Abstract
In glycopeptide-resistant enterococci and staphylococci, high-level resistance is achieved by replacing the C-terminal d-alanyl-d-alanine of lipid II with d-alanyl-d-lactate, thus reducing glycopeptide affinity for cell wall targets. Reorganization of the cell wall in these organisms is directed by the vanHAX gene cluster. Similar self-resistance mechanisms have been reported for glycopeptide-producing actinomycetes. We investigated glycopeptide resistance in Nonomuraea sp. ATCC 39727, the producer of the glycopeptide A40926, which is the precursor of the semisynthetic antibiotic dalbavancin, which is currently in phase III clinical trials. The MIC of Nonomuraea sp. ATCC 39727 toward A40926 during vegetative growth was 4 μg/ml, but this increased to ca. 20 μg/ml during A40926 production. vanHAX gene clusters were not detected in Nonomuraea sp. ATCC 39727 by Southern hybridization or by PCR with degenerate primers. However, the dbv gene cluster for A40926 production contains a gene, vanY (ORF7), potentially encoding an enzyme capable of removing the terminal d-Ala residue of pentapeptide peptidoglycan precursors. Analysis of UDP-linked peptidoglycan precursors in Nonomuraea sp. ATCC 39727 revealed the predominant presence of the tetrapeptide UDP-MurNAc-l-Ala-d-Glu-meso-Dap-d-Ala and only traces of the pentapeptide UDP-MurNAc-l-Ala-d-Glu-meso-Dap-d-Ala-d-Ala. This suggested a novel mechanism of glycopeptide resistance in Nonomuraea sp. ATCC 39727 that was based on the d,d-carboxypeptidase activity of vanY. Consistent with this, a vanY-null mutant of Nonomuraea sp. ATCC 39727 demonstrated a reduced level of glycopeptide resistance, without affecting A40926 productivity. Heterologous expression of vanY in a sensitive Streptomyces species, Streptomyces venezuelae, resulted in higher levels of glycopeptide resistance.
Actinomycetes are Gram-positive mycelial bacteria with a complex life cycle that consists of vegetative growth followed by the formation of aerial hyphae and ultimately spore formation, the last allowing both dispersal and persistence under unfavorable conditions. The onset of morphological differentiation generally coincides with the production of secondary metabolites, including many antibiotics of immense clinical and commercial importance. Antibiotic-producing actinomycetes must possess mechanisms to avoid suicide by their own toxic products. Several such resistance mechanisms have evolved, including target modification, antibiotic inactivation or sequestration, and efflux mechanisms. Microorganisms produce secondary metabolites mainly during the stationary phase of growth, and resistance genes are often coregulated with those for antibiotic production (8, 28). Glycopeptide antibiotics are frequently used to treat life-threatening infections caused by multidrug-resistant Gram-positive pathogens. Vancomycin, introduced into clinical practice in 1958, is produced by the actinomycete Amycolatopsis orientalis, whereas teicoplanin, used in European hospitals since 1988, is made by Actinoplanes teichomyceticus (23). Both compounds inhibit the late stages of peptidoglycan assembly by forming complexes with the d-alanyl-d-alanine (d-Ala-d-Ala) C termini of the peptidoglycan precursors on the external side of the cell membrane (29). The formation of these complexes prevents the cross-linking reaction catalyzed by transglycosylases, d,d-transpeptidases, and d,d-carboxypeptidases, thus hampering cell wall assembly. In vancomycin-resistant enterococci (VRE) and staphylococci (VRSA), resistance to glycopeptides is mediated by the synthesis of modified peptidoglycan precursors (29). The synthesis of the alternative peptidoglycan precursors requires the coordinate action of several enzymes encoded by the van genes (29). vanH encodes a dehydrogenase (α-keto acid reductase) that reduces pyruvate to d-lactate (d-Lac). vanA encodes a ligase that catalyzes the formation of the d-Ala-d-Lac depsipeptide, which replaces the dipeptide d-Ala-d-Ala in peptidoglycan synthesis. Glycopeptide resistance also requires the presence of the d,d-dipeptidase VanX, which selectively removes the intracellular pool of d-Ala-d-Ala produced by the native enterococcal ligase, ensuring that the d-Ala-d-Lac depsipeptide is incorporated into the peptidoglycan precursor. VanR and VanS constitute a two-component regulatory system that activates transcription of the vanHAX gene cluster (17, 29).
