Abstract
The combination of sulfadoxine-pyrimethamine is recommended for use as intermittent preventive treatment of malaria during pregnancy and is deployed in Africa. The emergence and the spread of resistant parasites are major threats to such an intervention. We have characterized the Plasmodium falciparum dhfr (pfdhfr) haplotypes and flanking microsatellites in 322 P. falciparum isolates collected from the Comoros Islands and Madagascar. One hundred fifty-six (48.4%) carried the wild-type pfdhfr allele, 19 (5.9%) carried the S108N single-mutation allele, 30 (9.3%) carried the I164L single-mutation allele, 114 (35.4%) carried the N51I/C59R/S108N triple-mutation allele, and 3 (1.0%) carried the N51I/C59R/S108N/I164L quadruple-mutation allele. Microsatellite analysis showed the introduction from the Comoros Islands of the ancestral pfdhfr triple mutant allele of Asian origin and its spread in Madagascar. Evidence for the emergence on multiple occasions of the I164L single-mutation pfdhfr allele in Madagascar was also obtained. Thus, the conditions required to generate mutants with quadruple mutations are met in Madagascar, representing a serious threat to current drug policy.
Despite the increasing financial support for the control of malaria (16, 31), malaria remains a major cause of morbidity and mortality in many developing countries in the tropical world (10). In the Indian Ocean region, where the burden of malaria is restricted to the Comoros Archipelago and Madagascar (33), various intervention strategies are currently being implemented (35). The use of effective and well-tolerated antimalarial drugs is the mainstay of the armory for the control and elimination of Plasmodium falciparum malaria. Artemisinin combination therapies (ACTs) are used for the first-line treatment of P. falciparum infections, and the antifolate sulfadoxine-pyrimethamine (SP) combination is recommended for the intermittent preventive treatment of malaria during pregnancy (IPTp) (3). Indeed, SP, effective in reducing placental malaria and low birth weight, acts as a competitive inhibitor of two enzymes in the parasite's folate synthesis pathway: dihydrofolate reductase (DHFR) and dihydropteroate synthetase (DHPS), respectively (9). Nevertheless, the emergence and the spread of SP-resistant parasites remain major threats that could render this intervention ineffective (19). Amino acid changes at positions 51, 59, 108, and 164 in the P. falciparum dhfr (pfdhfr) gene are strongly associated with pyrimethamine treatment failures (6). Field surveys and experimental studies suggest a stepwise process of accumulation of mutations. The single S108N mutation confers increased in vitro resistance to pyrimethamine (∼20-fold), and subsequent mutations at position 51 (N51I) or 59 (C59R) further increase it (11). Parasites with a triple-mutation allele (51I/59R/108N) have markedly reduced in vitro susceptibility to pyrimethamine, and the presence of the triple-mutation allele increases the risk of SP therapeutic failure. Finally the quadruple-mutation allele, which carries an additional mutation at position 164 (I164L), is highly resistant to pyrimethamine, abrogating the clinical efficacy of SP, as observed in Southeast Asia and South America (26).
Recent progress in molecular population genetic studies has greatly facilitated our understanding of the emergence and geographical spread of drug-resistant lineages. In particular, it has been demonstrated that the emergence and dissemination of pyrimethamine-resistant parasites in Africa in the 1990s resulted from the migration of a few resistant mutants from Southeast Asia (29). Indeed, analysis of the microsatellite regions flanking the P. falciparum pfdhfr gene has clearly revealed that in Africa, the pfdhfr triple-mutation allele (I51/R59/N108) associated with pyrimethamine resistance harbored microsatellite haplotypes identical to those found in Southeast Asia (12, 13, 24, 29).
In the context of the Indian Ocean, SP resistance has been widely reported in the Comoros Islands (23, 25, 28, 32), whereas SP is still effective in Madagascar (18). However, recent studies performed in Madagascar have shown that the situation is deteriorating and have demonstrated the introduction of P. falciparum multidrug-resistant parasites into Madagascar from the Comoros Islands (17), the rapid rise in the frequency of P. falciparum parasites with both pfdhfr and dhps mutations, and the alarming emergence of the single pfdhfr 164L allele from isolates collected during the last 3 years (2).
In order to better understand the origin of the SP-resistant genotypes circulating in the region and determine the importance of gene flow in parasite populations with regard to SP resistance between Africa, the Comoros Islands, and Madagascar, we have characterized the pfdhfr genotype and flanking microsatellite haplotypes of a collection of P. falciparum samples from these areas. Our results confirm that pyrimethamine resistance in Madagascar is essentially related to the introduction from the Comoros Islands of the ancestral pfdhfr triple-mutation allele of Asian origin. Interestingly, however, the I164L single-mutation pfdhfr allele was observed in multiple lineages in areas restricted to the Southeast Madagascar, suggesting local pressure to generate this allele. The coexistence in the same transmission area of mutants with triple mutations and the single I164L mutation indicates that the local emergence of a mutant with quadruple mutations is a likely event that deserves reinforced surveillance.
MATERIALS AND METHODS
Collection of P. falciparum isolates.
