous mycobacteria. Recently, a novel macrophage antimicrobial mechanism mediated by ubiquitin derived peptides has been described for both M. tuberculosis [46••] and M. marinum [47] (Fig. 2). Interestingly, while both organisms appear to be targeted for lysosomal degradation by cell-surface ubiquitination, destruction of M. tuberculosis is enhanced by autophagy [46••] while destruction of M. marinum is autophagy independent [47]. Further suggesting an important role for ubiquitin in mycobacterial infection, when M. smegmatis infected Drosophila S2 cells were depleted of the ESCRT machinery, M. smegmatis colocalized with ubiquitin and were readily killed [48].
Figure 2. Mycobacterial killing by Ubiquitinated Peptides.
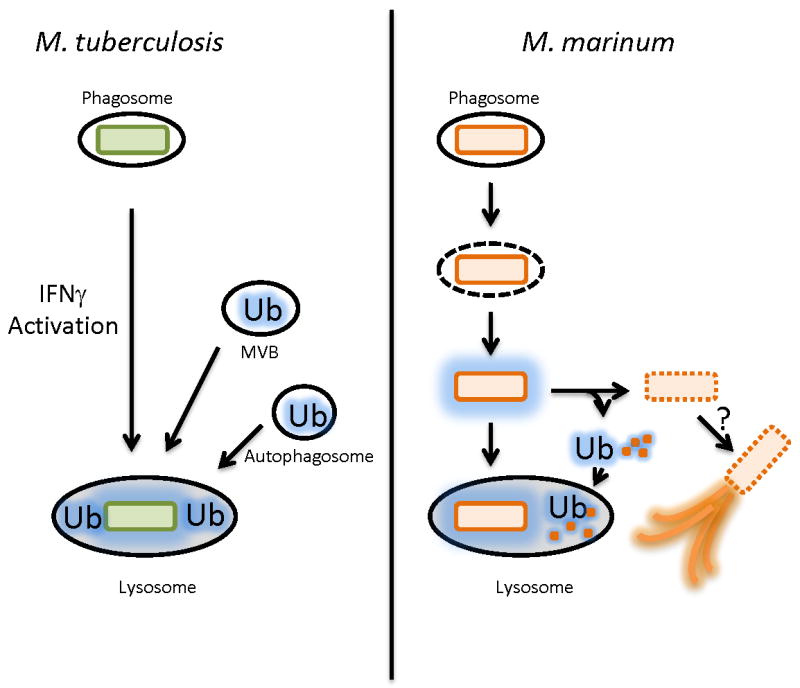
Left, Model for killing of M. tuberculosis by ubiquitin peptides by macrophages. In IFNγ activated macrophages, the M. tuberculosis phagosome is thought to fuse with ubiquitin (Ub) -containing vesicles including MVBs (multivesicular bodies generated by the ESCRT complex) and LC-3 positive (red line) autophagosomes, and the ubiquitin coated bacteria are targeted to the lysosome. Right, M. marinum differs from M. tuberculosis in that a population of M. marinum escape the phagosome. Cytosolic M. marinum are coated with ubiquitin in an autophagy-independent manner. The coated M. marinum have two possible fates. First, they can be targeted to the lysosome. Second, the bacteria can shed the ubiquitin coated cell wall (which itself is sent to the lysosome). It is possible that this population then forms actin tails, the actin tail population is not ubiquitinated.
How might ubiquitination kill mycobacteria? A ubiquitin-derived peptide, Ub2 was shown to impair the membrane integrity of both M. smegmatis and M. tuberculosis [49] (Fig. 2). While screening for M. smegmatis strains resistant to Ub2, Purdy et al. found that mycobacterial porins mediate Ub2 resistance by decreasing cell wall permeability. Surprisingly, Ub2 resistance was independent of porin function. In a complementary study, M. smegmatis mutants bearing deletions in one or more porins (including MspA) were found to be more resistant to killing by J774 macrophages and also to nitric oxide in vitro [50]. Thus, a plausible model is that host ubiquitination alters mycobacterial membrane permeability, allowing nitric oxide to kill more readily.