Marshall et al. (27) reported that many glycopeptide-producing actinomycetes possess genes with homology to the van genes of VRE. Cloning and sequencing of gene clusters for the biosynthesis of the glycopeptide antibiotics balhimycin (bal), chloroeremomycin (cep), A40926 (dbv), A47934 (sta), and teicoplanin (tcp) revealed the presence of vanHAX genes in both the sta and tcp gene clusters (10). In A. teichomyceticus (24, 33) and in the A47934 producer Streptomyces toyocaensis NRRL 15009 (30), vanHAX homologues are located at the 5′ ends of the tcp and sta gene clusters. More recently, constitutive expression of vanHAX genes was reported for A. teichomyceticus (4), while inducible expression of the same genes was found in the non-glycopeptide producer Streptomyces coelicolor (18, 19).
Nonomuraea sp. ATCC 39727 produces the teicoplanin-like glycopeptide antibiotic A40926 (12). A40926 is the precursor of the semisynthetic derivative dalbavancin that is currently in phase III clinical trials (7). The gene cluster for A40926 biosynthesis (dbv) does not possess homologues of vanHAX but contains vanY (ORF7) (34), homologues of which were identified previously in enterococci (11). vanY encodes a putative carboxypeptidase that could potentially cleave the d-Ala-d-Ala termini of peptidoglycan precursors outside the cell membrane (1, 2). A potential role for vanY in glycopeptide resistance in the A40926 producer was recently reinforced by the preferential accumulation of the tetrapeptide UDP precursor (instead of the pentapeptide) during cell wall synthesis in Nonomuraea sp. ATCC 39727 (25). In this paper, we describe the glycopeptide resistance phenotype of Nonomuraea sp. ATCC 39727 and investigate the role of vanY in glycopeptide resistance.
MATERIALS AND METHODS
Strains and cultural conditions.
The A40926 producer Nonomuraea sp. ATCC 39727 was maintained as a lyophilized master cell bank (MCB). The mycelium from the MCB was streaked on slants of salt medium (SM) ([in g/liter] glucose, 10; Bacto-peptone, 4; Bacto-yeast extract, 4; MgSO4·7H2O, 0.5; KH2PO4, 2; K2HPO4, 4; deionized water up to 1 liter) solidified with agar (15 g/liter). After growth, the mycelium from a slant was homogenized in 10 ml of physiological solution (0.9% [wt/vol] NaCl), inoculated into liquid SM, grown for 96 h at 28°C with aeration, and stored as a working cell bank (WCB) in 1.5-ml cryo-vials at −80°C. The WCB was used to inoculate all of the cultures described here. Unless otherwise specified, cultures were grown in media (SM or SM/agar) in which production of A40926 did not occur (as determined by high-pressure liquid chromatography [HPLC] analysis of culture extracts; see below). These cultures/media are defined as “vegetative” to distinguish them from cultures/media that support A40926 production.
Streptomyces venezuelae ATCC 10595 (37) was maintained as spores in 10% glycerol (22). Agar medium used for its propagation was MYM (37). Media and culture conditions for Escherichia coli were described in reference 9. E. coli DH5α (Invitrogen) was used as a host for plasmid constructions. E. coli ET12567/pUZ8002 (13) was the nonmethylating plasmid donor strain for intergeneric conjugation with Nonomuraea sp. ATCC 39727, as described in the work of G. L. Marcone, L. Foulston, E. Binda, F. Marinelli, M. J. Bibb, and F. Beltrametti (submitted for publication). Carbenicillin (Carb; 100 μg/ml), apramycin (Apra; 50 μg/ml), chloramphenicol (Cm; 25 μg/ml), and kanamycin (Kan; 50 μg/ml), all from Sigma, were added to growth media when required. l-Arabinose (10 mM final concentration; Sigma) was added as indicated to SOB medium (14) from a 1 M sterile (filtered) stock solution to induce genes under the control of the pBAD promoter (9).
Vectors.
Cosmid A40Y (G. L. Marcone et al., submitted for publication), derived from SuperCos1 (Stratagene), includes 22 kb of the dvb cluster (34) and 8 kb from SuperCos1. It does not contain an origin of replication for actinomycetes. pIJ86 (gift from Mervyn Bibb) was used as a multicopy vector for heterologous expression in Streptomyces venezuelae.
E. coli BW25113 [Δ(araD-araB)567 ΔlacZ4787(::rrnB-4) lacIp-4000(lacIq) λ− rpoS369(Am) rph-1 Δ(rhaD-rhaB)568 hsdR514] (9) was used to propagate the recombination plasmid pIJ790 [oriR101 repA101(Ts) araBp-gam-bet-exo] and cosmid A40Y. BW25113 was grown in SOB (14) or LB (Sigma). E. coli DH5α was used to propagate pIJ773 [pBluescript KS(+), aac(3)IV, oriT (RK2), and FRT sites] (13).