Blood samples were collected from P. falciparum-infected patients seeking treatment for malaria at health government centers in the Comoros Islands and Madagascar. Patients with fever (axillary temperature ≥ 37.5°C) were screened by a rapid diagnostic test (RDT), based on the detection of Plasmodium-specific lactate dehydrogenase (pLDH; OptiMAL-IT; DiaMed AG, Cressier sur Morat, Switzerland). For each patient with a positive RDT result and after informed consent had been obtained, blood samples either were collected from a finger prick and placed onto filter paper or were collected by venipuncture and placed into EDTA-containing tubes. The patients were then promptly treated according to the national malaria policy with a combination of artemether plus lumefantrine (Coartem; Novartis, Basel, Switzerland) in the Comoros Islands (17) and a combination of artesunate plus amodiaquine (Arsucam, Sanofi-Aventis, France, Paris) in Madagascar (18). The study protocol was reviewed and approved by the Ethics Committee of the Ministry of Health of Madagascar (approval number 007/SANPF/2007; registration number ISRCTN36517335). Informed written consent was provided by all patients or their parents or guardians before inclusion in the study.
The collection of clinical isolates from the Comoros Islands was performed in May and June 2006 during a 2-month survey at six sites: Grande Comore Island (Moroni and Foumboni), Anjouan Island (Pomoni and Domoni), and Mohéli Island (Fomboni and Wanani). Isolates from Madagascar were collected between 2006 and 2008 during in vivo tests or were obtained from sites involved in the national network for the surveillance of malaria resistance (2). Venous blood samples collected in EDTA-containing tubes were transported to Antananarivo, Madagascar, at +4°C within 24 to 48 h of collection. Giemsa-stained blood smears were examined to check for monoinfection with P. falciparum and determination of the parasite density. The samples were stored at −20°C before genomic DNA extraction.
Additional isolates of P. falciparum from symptomatic P. falciparum-infected travelers returning to France from various African countries from 1997 to 2007 were obtained from the National Reference Centre for Malaria (NRCM), Paris, France. These samples were previously genotyped for pfdhfr and were found to have triple mutations (N51I, C59R, S108N) (5, 8, 12, 22). Reference strains from ATCC (Manassas, VA) carrying the wild-type pfdhfr allele (strain 3D7 from Africa) or the triple-mutation-type pfdhfr allele (strain W2 from Indochina and strain FCM29 from Cameroon) were also analyzed. The haplotypes of the microsatellites obtained from these samples were compared to those from the Indian Ocean.
DNA extraction.
Parasite DNA was extracted from blood spots by the use of Instagene Matrix resin (Bio-Rad, Marnes la Coquette, France), according to the manufacturer's instructions, or directly from 100 μl of infected blood, by using the phenol-chloroform method (27). The parasite species was confirmed by using real-time PCR, as described by de Monbrison et al. (7).
pfdhfr genotyping.
pfdhfr was amplified by a nested PCR approach. The PCR products were directly sequenced with an ABI Prism BigDye Terminator cycle sequencing ready reaction kit run on a 3730 xl genetic analyzer (Applied Biosystems, Courtaboeuf, France), as reported previously (2). Sequences of insufficient quality were either resequenced or rejected. The pfdhfr haplotypes for drug resistance markers were reconstructed from full sequences presenting an unambiguous single allele signal at each nucleotide position.
Microsatellite haplotyping.
Five microsatellite markers flanking the pfdhfr gene were used to determine the evolutionary history of the pyrimethamine resistance-conferring alleles. The number of AT repeats was assessed at 6.58, 4.58, and 1.14 kb upstream and 1.24 and 5.04 kb downstream of the pfdhfr gene, located on chromosome 4. Microsatellite polymorphism was analyzed by use of a nested PCR strategy. The first round of PCR amplification was performed with a 25-μl reaction mixture containing 0.5 μl DNA, 0.4 μM each primer, 250 μM each deoxynucleoside triphosphate (dNTP), 2.5 mM MgCl2, and 1.25 U TaKaRa DNA polymerase (ExTaq; Takara Bio Inc., Japan) under the following conditions: 94°C for 5 min, followed by 30 cycles of 94°C for 40 s, 50 to 53°C for 40 s, and 72°C for 60 s and a final extension at 72°C for 10 min. Nested PCR amplifications were performed in 55 μl reaction buffer with 2 μl of the primary PCR products, 0.4 μM each primer, 250 μM each dNTP, 2.5 mM MgCl2ç and 1.25 U TaKaRa DNA polymerase and the amplification conditions provided above for 30 cycles (see Table S1 in the supplemental material).
After purification by filtration with a NucleoFast 96 PCR plate (Macherey-Nagel, Düren, Germany), sequencing reactions were performed for both strands by using the ABI Prism BigDye Terminator cycle sequencing ready reaction kit run on a 3730 xl genetic analyzer (Applied Biosystems). Electrophoregrams were visualized and analyzed with CEQ2000 genetic analysis system software (Beckman Coulter). Nucleotide sequences were compared to the strain 3D7 pfdhfr sequence (GenBank accession number AL844503). As for pfdhfr genotyping, sequences of insufficient quality were either resequenced or rejected. Microsatellite haplotypes were reconstructed from the sequence presenting an unambiguous single signal at all nucleotide positions.
Haplotypes harboring an association of 8-13-17-16-15 AT repeats at microsatellite positions 6.58, 4.58, and 1.14 kb upstream and 1.24 and 5.04 kb downstream of the pfdhfr gene and those with one different microsatellite marker at the periphery (−6.58 kb, −4.58 kb, or +5.04 kb) were designated the “Southeast Asian haplotype” (SEA). Haplotypes with one different microsatellite marker just upstream or downstream of the pfdhfr coding region from the “SEA haplotype” were designated SEA-1. Those displaying variations at least at two microsatellite loci from the SEA haplotype were designated “local” (LOC).