Conclusions
Given the dire need to understand the basic biology of mycobacterial pathogenesis, it is important that researchers use all available tools to study M. tuberculosis. We have focused on a small sample of recent studies in which mycobacterial models have significantly contributed to understanding M. tuberculosis. Despite the obvious utility of these models, it is crucial to remember that model organisms such as BCG, M. smegmatis and M. marinum are themselves unique species that have adapted to distinct environments (Table 1). It will be interesting to compare the findings in these models to similar studies in M. tuberculosis. This assessment is invaluable, as direct comparison of model organisms and M. tuberculosis will reveal both shared and divergent pathways. Ultimately, this information will provide a better understanding of how M. tuberculosis has evolved to be such a highly successful human pathogen.
Table1.
A comparison of three mycobacterial models to M. tuberculosis. In this table, we highlight the similarities, differences between M. smegmatis, M. marinum, BCG and M. tuberculosis.
| Species | Genome Size (MB) | Growth Rate | Primary Host | Category | Major Advantages | Major Disadvantages |
|---|---|---|---|---|---|---|
| M. smegmatis | 7.0 |
|
n/a | 1 |
|
|
| M. marinum | 6.6 |
|
ectotherms | 2 |
|
|
| M. bovis (BCG) | 4.3 |
|
cattle humans | 2 |
|
|
| M. tuberculosis | 4.4 |
|
humans | 3 |
|
|
Acknowledgments
We would like to thank Dr. Matthew Champion and Dr. Shaun Lee for the critical reading of this manuscript. MUS is grateful to the National Institutes of Health [grant K08 AI076632] for support.
References and Annotations
- 1.Alderton H, Smith D. Safety in the laboratory. In: Parish T, Stoker NG, editors. Methods in molecular medicine: Mycobacterium tuberculosis protocols. Humana Press; 2001. pp. 367–383. [DOI] [PubMed] [Google Scholar]
- 2.Tobin DM, Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol. 2008;10:1027–1039. doi: 10.1111/j.1462-5822.2008.01133.x. [DOI] [PubMed] [Google Scholar]
- 3.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 4.van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat Methods. 2007;4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 5.van Kessel JC, Hatfull GF. Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering: characterization of antimycobacterial drug targets. Mol Microbiol. 2008;67:1094–1107. doi: 10.1111/j.1365-2958.2008.06109.x. [DOI] [PubMed] [Google Scholar]
- 6••.Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]; Genes that are essential for M. tuberculosis growth, either in in vitro or in vivo, are thought to make up about 20% of the M. tuberculosis genome. Understanding how these 800 or so genes function during infection is important, but technically challenging due to their essential nature. This paper uses the Tet-inducible promoter systems to deplete the essential genes that encode for the core mycobacterial proteasome during mouse infection. They find that the proteasome is required for M. tuberculosis persistence during the chronic phase of infection in mice.
- 7.Guo XV, Monteleone M, Klotzsche M, Kamionka A, Hillen W, Braunstein M, Ehrt S, Schnappinger D. Silencing Mycobacterium smegmatis by using tetracycline repressors. J Bacteriol. 2007;189:4614–4623. doi: 10.1128/JB.00216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Klotzsche M, Ehrt S, Schnappinger D. Improved tetracycline repressors for gene silencing in mycobacteria. Nucleic Acids Res. 2009;37:1778–1788. doi: 10.1093/nar/gkp015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Although Tet-ON and Tet-OFF inducible systems allow tight regulation of some mycobacterial genes, they show weak or no regulation of other mycobacterial genes. In this paper, the authors present updated versions of the Tet-ON and Tet-OFF inducible promoter systems, resulting in 50 -fold better repression of reporter genes in both M. smegmatis and BCG.
- 9•.Chang Y, Mead D, Dhodda V, Brumm P, Fox BG. One-plasmid tunable Co-expression for mycobacterial protein-protein interaction studies. Protein Sci. 2009 doi: 10.1002/pro.242. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this paper, the authors construct a single plasmid containing a Tet-inducible promoter, and a constitutive promoter allowing for controlled co-expression of two genes. They optimize this system in M. smegmatis using Tet-inducible expression of gfp and constitutive expression of rfip from G. atermophilus as reporters. They then apply this technology to studying the M. tuberculosis fatty acid desaturase complex in M smegmatis.
- 10•.Rigel NW, Braunstein M. A new twist on an old pathway--accessory Sec [corrected] systems. Mol Microbiol. 2008;69:291–302. doi: 10.1111/j.1365-2958.2008.06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review comprehensively addresses the accessory Sec secretion systems in both mycobacteria and Gram-positive pathogens, highlighting the similarities and differences. They also review the accessory Sec systems relative to virulence.