DNA extraction, Southern hybridization, and PCR amplification.
For DNA extraction, 2 ml WCB of Nonomuraea sp. ATCC 39727 was inoculated into 100 ml of SM in a 500-ml baffled flask containing 5-ml sterile glass beads (diameter, 4 mm) and incubated at 28°C for 72 h on a rotary shaker at 200 rpm. The mycelium was recovered by centrifugation at 3,250 × g for 15 min.
Total DNA was isolated by the Kirby procedure (22), digested, and analyzed by Southern hybridization according to standard methods (31). Probes from van genes were obtained by PCR amplification of the A. teichomyceticus ATCC 31121 vanHAX (vanHatAatXat) genes. vanHat was amplified with primers vanHFw (primer coordinate 12057 5′-AGCGCGGACGCCATCCGT-3′ 12075) and vanHRev (13256 5′-CCCATGCTGTTGTTCCCT-3′ 13238); vanAat with primers vanAFw (13269 5′-CGGCATCATCTTCGGCG-3′ 13286) and vanARev (14273 5′-AGGGCCAGCGACACGAT-3′ 14256); and vanXat with primers vanXFw (14301 5′-TTCGCGTTCGTGGACGAG-3′ 14319) and vanXRev (14897 5′-CTACACGATCGGAAAATCAAA-3′ 14876). The primer coordinates given above correspond to those of GenBank accession no. AJ605139.
For van gene mining, PCR amplifications were performed using primer pairs and conditions reported by Marshall et al. (27), as well as the primer pairs described above for the A. teichomyceticus van genes.
Replacement of vanY.
vanY (dbv ORF7) was replaced with an apramycin resistance gene [aac(3)IV] using PCR-targeting mutagenesis (13). For gene replacement, two long PCR primers (58 nucleotides [nt] and 59 nt) were made. Each of them possessed at the 5′ end 39 nt matching the Nonomuraea sp. ATCC 39727 sequence adjacent to vanY (shown in bold in the following primer sequences) and a 3′ sequence (19 nt or 20 nt) matching the right or left end of the disruption cassette. The oligonucleotides used were 59nt Fw (5′-AGAACTGGTGTCAGATCACCAGACTGGAGGAGAGGGATGATTCCGGGGATCCGTCGACC-3′) and 58nt Rev (5′-CCGGGGACCGATCCCCGCGGCGACCCCGTGCCCTAGC TATGTAGGCTGGAGCTGCTTC-3′). The aac(3)IV cassette was amplified by PCR using pIJ773 as a template and the above primers. The resulting PCR product was purified using a gel extraction kit (Qiagen). To induce the replacement of the targeted gene with the apramycin resistance gene cassette by λ red recombination, cosmid A40Y, carrying vanY, was introduced into E. coli BW25113/pIJ790, and the resulting transformant (E. coli BW25113/pIJ790/A40Y) was cultured in SOB-Mg2+ medium containing carbenicillin (50 μg/ml), kanamycin (50 μg/ml), chloramphenicol (25 μg/ml), and 20 mM l-arabinose at 30°C to an optical density at 600 nm (OD600) of 0.6. Electrocompetent cells were generated from the induced culture, and the purified aac(3)IV cassette was introduced by electroporation. Transformants were selected on LB agar plates containing apramycin (50 μg/ml) and carbenicillin (100 μg/ml) incubated overnight at 37°C. Gene replacement in the mutated cosmid (A40ΔY) (G. L. Marcone et al., submitted for publication) was confirmed by restriction enzyme analysis and by PCR using the following oligonucleotides, which anneal 100 to 200 bp upstream and downstream of the 39-bp recombination sites: vanY Fw (5′-GTGGCACTTCCGCTGCTAGT-3′) and vanY rev (5′-CTGGAGTTCGTCTTCCGCTA-3′). Confirmed mutant cosmids were introduced into E. coli ET12567/pUZ8002 (methylation-negative strain) (13) by electroporation and then transferred into the mycelium of Nonomuraea sp. ATCC 37927 by conjugation (G. L. Marcone et al., submitted for publication). Exconjugants were selected by culturing on MV0.1 agar plates supplemented with 10 mM MgCl2 at 30°C for 24 h and then overlaying the plates with apramycin (25 μg/ml) and nalidixic acid (20 μg/ml). Colonies that developed on the plates were transferred to MV0.1 agar plates containing apramycin (25 μg/ml) and incubated at 30°C for 10 to 15 days (G. L. Marcone et al., submitted for publication). Replacement of the entire coding sequence (591 bp) of vanY with aac(3)IV (1,377 bp) was confirmed by sequencing. Nonomuraea sp. ATCC 39727 ΔvanY was maintained and cultivated as described for the wild-type strain.