Statistical analysis.
The expected heterozygosity (He) for estimation of the genetic variation for each microsatellite locus was calculated as [n/(n − 1)][1 − ∑pi2], where n is the number of isolates sampled and pi2 is the frequency of the ith allele, as determined by the use of Genetix software. The heterozygosity of microsatellites flanking each of the five pfdhfr alleles studied was calculated separately for each country.
RESULTS
pfdhfr genotype.
Among a total of 592 selected samples collected from 2006 to 2008 from the Comoros Islands and Madagascar, the pfdhfr gene and microsatellite loci of 322 (54.4%) were successfully amplified and the strains were included in the analysis. One hundred fifty-six (48.4%) of them carried the wild-type pfdhfr allele, 19 (5.9%) carried the S108N single-mutation allele, 30 (9.3%) carried the I164L single-mutation allele, 114 (35.4%) carried the N51I/C59R/S108N triple-mutation allele, and 3 (1.0%) carried the N51I/C59R/S108N/I164L quadruple-mutation allele. The spatiotemporal distribution of the various alleles is given in Table 1.
TABLE 1.
Distribution of the 322 P. falciparum pfdhfr alleles from the Comoros Islands and Madagascar collected in 2006 and 2007
| Country and site | No. of isolates with the following pfdhfr haplotypea: |
||||
|---|---|---|---|---|---|
| NCSI | NCNI | NCSL | IRNI | IRNL | |
| Comoros Islands | |||||
| Anjouanb | 13 | 4 | 0 | 10 | 0 |
| Grande Comoreb | 16 | 4 | 0 | 19 | 3 |
| Mohélib | 16 | 4 | 0 | 15 | 0 |
| Total | 45 | 12 | 0 | 44 | 3 |
| Madagascar | |||||
| North | |||||
| Antsirananac | 0 | 0 | 1 | 2 | 0 |
| Antsohihyc | 1 | 0 | 0 | 0 | 0 |
| Andapac | 8 | 1 | 0 | 0 | 0 |
| Northwest | |||||
| Mahajungac | 7 | 0 | 0 | 3 | 0 |
| Maevatananab,c | 17 | 1 | 0 | 16 | 0 |
| West-central highlands, Tsiroanomandidyb,c | 5 | 1 | 0 | 17 | 0 |
| East-central highlands, Moramangab,c | 17 | 0 | 0 | 4 | 0 |
| South-central highlands, Ihosyb,c | 14 | 0 | 1 | 3 | 0 |
| Central west | |||||
| Miandrivazob,c | 16 | 2 | 0 | 16 | 0 |
| Morondavac | 8 | 1 | 3 | 0 | 0 |
| Central east, Toamasinac | 2 | 0 | 1 | 0 | 0 |
| Southwest | |||||
| Ejedab,c | 6 | 1 | 1 | 2 | 0 |
| Tuléarc | 3 | 0 | 1 | 0 | 0 |
| Southeast | |||||
| Manakarac | 0 | 0 | 1 | 0 | 0 |
| Farafanganac | 7 | 0 | 21 | 7 | 0 |
| Total | 111 | 7 | 30 | 70 | 0 |
The amino acids conferring resistance are shown in underlined boldface. The four letter codes show the amino acid residues at positions 51, 59, 108, and 164, respectively.
Collection year, 2006.
Collection year, 2007.
Polymorphisms in microsatellite haplotypes.
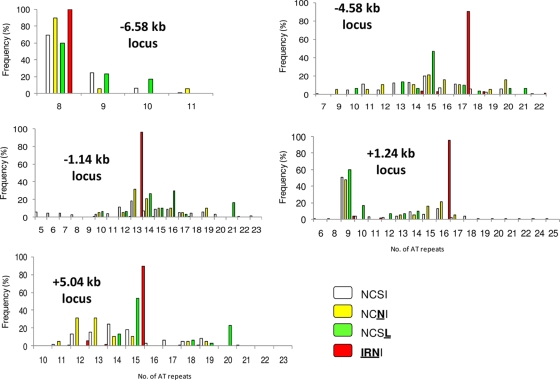
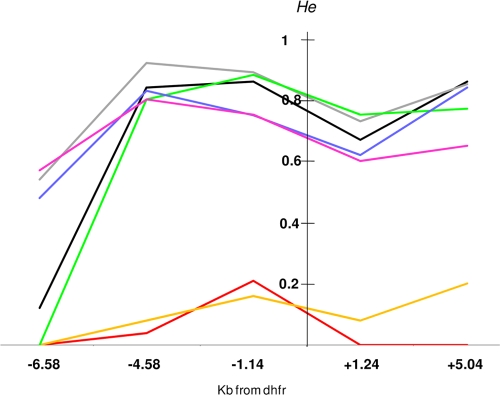
The polymorphisms of five microsatellite markers flanking the wild-type coding sequence and mutant-type alleles (with a single mutation to triple mutations) are shown in Fig. 1. The locus at −6.58 kb had 4 alleles with 8 to 11 AT repeats, the locus at −4.58 kb had 19 alleles with 5 to 23 AT repeats, the locus at −1.14 kb had 15 alleles with 7 to 22 AT repeats, the locus at +1.24 kb had 18 alleles with 6 to 25 AT repeats, and the locus at +5.04 kb had 14 alleles with 10 to 23 AT repeats. The microsatellite markers were highly polymorphic for parasites carrying the wild-type sequence. In contrast, triple-mutation pfdhfr alleles displayed a restricted microsatellite polymorphism at each locus. The expected He at each microsatellite locus of the Comorian and Malagasy isolates is shown in Fig. 2. In isolates carrying the wild-type sequence, He was high (0.67 to 0.92) at all five loci located between 4.58 kb upstream and 5.04 kb downstream of pfdhfr, except at the monomorphic locus at −6.58 kb. In isolates carrying the pfdhfr allele with a single mutation (S108N or I164L), He was also high at the four polymorphic loci (0.62 to 0.88 for S108N isolates and 0.60 to 0.80 for I164L isolates). In contrast, those isolates carrying the triple mutations in pfdhfr had very low He values (0 to 0.21) at all microsatellite loci, indicating limited diversity.