- 11.Gibbons HS, Wolschendorf F, Abshire M, Niederweis M, Braunstein M. Identification of two Mycobacterium smegmatis lipoproteins exported by a SecA2-dependent pathway. J Bacteriol. 2007;189:5090–5100. doi: 10.1128/JB.00163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou JM, D'Lima NG, Rigel NW, Gibbons HS, McCann JR, Braunstein M, Teschke CM. ATPase activity of Mycobacterium tuberculosis SecA1 and SecA2 proteins and its importance for SecA2 function in macrophages. J Bacteriol. 2008;190:4880–4887. doi: 10.1128/JB.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rigel NW, Gibbons HS, McCann JR, McDonough JA, Kurtz S, Braunstein M. The Accessory SecA2 System of Mycobacteria Requires ATP Binding and the Canonical SecA1. J Biol Chem. 2009;284:9927–9936. doi: 10.1074/jbc.M900325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Ranganathan UD, Larsen MH, Kim J, Porcelli SA, Jacobs WR, Jr, Fennelly GJ. Recombinant pro-apoptotic Mycobacterium tuberculosis generates CD8(+) T cell responses against human immunodeficiency virus type 1 Env and M. tuberculosis in neonatal mice. Vaccine. 2009 doi: 10.1016/j.vaccine.2009.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]; Because BCG can cause disease in immuno-compromised individuals it is unsafe as a vaccine in HIV-infected infants. A safer alternative may be the use of attenuated strains of M. tuberculosis. Here the authors evaluate the immunogenicity in mice of multiple potential vaccine strains, including a M. tuberculosis strain with deletions in the lysA and secA2 genes, which also expresses the HIV envelope antigen construct. They conclude that this combination may be a promising new vaccine platform.
- 15.Gey Van Pittius NC, Gamieldien J, Hide W, Brown GD, Siezen RJ, Beyers AD. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2001;2:RESEARCH0044. doi: 10.1186/gb-2001-2-10-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Simeone R, Bottai D, Brosch R. ESX/type VII secretion systems and their role in host-pathogen interaction. Curr Opin Microbiol. 2009;12:4–10. doi: 10.1016/j.mib.2008.11.003. [DOI] [PubMed] [Google Scholar]; This recent review focuses on the role of the ESX-1 system in virulence and host-pathogen interactions.
- 17.Coros A, Callahan B, Battaglioli E, Derbyshire KM. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol Microbiol. 2008;69:794–808. doi: 10.1111/j.1365-2958.2008.06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiGiuseppe Champion PA, Champion MM, Manzanillo P, Cox JS. ESX-1 secreted virulence factors are recognized by multiple cytosolic AAA ATPases in pathogenic mycobacteria. Mol Microbiol. 2009;73:950–962. doi: 10.1111/j.1365-2958.2009.06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaughlin B, Chon JS, MacGurn JA, Carlsson F, Cheng TL, Cox JS, Brown EJ. A mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog. 2007;3:e105. doi: 10.1371/journal.ppat.0030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, Laine O, Masciocchi M, Manoranjan J, Smith J, Du SJ, Edwards N, Zhu X, Fenselau C, Gao LY. A unique Mycobacterium ESX-1 protein co-secretes with CFP-10/ESAT-6 and is necessary for inhibiting phagosome maturation. Mol Microbiol. 2007;66:787–800. doi: 10.1111/j.1365-2958.2007.05959.x. [DOI] [PubMed] [Google Scholar]
- 21••.Carlsson F, Joshi SA, Rangell L, Brown EJ. Polar localization of virulence-related Esx-1 secretion in mycobacteria. PLoS Pathog. 2009;5:e1000285. doi: 10.1371/journal.ppat.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides the first visualization of the ESX-1 system in the mycobacterial cell, and interestingly shows that this system is localized to the cell poles. Moreover, they identify a novel ESX-1 substrate in M. marinum, Mh3864, which remains cell wall associated following secretion from the cell.