Heterologous expression of vanY in Streptomyces venezuelae.
The vanY open reading frame was amplified from Nonomuraea sp. ATCC 39727 genomic DNA using Expand High Fidelity polymerase (Roche) and vanY86Fw (5′-ATGGATCCCAGACTGGAGGAGAGGGATG-3′) and vanY86Rev (5′-GATAAGCTTCGATCCTGGAGTTCGTCTTC-3′) oligonucleotide primers that introduced BamHI and HindIII restriction sites (in bold), respectively, into the PCR product, allowing insertion into the multiple cloning site of the multicopy expression vector pIJ86. The PCR product was purified, digested with BamHI and HindIII, and ligated with pIJ86 that had similarly been digested to produce pIJ86ΩvanY. pIJ86ΩvanY, with vanY transcribed from the strong constitutive ermE* promoter (22) was used to transform E. coli ET12567/pUZ8002. Conjugations were performed according to standard protocols (22) using E. coli ET12567/pUZ8002/pIJ86ΩvanY as the donor and S. venezuelae as the recipient. The resulting transconjugants were selected on MYM agar medium, containing apramycin and nalidixic acid.
Determination of MICs and PAPs.
MICs were extrapolated from population analysis profile experiments (PAPs) as the concentrations inhibiting growth of 99.99% of the colonies growing on solid media. Mycelia for PAPs of Nonomuraea sp. ATCC 39727 and its ΔvanY mutant were prepared as follows. Cryo-vials of the WCB were thawed at room temperature, and 2 ml was used to inoculate 100 ml of SM medium. Strains were grown to exponential phase (approximately 48 h) at 28°C with shaking. The mycelium was harvested by centrifugation, suspended in physiological solution (0.9% [wt/vol] NaCl), and fragmented by sonication with a Vibracell Albra sonicator 400 W model as previously described (3, 6). The mycelium was stored either in 1-ml aliquots at −80°C or used immediately. Sonicated hyphae were then seeded onto SM agar plates supplemented with different glycopeptide concentrations. Up to 107 CFU were spread per plate and incubated at 28°C for 15 days. After incubation, colonies were counted and the surviving fraction of the population for each antibiotic concentration was determined.
PAPs for S. venezuelae and its derivatives were determined similarly, but with 107 spores of each strain plated on MYM agar containing appropriate levels of antibiotics and then incubated for 3 days.
Induction of glycopeptide resistance was investigated with Nonomuraea sp. ATCC 39727 cultures grown as described above and then incubated for 24 h with sub-MIC concentrations of vancomycin (12 μg/ml), teicoplanin (0.25 μg/ml), or A40926 (0.5 μg/ml). The MICs and PAPs for the different glycopeptides were then determined as described above. Minimal bactericidal concentrations (MBC) were estimated as follows. The sonicated mycelium was inoculated into a set of SM medium flasks and allowed to grow for a time interval corresponding to three generations. Increasing antibiotic concentrations were added to the flasks, and incubation was allowed for a further 24 h. The mycelium was then washed with physiologic solution to eliminate exogenous glycopeptide, diluted, and plated on SM agar plates without antibiotic. Colonies were allowed to grow at 28°C for ca. 15 days and then the surviving fraction corresponding to each original antibiotic concentration was determined. Antibiotics that gave less than 0.1% survival under the conditions described above were considered bactericidal.
A40926 production.
Fermentations for A40926 production by Nonomuraea sp. ATCC 39727 and from its ΔvanY mutant were carried out with slight modification of the protocols described previously (3, 6). In brief, one vial of the WCB was inoculated into 100 ml of vegetative medium E25 (5) in 500-ml baffled flasks. Strains were grown for 72 to 96 h on a rotary shaker at 200 rpm and 28°C. Fermentation was started by adding a 10% inoculum from the vegetative medium flask into production medium T/2 (5) in a 2-liter working volume P-100 Applikon glass reactor (height, 25 cm; diameter, 13 cm) equipped with an AD1030 Biocontroller and AD1032 motor. Cultivations in fermentors were carried out at 30°C, with stirring at 500 to 700 rpm (corresponding to 1.17 to 1.64 m/s of tip speed) and 2 liters/min aeration rate. Foam production was controlled by the addition of Hodag antifoam through an antifoam sensor. A total of 25 ml of each culture was extracted every day for 1 week of fermentation. Biomass production was estimated as dry weight after 24-h incubation in an 80°C oven. Mycelia from fermentor cultures were processed for MIC determination as described above.