FIG. 1.
Distribution and prevalence of the five microsatellite (AT repeat) loci flanking the pfdhfr gene in 322 Plasmodium falciparum isolates collected from the Comoros Islands and Madagascar in 2006 and 2007. The amino acids conferring resistance are shown in underlined boldface.
FIG. 2.
Expected He of five microsatellite markers around pfdhfr alleles in P. falciparum collected in the Comoros Islands and Madagascar in 2006 and 2007.
Microsatellite haplotypes.
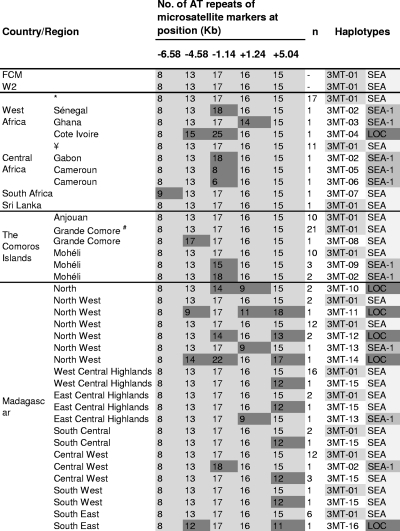
Thirty-five additional samples carrying the pfdhfr allele with a triple mutation (N51I/C59R/S108N) collected in different African countries (20 samples from West Africa, 14 samples from Central Africa, and 1 sample from South Africa) and 1 sample collected from Sri Lanka were selected from CNRP (Centre National de Référence du Paludisme). The microsatellite haplotypes of those samples were used to determine the importance of gene flow in parasite populations with regard to SP resistance between Africa, the Comoros Islands, and Madagascar. Descriptions of the five-locus microsatellite haplotypes of the wild-type allele and mutant-type alleles (a single mutation to quadruple mutations) are given in Fig. 3 and Tables S2 and S3 in the supplemental material and show large differences in haplotype diversity between allele groups. We observed 135 distinct haplotypes (WT1 to WT135; see Table S3 in the supplemental material) for 156 wild-type alleles, 15 haplotypes (108-1 to 108-15; see Table S2 in the supplemental material) for 19 isolates carrying the S108N single-mutation allele, 14 haplotypes (164-1 to 164-14; see Table S2 in the supplemental material) for 30 isolates with the I164L single-mutation allele, and only 16 different haplotypes (3MT-1 to 3MT-16; Fig. 3) for 114 isolates with triple-mutation alleles from Africa and the Indian Ocean samples.
FIG. 3.
P. falciparum pfdhfr flanking microsatellite haplotypes from isolates carrying triple mutant pfdhfr allele (51I/59R/108N) collected in the Comoros Islands and Madagascar in 2006 and 2007. *, isolates from Benin, Burkina Faso, Ivory Coast, Gambia, Guinea, Mali, Mauritania, Niger, Senegal, Sierra Leone, and Togo; ¥, isolates from Cameroon, the Central African Republic, and Congo; #, including the IRNL mutant with quadruple mutations (n = 3). The numbers of AT repeats in microsatellite markers (indicated in light gray boxes) corresponded to the number of AT repeats found in the SEA. Those displaying variations in the number of AT repeats are indicated in dark gray boxes. Haplotypes are numbered from 3MT-01 to 3MT-16. Haplotypes harboring an association of 8-13-7-16-15 AT repeats and those with one different microsatellite marker at the periphery (at the locus at −6.58 kb, −4.58 kb, or +5.04 kb) are designated SEA. Haplotypes with one different microsatellite marker just upstream or downstream of the pfdhfr-coding region are collectively designated SEA-1. Those displaying variations from the SEA haplotypes at least at two microsatellite loci are designated LOC.
All except 2 wild-type allele haplotypes were unique in the Comoros Islands (37 haplotypes/45 samples) and in Madagascar (98 haplotypes/111 samples). Two haplotypes were shared between the two countries (WT-01 between Anjouan and the central highlands and WT-17 between Grande Comore, the east-central highlands, and the north). Among the Comorian isolates, six haplotypes were shared between the islands: three haplotypes between Anjouan and Mohéli (WT-03, WT-09, and WT-10), two between Anjouan and Grand Comore (WT-05 and WT-15), and one between Grande Comore and Mohéli (WT-24). In Madagascar, four haplotypes were found in different regions: WT-51 (northwest, central west, and southwest), WT-52 (northwest and southwest), WT-83 (east-central highlands and southeast), and WT-88 (south-central highlands and central west).