- 22.Flint JL, Kowalski JC, Karnati PK, Derbyshire KM. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc Natl Acad Sci U S A. 2004;101:12598–12603. doi: 10.1073/pnas.0404892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maciag A, Dainese E, Rodriguez GM, Milano A, Provvedi R, Pasca MR, Smith I, Palu G, Riccardi G, Manganelli R. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J Bacteriol. 2007;189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maciag A, Piazza A, Riccardi G, Milano A. Transcriptional analysis of ESAT-6 cluster 3 in Mycobacterium smegmatis. BMC Microbiol. 2009;9:48. doi: 10.1186/1471-2180-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, Fortune SM, Moody DB, Rubin EJ. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper thoroughly investigates the ESX-3 system in both M. smegmatis and BCG. The authors demonstrate that mycobacteria lacking ESX-3 are deficient in iron acquisition, and demonstrate that this is due to the synthesis of mycobactins that fail to bind iron. They provide the first indication that ESX-3 is a functional secretion system by showing ESX-3 dependent secretion of the ESAT-6 ortholog, EsxH. Finally, they demonstrate that ESX-3 is important for growth of BCG in macrophages.
- 26.Serafini A, Boldrin F, Palu G, Manganelli R. Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J Bacteriol. 2009;191:6340–6344. doi: 10.1128/JB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Bottai D, Brosch R. Mycobacterial PE, PPE and ESX clusters: novel insights into the secretion of these most unusual protein families. Mol Microbiol. 2009;73:325–328. doi: 10.1111/j.1365-2958.2009.06784.x. [DOI] [PubMed] [Google Scholar]; This review focuses on the PE, PPE and PE_GRS genes in mycobacteria, and the recent discovery that some of these proteins are secreted by the ESX-5 system in M marinum.
- 28•.Abdallah AM, Savage ND, van Zon M, Wilson L, Vandenbroucke-Grauls CM, van der Wel NN, Ottenhoff TH, Bitter W. The ESX-5 secretion system of Mycobacterium marinum modulates the macrophage response. J Immunol. 2008;181:7166–7175. doi: 10.4049/jimmunol.181.10.7166. [DOI] [PubMed] [Google Scholar]; In this study, the authors evaluate the role of ESX-5 during M. marinum infection of macrophages. They find that unlike ESX-1, ESX-5 is not required for phagosomal escape of M. marinum into the cytosol. Rather, ESX-5 seems to be involved in the supression of proinflammatory cytokines, and in the induction of IL-1β by M. marinum. Moreover, they find that ESX-5 is involved in suppressing innate immmune cytokine secretion in a TLR-dependent manner.
- 29•.Abdallah AM, Verboom T, Weerdenburg EM, Gey van Pittius NC, Mahasha PW, Jimenez C, Parra M, Cadieux N, Brennan MJ, Appelmelk BJ, et al. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol. 2009;73:329–340. doi: 10.1111/j.1365-2958.2009.06783.x. [DOI] [PubMed] [Google Scholar]; Using proteomics, the authors evaluate the ESX-3 secretome. Interestingly, they find that ESX-5 is required for the secretion of PE_PGRS and PPE proteins, as well as ESAT-6 like proteins. The PE_PGRS and PPE proteins represent a family of proteins unique to mycobacteria, that seem to be expanded in the genomes of pathogenic mycobacteria in particular.
- 30.Rustad TR, Sherrid AM, Minch KJ, Sherman DR. Hypoxia: a window into Mycobacterium tuberculosis latency. Cell Microbiol. 2009;11:1151–1159. doi: 10.1111/j.1462-5822.2009.01325.x. [DOI] [PubMed] [Google Scholar]
- 31.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiloh MU, Manzanillo P, Cox JS. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe. 2008;3:323–330. doi: 10.1016/j.chom.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A, Deshane JS, Crossman DK, Bolisetty S, Yan BS, Kramnik I, Agarwal A, Steyn AJ. Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J Biol Chem. 2008;283:18032–18039. doi: 10.1074/jbc.M802274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Lin MY, Reddy TB, Arend SM, Friggen AH, Franken KL, van Meijgaarden KE, Verduyn MJ, Schoolnik GK, Klein MR, Ottenhoff TH. Cross-Reactive Immunity to Mycobacterium tuberculosis DosR Regulon-Encoded Antigens in Individuals Infected with Environmental, Nontuberculous Mycobacteria. Infect Immun. 2009;77:5071–5079. doi: 10.1128/IAI.00457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; Analysis of all sequenced mycobacterial genomes reveals that many genes of the DosR regulon are conserved amongst MTB complex and non-tuberculous mycobacteria.