A40926 was extracted by mixing 1 volume of mycelium and 3 volumes of borate buffer (100 mM H3BO3, 100 mM NaOH, pH 12). Samples were then centrifuged (16,000 × g for 15 min) and incubated for 1 h at 50°C. The glycopeptide-containing supernatant was filtered through a Durapore membrane filter (0.45 μm) (Millipore). Glycopeptide production was estimated by HPLC performed on a 5-μm-particle-size Ultrasphere ODS (Beckman) column (4.6 by 250 mm) eluted at a flow rate of 1 ml/min with a 26-min linear gradient from 25% to 37% of phase B. Phase A was 20 mM HCOONH4 (pH 4.5)-CH3CN (95:5 [vol/vol]), and Phase B was 20 mM HCOONH4 (pH 4.5)-CH3CN (5:95 [vol/vol]) mixture. Chromatography was performed with a VWR Hitachi diode array L-2455 HPLC system with detection at 254 nm. Pure samples of A40926 were used as an internal standard (3, 6).
Peptidoglycan precursor extraction and analysis.
Extraction of UDP-linked peptidoglycan precursors was performed as already described (4, 25). In brief, mycelium was harvested by centrifugation, suspended in distilled water at a final concentration of 0.1 g of fresh weight per ml of water, and boiled for 20 min. The mycelium was then brought to room temperature and incubated in ice prior to centrifugation at 39,000 × g for 1 h. The supernatant was filtered through a 0.22-μm membrane, lyophilized, and then dissolved in 0.1 volume of water adjusted to pH 3 with formic acid. The samples were analyzed by reversed-phase HPLC and electrospray ionization mass spectrometry (ESI-MS) on an LCQ-Deca spectrometer equipped with an ion-trap analyzer (Thermo Finnigan, San Jose, CA) as described previously (4, 25).
RESULTS AND DISCUSSION
Resistance to glycopeptides in Nonomuraea sp. ATCC 39727.
There are few detailed studies of resistance phenotypes in glycopeptide-producing microorganisms. In mycelial actinomycetes, the standard method used in unicellular bacteria to determine MICs is compromised by the formation of multicellular aggregates and by the coexistence of cells in different physiological states (e.g., vegetative mycelium, aerial mycelium, and spores).
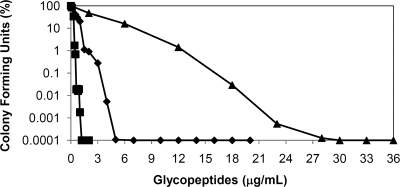
In our experiments with Nonomuraea sp. ATCC 39727, the most reliable method for MIC determination was the population analysis profile (PAP), which has been used to identify subpopulations or small differences in resistance levels. The MICs under these conditions are those concentrations which inhibit the growth of 99.99% of the population (15, 16). Under these conditions, Nonomuraea sp. ATCC 39727 showed moderate resistance to glycopeptides, with MIC values of 4, 0.9, and 19.5 μg/ml for A40926, teicoplanin, and vancomycin, respectively, as shown in Fig. 1. These levels of glycopeptide resistance are markedly lower than those reported for the glycopeptide producer A. teichomyceticus (MIC values of 20, 25, and 90 μg/ml for A40926, teicoplanin, and vancomycin, respectively) (4) and for the nonproducer S. coelicolor (MICs of 0.5 and 100 μg/ml for teicoplanin and vancomycin, respectively [19]), which contains a complete set of van genes. When MICs were determined after growth of Nonomuraea sp. ATCC 39727 in the presence of subinhibitory concentrations of A40926 (0.5 μg/ml), teicoplanin (0.25 μg/ml), or vancomycin (12 μg/ml), induction of A40926 resistance was observed but only in the presence of A40926. No changes in A40926 MICs were observed in the presence of the other glycopeptides (Table 1). These data suggest that A40926 is able to specifically induce self-resistance in the producing organism. Induction of resistance could be coregulated with the onset of antibiotic production or induced by increasing levels of A40926 biosynthesis. While the presence of subinhibitory concentrations of vancomycin in the PAP plates did not increase teicoplanin or A40926 resistance, both A40926 and teicoplanin induced an increase in vancomycin resistance (Table 1). This resistance profile is different from the vancomycin-inducible resistance described for S. coelicolor, in which vanHAX expression is induced by vancomycin but not by teicoplanin (19). It also differs from the regulation of glycopeptide resistance described for A. teichomyceticus, in which the vanHAX genes are transcribed constitutively (4). The differential responses of actinomycetes, enterococci, and staphylococci to glycopeptides belonging to the vancomycin or teicoplanin classes pose questions about the nature and specificity of the inducer and its putative receptor, which have not been resolved (21). The two classes of glycopeptides, while sharing the same molecular target (the d-Ala-d-Ala C terminus of the peptidoglycan precursor), interact in a different manner with it. Vancomycin is known to bind cooperatively to the target in the form of dimers, whereas teicoplanin and A40926 are localized at the site of action by their lypophilic tails, which anchor them to the cell membrane (21). These distinct structural features and the consequent differences in localization and local concentration of glycopeptides at the target site may be responsible for the differences in their abilities to induce resistance or to inhibit growth in susceptible strains.