No haplotype from isolates carrying the S108N allele was shared between countries or regions. Among the Malagasy isolates carrying the I164L allele, 14 different haplotypes were identified, suggesting the absence of a clonal expansion. The spread of this allele was limited to the areas of initial detection (164-2 in the west-central highlands; 164-5, 164-6, and 164-7/164-8 in the southeast) or to nearby regions (164-3 in the south-central highlands and southwest; 164-4 in the east-central highlands and southeast).
In the Comoros Islands, four microsatellite haplotypes associated with the triple-mutation allele were observed among the 44 isolates studied (Fig. 3). The SEA haplotype was the most prevalent (89%). The two other haplotypes (3MT-02 and 3MT-09) identified in Mohéli had a minor variation (one microsatellite locus change in the locus at −1.14 kb) from the SEA haplotype. In addition, the haplotype associated with the quadruple mutation was identical to the SEA haplotype. In Madagascar, 9 microsatellite haplotypes from the triple-mutation allele were observed among 70 isolates. The SEA haplotype was also the most prevalent (86%) and was observed in all the regions where samples were collected except the north. Two additional haplotypes with a minor variation from the SEA haplotype at one locus (3MT-13 with a change in the locus at +1.24 kb and 3MT-02 with a change in the locus at −1.14 kb) were identified (3/70, 4%). Only five haplotypes were considered the local haplotype. The local haplotype was very uncommon (10%) and was restricted to the north (3MT-10, n = 2), the northwest (3MT-11, n = 1; 3MT-12, n = 2l and 3MT-14, n = 1), and the southeast (3MT-16, n = 1).
DISCUSSION
To further document the rapid rise in the frequency of point mutations in the pfdhfr gene associated with pyrimethamine resistance that we reported previously (2), we have analyzed five microsatellite loci flanking the pfdhfr gene on chromosome 4 of wild-type and mutant-type alleles in a total of 357 P. falciparum isolates from the Comoros Islands (n = 94), Madagascar (n = 218), and other areas of Africa (n = 37). Consistent with the data from Southeast Asia (21), South America (14), and Africa (12, 13, 24, 29), we found evidence for the selective sweep of the pfdhfr triple-mutation allelic form. Indeed, most of the isolates carrying the triple-mutation allele from Africa (77%), the Comoros Islands (89%), and Madagascar (86%) shared the same SEA haplotype (the association of 8-13-17-16-15 AT repeats at microsatellite positions 6.58, 4.58, and 1.14 kb upstream and 1.24 and 5.04 kb downstream from pfdhfr, respectively) or a haplotype with one different microsatellite marker at the periphery (the locus at −6.58 kb, −4.58 kb, or +5.04 kb), further highlighting the importance of gene flow of the P. falciparum pyrimethamine-resistant populations between Asia, Africa, and the Indian Ocean. Moreover, most of the remaining haplotypes identified in our study presented only a minor variation at one locus compared with the sequence of the SEA haplotype (97% for African haplotypes, 100% for the Comorian haplotypes, and 90% for Malagasy haplotypes), consistent with the limited local evolution of the SEA pfdhfr mutant with triple mutations imported from Southeast Asia rather than de novo emergence from an indigenous lineage. Unlike Maiga et al. (12), the pfdhfr quadruple-mutation allele, observed here in three Comorian isolates, displayed the same flanking microsatellite signatures as the pfdhfr triple-mutation genotype; i.e., it was identical to the quadruple-mutation allele that arose in Southeast Asia (21), indicating that this Southeast Asian allele had spread to the Comoros Islands. These findings point to the existence of an efficient westward gene flow route across Asia, resulting in import into the Comoros (and other East African areas) of the pfdhfr triple and quadruple mutants and of the CVIET P. falciparum crt (pfcrt) chloroquine resistance-conferring allele (4).
The data reported here add support to our recent findings demonstrating the invasion of multidrug-resistant parasites into Madagascar from the Comoros Islands (17) and confirm the hypothesis that the Comoros Islands is a port of entry of antimalarial drug-resistant malaria parasites into the southwestern Indian Ocean. We have witnessed the rapid spread of the mutant with the pfdhfr triple mutation of the SEA lineage since 2006 along the north-to-south axis and its current widespread distribution in Madagascar. The present data, along with data from our previous studies (17, 18), show that the invasion of parasites harboring the pfdhfr triple-mutation allele into Madagascar is probably a recent event which is still in progress and confirm that gene flow is the major force driving this haplotype across continents and countries (1). Because of the massive use of SP in IPTp and because of human population movements, the prevalence of the pfdhfr triple-mutation allele may continue to increase in Madagascar, as it is not yet as high as the prevalence in the Comoros Islands or many other African countries. This is of major concern, in view of the excellent fitness of mutants with the pfdhfr triple mutation, even under conditions of low rates of pyrimethamine usage (30).
In addition to the haplotypes closely related to the triple-mutation SEA allele, we identified local haplotypes in African (Ivory Coast) and Malagasy mutant isolates, suggesting that the local evolutionary history may come into play as well and as documented elsewhere in Africa (12, 13, 15, 30) that pfdhfr triple-mutation alleles may also have indigenous multilineage origins. Absent from the Comorian isolates, the local haplotypes had a modest prevalence in Madagascar (10%), especially in the north of the country. In agreement with the findings of McCollum et al. (15), we hypothesize that the emergence of these additional novel haplotypes is favored by the combination of multiple local factors, such as the transmission level, the genetic diversity of the P. falciparum population, and the antifolate drug pressure.