- 35.Gupta RK, Thakur TS, Desiraju GR, Tyagi JS. Structure-based design of DevR inhibitor active against nonreplicating Mycobacterium tuberculosis. J Med Chem. 2009;52:6324–6334. doi: 10.1021/jm900358q. [DOI] [PubMed] [Google Scholar]
- 36••.Ghosh J, Larsson P, Singh B, Pettersson BM, Islam NM, Sarkar SN, Dasgupta S, Kirsebom LA. Sporulation in mycobacteria. Proc Natl Acad Sci U S A. 2009;106:10781–10786. doi: 10.1073/pnas.0904104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37••.Anuchin AM, Mulyukin AL, Suzina NE, Duda VI, El-Registan GI, Kaprelyants AS. Dormant forms of Mycobacterium smegmatis with distinct morphology. Microbiology. 2009;155:1071–1079. doi: 10.1099/mic.0.023028-0. [DOI] [PubMed] [Google Scholar]; Combined, both articles demonstrate that non-tuberculous mycobacteria (BCG, M. marinum and M. smegmatis) grown into stationary phase or exposed to nutrient starvation form spore-like structures with biologic characteristics of spores. Microscopy shows that resuscitation by exposure to fresh medium is accompanied by apparent germination.
- 38.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A. 2005;102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukamolova GV, Kaprelyants AS, Young DI, Young M, Kell DB. A bacterial cytokine. Proc Natl Acad Sci U S A. 1998;95:8916–8921. doi: 10.1073/pnas.95.15.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biketov S, Potapov V, Ganina E, Downing K, Kana BD, Kaprelyants A. The role of resuscitation promoting factors in pathogenesis and reactivation of Mycobacterium tuberculosis during intra-peritoneal infection in mice. BMC Infect Dis. 2007;7:146. doi: 10.1186/1471-2334-7-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kana BD, Gordhan BG, Downing KJ, Sung N, Vostroktunova G, Machowski EE, Tsenova L, Young M, Kaprelyants A, Kaplan G, et al. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol Microbiol. 2008;67:672–684. doi: 10.1111/j.1365-2958.2007.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell-Goldman E, Xu J, Wang X, Chan J, Tufariello JM. A Mycobacterium tuberculosis Rpf double-knockout strain exhibits profound defects in reactivation from chronic tuberculosis and innate immunity phenotypes. Infect Immun. 2008;76:4269–4281. doi: 10.1128/IAI.01735-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hett EC, Chao MC, Steyn AJ, Fortune SM, Deng LL, Rubin EJ. A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Mol Microbiol. 2007;66:658–668. doi: 10.1111/j.1365-2958.2007.05945.x. [DOI] [PubMed] [Google Scholar]
- 44•.Hett EC, Chao MC, Deng LL, Rubin EJ. A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLoS Pathog. 2008;4:e1000001. doi: 10.1371/journal.ppat.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Depletion of RipA in M. smegmatis reversibly inhibited cell growth and resulted in the formation of filamentous bacteria potentially resulting from defective cell division. This phenotype could be complemented by ripA from M tuberculosis.
- 45.Gao LY, Pak M, Kish R, Kajihara K, Brown EJ. A mycobacterial operon essential for virulence in vivo and invasion and intracellular persistence in macrophages. Infect Immun. 2006;74:1757–1767. doi: 10.1128/IAI.74.3.1757-1767.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first paper to directly show ubiquitin-derived peptides are bactericidal to intracellular mycobacteria. This identifies another component of the antimicrobioal repertoire of macrophages and may explain one mechanism by which autophagous cells kill bacteria.
- 47.Collins CA, De Maziere A, van Dijk S, Carlsson F, Klumperman J, Brown EJ. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog. 2009;5:e1000430. doi: 10.1371/journal.ppat.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Philips JA, Porto MC, Wang H, Rubin EJ, Perrimon N. ESCRT factors restrict mycobacterial growth. Proc Natl Acad Sci U S A. 2008;105:3070–3075. doi: 10.1073/pnas.0707206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Purdy GE, Niederweis M, Russell DG. Decreased outer membrane permeability protects mycobacteria from killing by ubiquitin-derived peptides. Mol Microbiol. 2009;73:844–857. doi: 10.1111/j.1365-2958.2009.06801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fabrino DL, Bleck CK, Anes E, Hasilik A, Melo RC, Niederweis M, Griffiths G, Gutierrez MG. Porins facilitate nitric oxide-mediated killing of mycobacteria. Microbes Infect. 2009;11:868–875. doi: 10.1016/j.micinf.2009.05.007. [DOI] [PubMed] [Google Scholar]