FIG. 1.
Population analysis profile of Nonomuraea sp. ATCC 39727. Colony forming units in the presence of A40926 (♦), teicoplanin (▪), and vancomycin (▴) are shown. Results are the average of three independent experiments in which the standard deviation (SD) was less than 5%.
TABLE 1.
Glycopeptide resistance after induction with subinhibitory concentrations of A40926, teicoplanin, or vancomycin
| Glycopeptide used for inductiona | MICb |
||
|---|---|---|---|
| A40926 | Teicoplanin | Vancomycin | |
| None | 4.0 | 0.9 | 20.0 |
| A40926 | 5.0 | 0.9 | 35.0 |
| Teicoplanin | 4.0 | 0.9 | 27.0 |
| Vancomycin | 4.0 | 0.9 | 20.0 |
Concentrations were as follows: A40926, 0.5 μg/ml; teicoplanin, 0.25 μg/ml; or vancomycin, 12 μg/ml.
MICs (μg/ml) are calculated from PAPs as those concentrations which inhibit the growth of 99.99% of the population. Results are the average of three independent experiments in which the standard deviation (SD) was less than 5%.
Self-resistance during A40926 production.
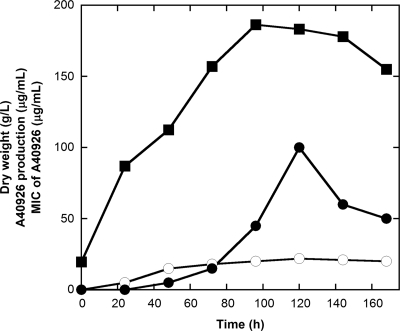
Resistance could be coregulated with endogenous antibiotic production in producing microorganisms, and an increase in self-resistance during antibiotic production has indeed been reported previously (8, 28). Figure 2 shows the correlation between growth, A40926 production, and MICs at different stages of Nonomuraea sp. ATCC 39727 during flask cultivation. The MIC for A40926 at the beginning of fermentation was approximately the same as that for vegetative cultures (5 μg/ml) and then increased progressively to 22 μg/ml during production phase (from 48 to 120 h of fermentation). Maximum A40926 productivity reached 100 μg/ml after 120 h of cultivation. Again, the A40926 producer differs from the teicoplanin producer A. teichomyceticus, whose MICs to teicoplanin during production phase did not change significantly (4).
FIG. 2.
A40926 resistance during fermentation of Nonomuraea sp. ATCC 39727. Symbols: ▪, dry weight; •, A40926 production; ○, MIC values. Results are the average of three independent experiments in which the SD was less than 5%.
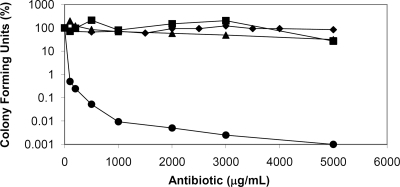
Notwithstanding the increase in resistance during A40926 production, the maximal MIC is about 5-fold lower than the maximal level of A40926 production. Given that glycopeptides are reported to exert a bactericidal action in pathogens (23), the minimal bactericidal concentrations (MBCs) of A40926, teicoplanin, and vancomycin were determined (Fig. 3). Nonomuraea sp. ATCC 39727, as previously reported for A. teichomyceticus (4), survived 24 h of exposure to A40926, teicoplanin, and vancomycin at 5,000 μg/ml, while the control antibiotic kanamycin showed bactericidal action at much lower concentrations. From these experiments we conclude that A40926 and the other tested glycopeptides are not bactericidal for Nonomuraea sp. ATCC 39727. That is, while the strain failed to grow in the presence of more than 5 and 20 μg/ml of A40926 during vegetative and production phases, respectively, it could resume growth once the antibiotic was removed. Although the maximum level of A40926 production (ca. 100 μg/ml) was much higher than the MIC, it occurred in stationary phase when little or no growth and cell wall biosynthesis presumably occurred. Thus, the level of A40926 resistance exceeds production levels during growth, falling below exogenous levels as the strain enters stationary phase, in which nonproliferating cells are tolerant to high concentrations of their own product. Interestingly, when Pootoolal and coworkers (30) knocked out the vanA-like gene that confers A47934 resistance in the producing strain, S. toyocaensis NRRL15009, the mutant retained the ability to produce the antibiotic, but its production was delayed by over 16 h, perhaps reflecting a mechanism that prevents A47934 production until cells have ceased growing and are predicted to be insensitive to antibiotic action.