The main result of the present study is the demonstration that the I164L mutation, which is so far unique to Madagascar, has appeared locally on multiple occasions on a wild-type background, as shown by the large diversity of flanking microsatellite haplotypes (a diversity comparable to that of the S108N single mutation). Lozovsky et al., using a transgenic bacterial system, did not find evidence for the diminished in vitro pyrimethamine susceptibility of the mutant with the pfdhfr I164L single mutation (11). If so, a parasite harboring such an allele is not predicted to be selected by antifolate therapeutic pressure. However, the distribution of I164L mutants with a relatively high prevalence in some sites of the southeast suggests some selective advantage with minioutbreaks. Additional work is needed to confirm this, as we cannot exclude the possibility that some additional genetic mechanism, such as the copy number polymorphism in the gene encoding GTP-cyclohydrolase I possibly influencing susceptibility to pyrimethamine, confers some advantageous effects on fitness to I164L mutant parasites which could explain their relative abundance and their increasing prevalence in the last 3 years (20, 34).
The coexistence of such I164L single-mutation alleles alongside mutants with triple mutations in Madagascar suggests that the epidemiological conditions are met for the local generation of quadruple mutants by de novo mutation or recombination, as has been suggested by McCollum et al. (15) for Kenyan isolates. Furthermore, since an SEA mutant with quadruple mutations has been observed in the Comoros Islands (although, fortunately, it is still rare), there is a substantial risk of the spread of pfdhfr quadruple-mutation alleles across Madagascar with SP treatment. This challenges the current recommendation of using SP for IPTp.
In conclusion, our data underscore the fact that the molecular mechanisms underlying antimalarial drug resistance are multifactorial and that the dispersal of drug resistance in Asia, South America, or Africa is fully comparable. It depends on many factors linked to the human hosts, vectors, and parasites. Although the spread of drug resistance-conferring alleles with particularly good fitness is common, the local emergence of drug resistance should not be ignored. Thus, specific studies are needed at the local level to follow the appearance of drug resistance and understand how the prevailing epidemiological conditions favor their spread. The results obtained here, which extend upon our previous findings (2), point to a clear threat to the efficacy of SP in Madagascar. Its life span is challenged both by the presence of mutants with triple pfdhfr mutations and by the risk of importation of the SEA mutant with quadruple mutations from the Comoros Islands and of the local generation of mutants with quadruple mutations. It is critical that the clinical efficacy of SP be carefully monitored in the Indian Ocean and that the evolution of its target genes in the region be documented to adjust accordingly the health care policies for IPTp and reduce the diffusion of resistant parasites.
Supplementary Material
Acknowledgments
We thank the patients and health care workers involved in the national network for the surveillance of malaria resistance in Madagascar (Réseau d'Etude de la Résistance [RER]) from which these samples were obtained and the staff of the Ministry of Health of Madagascar and of the Comoros Islands for their collaboration.
This study was supported by grants from the Institut de Médecine et d'Epidémiologie Appliquée (IMEA), Fondation Léon M'Ba, Paris, France, and the Genomics Platform, Pasteur Génopole, Pasteur Institute, France. Sample collection in Madagascar and the Comoros Islands was funded by the FSP/RAI 2001-168 project (French Ministry of Foreign Affairs) and the Global Fund to Fight AIDS, Tuberculosis and Malaria, round 3 (Community Action to Roll Back Malaria, grant no. MDG-304-G05-M) and in France by the Institut de Veille Sanitaire, French Ministry of Health. Valérie Andriantsoanirina is a graduate Ph.D. student funded by the Institut Pasteur de Madagascar (Bourse Girard) and the Direction des Affaires Internationales (Institut Pasteur).
Footnotes
Published ahead of print on 22 March 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Anderson, T. J., and C. Roper. 2005. The origins and spread of antimalarial drug resistance: lessons for policy makers. Acta Trop. 94:269-280. [DOI] [PubMed] [Google Scholar]
- 2.Andriantsoanirina, V., A. Ratsimbasoa, C. Bouchier, M. Jahevitra, S. Rabearimanana, R. Radrianjafy, V. Andrianaranjaka, T. Randriantsoa, M. A. Rason, M. Tichit, L. P. Rabarijaona, O. Mercereau-Puijalon, R. Durand, and D. Menard. 2009. Plasmodium falciparum drug resistance in Madagascar: facing the spread of unusual pfdhfr and pfmdr-1 haplotypes and the decrease of dihydroartemisinin susceptibility. Antimicrob. Agents Chemother. 53:4588-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aponte, J. J., D. Schellenberg, A. Egan, A. Breckenridge, I. Carneiro, J. Critchley, I. Danquah, A. Dodoo, R. Kobbe, B. Lell, J. May, Z. Premji, S. Sanz, E. Sevene, R. Soulaymani-Becheikh, P. Winstanley, S. Adjei, S. Anemana, D. Chandramohan, S. Issifou, F. Mockenhaupt, S. Owusu-Agyei, B. Greenwood, M. P. Grobusch, P. G. Kremsner, E. Macete, H. Mshinda, R. D. Newman, L. Slutsker, M. Tanner, P. Alonso, and C. Menendez. 2009. Efficacy and safety of intermittent preventive treatment with sulfadoxine-pyrimethamine for malaria in African infants: a pooled analysis of six randomised, placebo-controlled trials. Lancet 374:1533-1542. [DOI] [PubMed] [Google Scholar]
- 4.Ariey, F., T. Fandeur, R. Durand, M. Randrianarivelojosia, R. Jambou, E. Legrand, M. T. Ekala, C. Bouchier, S. Cojean, J. B. Duchemin, V. Robert, J. Le Bras, and O. Mercereau-Puijalon. 2006. Invasion of Africa by a single pfcrt allele of South East Asian type. Malar. J. 26:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aubouy, A., S. Jafari, V. Huart, F. Migot-Nabias, J. Mayombo, R. Durand, M. Bakary, J. Le Bras, and P. Deloron. 2003. DHFR and DHPS genotypes of Plasmodium falciparum isolates from Gabon correlate with in vitro activity of pyrimethamine and cycloguanil, but not with sulfadoxine-pyrimethamine treatment efficacy. J. Antimicrob. Chemother. 52:43-49. [DOI] [PubMed] [Google Scholar]
- 6.Cowman, A. F., M. J. Morry, B. A. Biggs, G. A. Cross, and S. J. Foote. 1988. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 85:9109-9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Monbrison, F., C. Angei, A. Staal, K. Kaiser, and S. Picot. 2003. Simultaneous identification of the four human Plasmodium species and quantification of Plasmodium DNA load in human blood by real-time polymerase chain reaction. Trans. R. Soc. Trop. Med. Hyg. 97:387-390. [DOI] [PubMed] [Google Scholar]
- 8.Durand, R., J. Eslahpazire, S. Jafari, J. F. Delabre, A. Marmorat-Khuong, J. P. di Piazza, and J. Le Bras. 2000. Use of molecular beacons to detect an antifolate resistance-associated mutation in Plasmodium falciparum. Antimicrob. Agents Chemother. 44:3461-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregson, A., and C. V. Plowe. 2005. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 57:117-145. [DOI] [PubMed] [Google Scholar]
- 10.Hay, S. I., C. A. Guerra, P. W. Gething, A. P. Patil, A. J. Tatem, A. M. Noor, C. W. Kabaria, B. H. Manh, I. R. Elyazar, S. Brooker, D. L. Smith, R. A. Moyeed, and R. W. Snow. 2009. A world malaria map: Plasmodium falciparum endemicity in 2007. PLoS Med. 6:e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozovsky, E. R., T. Chookajorn, K. M. Brown, M. Imwong, P. J. Shaw, S. Kamchonwongpaisan, D. E. Neafsey, D. M. Weinreich, and D. L. Hartl. 2009. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc. Natl. Acad. Sci. U. S. A. 106:12025-12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiga, O., A. A. Djimde, V. Hubert, E. Renard, A. Aubouy, F. Kironde, B. Nsimba, K. Koram, O. K. Doumbo, J. Le Bras, and J. Clain. 2007. A shared Asian origin of the triple-mutant dhfr allele in Plasmodium falciparum from sites across Africa. J. Infect. Dis. 196:165-172. [DOI] [PubMed] [Google Scholar]
- 13.McCollum, A. M., L. K. Basco, R. Tahar, V. Udhayakumar, and A. A. Escalante. 2008. Hitchhiking and selective sweeps of Plasmodium falciparum sulfadoxine and pyrimethamine resistance alleles in a population from central Africa. Antimicrob. Agents Chemother. 52:4089-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCollum, A. M., K. Mueller, L. Villegas, V. Udhayakumar, and A. A. Escalante. 2007. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob. Agents Chemother. 51:2085-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCollum, A. M., A. C. Poe, M. Hamel, C. Huber, Z. Zhou, Y. P. Shi, P. Ouma, J. Vulule, P. Bloland, L. Slutsker, J. W. Barnwell, V. Udhayakumar, and A. A. Escalante. 2006. Antifolate resistance in Plasmodium falciparum: multiple origins and identification of novel dhfr alleles. J. Infect. Dis. 194:189-197. [DOI] [PubMed] [Google Scholar]
- 16.McCoy, D., G. Kembhavi, J. Patel, and A. Luintel. 2009. The Bill & Melinda Gates Foundation's grant-making programme for global health. Lancet 373:1645-1653. [DOI] [PubMed] [Google Scholar]
- 17.Menard, D., A. E. Randrianarivo-Solofoniaina, B. S. Ahmed, M. Jahevitra, V. Andriantsoanirina, J. R. Rasolofomanana, and L. P. Rabarijaona. 2007. Drug-resistant malaria parasites introduced into Madagascar from Comoros Islands. Emerg. Infect. Dis. 13:1759-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menard, D., A. Ratsimbasoa, M. Randrianarivelojosia, L. P. Rabarijaona, L. Raharimalala, O. Domarle, L. Randrianasolo, A. Randriamanantena, M. Jahevitra, V. Andriantsoanirina, M. A. Rason, R. Raherinjafy, E. Rakotomalala, L. Tuseo, and A. Raveloson. 2008. Assessment of the efficacy of antimalarial drugs recommended by the National Malaria Control Programme in Madagascar: up-dated baseline data from randomized and multi-site clinical trials. Malar. J. 7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mita, T., K. Tanabe, and K. Kita. 2009. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol. Int. 58:201-209. [DOI] [PubMed] [Google Scholar]