FIG. 3.
Minimal bactericidal concentrations (MBCs) of A40926 (♦), teicoplanin (▪), and vancomycin (▴). The glycopeptides do not show bactericidal activity against Nonomuraea sp. ATCC 39727, in contrast to the nonglycopeptide control antibiotic kanamycin (•). Results are the average of three independent experiments in which the SD was less than 5%.
van genes in Nonomuraea sp. ATCC 39727.
In Nonomuraea sp. ATCC 39727, A40926 biosynthesis, regulation, and export are encoded by the dbv gene cluster (34). The results reported above suggested that A40926 self-resistance might be coregulated with A40926 production. However, analysis of the dbv cluster failed to reveal homologues of vanHAX (34). Consequently, we screened the Nonomuraea sp. ATCC 39727 genome for the presence of vanHAX genes by Southern hybridization and by PCR amplification. Degenerate primers (described in reference 27) and the vanHatAatXat genes of A. teichomyceticus (32) were used as probes; the latter were chosen because of the taxonomical closeness of A. teichomyceticus and Nonomuraea sp. ATCC 39727, which both produce chemically similar glycopeptides. No van gene homologues were identified (data not shown). Moreover, data from genome sequencing of Nonomuraea sp. ATCC 39727 indicate a lack of significant homology with van genes from A. teichomyceticus and enterococci (Pietro Alifano, personal communication). However, the dbv cluster does contain a homologue of vanY (ORF7), a putative d-alanyl-d-alanine carboxypeptidase (34), that might play a role in glycopeptide resistance. Analysis of VanY (196 amino acids, protein identification no. [id] CAD91202.1) revealed that, despite the low percentage of overall sequence identity between VanY from high-GC-content Nonomuraea sp. ATCC 39727 and low-GC Enterococcus faecalis (32%) or Enterococcus faecium (27%), the key active site residues (SXHXXGXAXD and EXXH) are conserved. This implies a similar mechanism of action and substrate binding in VanY from Nonomuraea sp. ATCC 39727 and in VanY proteins belonging to the Zn2+-dependent d,d-carboxypeptidases previously described for enterococci (1, 2).
Analysis of cell wall precursors in Nonomuraea sp. ATCC 39727.
In vanA and vanB enterococci, VanY contributes to high-level vancomycin resistance in the presence of the vanHAX cluster by cleaving the C-terminal residue of peptidoglycan precursors ending in d-Ala-d-Ala or d-Ala-d-Lac but not the dipeptide d-Ala-d-Ala (1, 2). However, the presence of VanY is not essential for vancomycin resistance; clinical isolates of VRE in which vanY has been disrupted by insertion elements are still vancomycin resistant. VanX and VanY could play distinct roles in conferring glycopeptide resistance by hydrolysis of d-Ala-d-Ala and by the removal of d-Ala from membrane-bound lipid intermediates, respectively (1, 2).
A potential role for VanY in glycopeptide resistance in Nonomuraea sp. ATCC 39727 was also indicated by the detection of peptidoglycan UDP-MurNAc-tetrapeptide precursors during cell wall biosynthesis (25). Using a liquid chromatography (LC)-MS method previously applied to the analysis of cell wall biosynthesis in A. teichomyceticus and in S. coelicolor (4), we blocked cell wall assembly in Nonomuraea sp. ATCC 39727 by the addition of ramoplanin, which inhibits peptidoglycan transglycosylases and leads to the accumulation of cell wall precursors (which are undetectable in untreated cells because of their rapid assembly into peptidoglycan) (20). LC-MS analysis of cytoplasmic pools of peptidoglycan precursors in ramoplanin-treated cells revealed two molecular species. One was attributable to a pentapeptide precursor (UDP-N-acetylmuramyl-l-Ala-d-Glu-m-Dap-d-Ala-d-Ala) identical to that described for S. coelicolor (18), whereas the other corresponded to the tetrapeptide peptidoglycan precursor UDP-N-acetylmuramyl-l-Ala-d-Glu-m-Dap-d-Ala lacking the terminal d-alanine. Quantification of the areas of the LC-MS peaks, corresponding to tetrapeptide and pentapeptide precursors, respectively (25), indicated that the tetrapeptide precursor predominated in nonproducing vegetative cultures (Table 2). The ratio between tetrapeptide and pentapeptide precursors increased following induction by A40926 and teicoplanin. During synthesis of A40926 in production medium, we detected the exclusive accumulation of the tetrapeptide precursor, while the pentapeptide was below the limit of detection (data not shown). These data suggested a possible mechanism for self-resistance in Nonomuraea sp. ATCC 39727 that is based on the elimination of the pentapeptide and its replacement with the tetrapeptide, which has a lower affinity for glycopeptides. Variations in the tetrapeptide/pentapeptide ratio correlated well with the resistance profile described above, which showed induction by A40926 and an increased level of resistance during A490926 production. No UDP-linked depsipeptide containing either d-Lac or d-Ser in the terminal position of the UDP precursor was detected in Nonomuraea sp. ATCC 39727, suggesting the absence of mechanisms of resistance based on the modification of the cell wall similar to those found in the van systems of enterococci (11), S. coelicolor (18), and A. teichomyceticus (4).