- 20.Nair, S., B. Miller, M. Barends, A. Jaidee, J. Patel, M. Mayxay, P. Newton, F. Nosten, M. T. Ferdig, and T. J. Anderson. 2008. Adaptive copy number evolution in malaria parasites. PLoS Genet. 4:e1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair, S., J. T. Williams, A. Brockman, L. Paiphun, M. Mayxay, P. N. Newton, J. P. Guthmann, F. M. Smithuis, T. T. Hien, N. J. White, F. Nosten, and T. J. Anderson. 2003. A selective sweep driven by pyrimethamine treatment in Southeast Asian malaria parasites. Mol. Biol. Evol. 20:1526-1536. [DOI] [PubMed] [Google Scholar]
- 22.Nsimba, B., S. Jafari-Guemouri, D. A. Malonga, A. M. Mouata, J. Kiori, F. Louya, D. Yocka, M. Malanda, R. Durand, and J. Le Bras. 2005. Epidemiology of drug-resistant malaria in Republic of Congo: using molecular evidence for monitoring antimalarial drug resistance combined with assessment of antimalarial drug use. Trop. Med. Int. Health 10:1030-1037. [DOI] [PubMed] [Google Scholar]
- 23.Parola, P., B. Pradines, F. Simon, M. P. Carlotti, P. Minodier, M. P. Ranjeva, S. Badiaga, L. Bertaux, J. Delmont, M. Morillon, R. Silai, P. Brouqui, and D. Parzy. 2007. Antimalarial drug susceptibility and point mutations associated with drug resistance in 248 Plasmodium falciparum isolates imported from Comoros to Marseille, France in 2004 2006. Am. J. Trop. Med. Hyg. 77:431-437. [PubMed] [Google Scholar]
- 24.Pearce, R., A. Malisa, S. P. Kachur, K. Barnes, B. Sharp, and C. Roper. 2005. Reduced variation around drug-resistant dhfr alleles in African Plasmodium falciparum. Mol. Biol. Evol. 22:1834-1844. [DOI] [PubMed] [Google Scholar]
- 25.Pettinelli, F., M. E. Pettinelli, P. Eldin de Pecoulas, J. Millet, D. Michel, P. Brasseur, and P. Druilhe. 2004. Short report: high prevalence of multidrug-resistant Plasmodium falciparum malaria in the French territory of Mayotte. Am. J. Trop. Med. Hyg. 70:635-637. [PubMed] [Google Scholar]
- 26.Plowe, C. V., J. G. Kublin, and O. K. Doumbo. 1998. P. falciparum dihydrofolate reductase and dihydropteroate synthase mutations: epidemiology and role in clinical resistance to antifolates. Drug Resist. Updat. 1:389-396. [DOI] [PubMed] [Google Scholar]
- 27.Rakotonirina, H., C. Barnadas, R. Raherijafy, H. Andrianantenaina, A. Ratsimbasoa, L. Randrianasolo, M. Jahevitra, V. Andriantsoanirina, and D. Menard. 2008. Accuracy and reliability of malaria diagnostic techniques for guiding febrile outpatient treatment in malaria-endemic countries. Am. J. Trop. Med. Hyg. 78:217-221. [PubMed] [Google Scholar]
- 28.Randrianarivelojosia, M., R. H. Raherinjafy, R. Migliani, O. Mercereau-Puijalon, F. Ariey, and S. A. Bedja. 2004. Plasmodium falciparum resistant to chloroquine and to pyrimethamine in Comoros. Parasite 11:419-423. [DOI] [PubMed] [Google Scholar]
- 29.Roper, C., R. Pearce, S. Nair, B. Sharp, F. Nosten, and T. Anderson. 2004. Intercontinental spread of pyrimethamine-resistant malaria. Science 305:1124. [DOI] [PubMed] [Google Scholar]
- 30.Sandefur, C. I., J. M. Wooden, I. K. Quaye, W. Sirawaraporn, and C. H. Sibley. 2007. Pyrimethamine-resistant dihydrofolate reductase enzymes of Plasmodium falciparum are not enzymatically compromised in vitro. Mol. Biochem. Parasitol. 154:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snow, R. W., C. A. Guerra, J. J. Mutheu, and S. I. Hay. 2008. International funding for malaria control in relation to populations at risk of stable Plasmodium falciparum transmission. PLoS Med. 5:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tall, A., L. P. Rabarijaona, V. Robert, S. A. Bedja, F. Ariey, and M. Randrianarivelojosia. 2007. Efficacy of artesunate plus amodiaquine, artesunate plus sulfadoxine-pyrimethamine, and chloroquine plus sulfadoxine-pyrimethamine in patients with uncomplicated Plasmodium falciparum in the Comoros Union. Acta Trop. 102:176-181. [DOI] [PubMed] [Google Scholar]
- 33.Tchen, J., A. Ouledi, J. F. Lepere, D. Ferrandiz, and J. L. Yvin. 2006. Epidemiology and prevention of malaria in the southwestern islands of the Indian Ocean. Med. Trop. (Mars.) 66:295-301. (In French.) [PubMed] [Google Scholar]
- 34.Volkman, S. K., P. C. Sabeti, D. DeCaprio, D. E. Neafsey, S. F. Schaffner, D. A. Milner, Jr., J. P. Daily, O. Sarr, D. Ndiaye, O. Ndir, S. Mboup, M. T. Duraisingh, A. Lukens, A. Derr, N. Stange-Thomann, S. Waggoner, R. Onofrio, L. Ziaugra, E. Mauceli, S. Gnerre, D. B. Jaffe, J. Zainoun, R. C. Wiegand, B. W. Birren, D. L. Hartl, J. E. Galagan, E. S. Lander, and D. F. Wirth. 2007. A genome-wide map of diversity in Plasmodium falciparum. Nat. Genet. 39:113-119. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization. 2008. World malaria report 2008. World Health Organization, Geneva, Switzerland.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.