TABLE 2.
Accumulation of UDP-linked cell wall precursors during Nonomuraea sp. ATCC 39727 growth in vegetative medium
| Inducer | UDP-muramyl pentapeptide (A)a | UDP-muramyl tetrapeptide (B) | B/A ratio |
|---|---|---|---|
| None | 15.4 × 105 | 49.2 × 105 | 3.2 |
| A40926 | 99.9 × 104 | 91.7 × 105 | 9.2 |
| Teicoplanin | 84 × 104 | 70.7 × 105 | 8.5 |
| Vancomycin | 48 × 104 | 28 × 105 | 5.83 |
A and B give the area values (calculated as intensity of total ion current per second) of the LC-MS peaks eluting at 12.56 and 13.40 min (25), corresponding to the tetrapeptide and pentapeptide precursors, respectively.
Analysis of a vanY mutant of Nonomuraea sp. ATCC 39727.
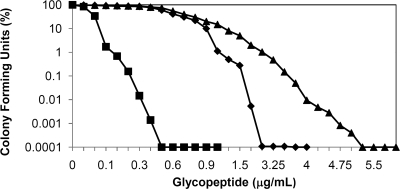
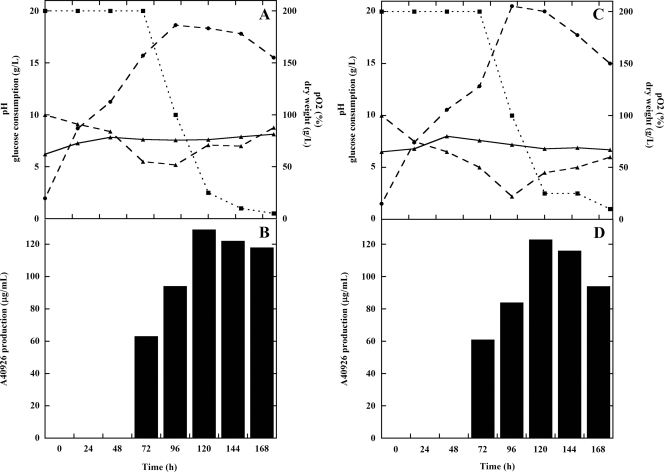
To confirm a role for vanY in glycopeptide resistance in Nonomuraea sp. ATCC 39727, we took two parallel approaches: deletion of the gene in the producing organism and heterologous expression of vanY (see below). To date, genetic manipulation of Nonomuraea sp. ATCC 39727 has been hampered by the lack of molecular tools, and only recently have methods for protoplast transformation and intergeneric conjugation from E. coli been developed (25, 35, 36; G. L. Marcone et al., submitted for publication). Inactivation of vanY was accomplished by PCR-targeting (13). A total of 55 independent apramycin-resistant Nonomuraea sp. ATCC 39727 exconjugants were obtained and screened for kanamycin sensitivity, revealing 18 putative double-crossover mutants, each of which was confirmed by PCR analysis. DNA sequencing of three of these confirmed the construction of the vanY mutant. Five of the 18 clones were then used to characterize the vanY resistance phenotype during vegetative growth. The MICs in these five independent ΔvanY clones for A40926, teicoplanin, and vancomycin were 2, 0.3, and 4 μg/ml, respectively (as shown by the PAPs reported in Fig. 4), showing a reduction in resistance compared to the parental strain (MICs for A40926, teicoplanin, and vancomycin of 4, 0.9, and 19.5 μg/ml, respectively). These results clearly indicated a role for VanY in self-resistance. Since antibiotic production and self-resistance are often coregulated in producing actinomycetes, we investigated if vanY inactivation had any effect on growth and on A40926 production in Nonomuraea sp. ATCC 39727. No effect was observed when five independent vanY mutants were compared with the wild-type strain in flask fermentations (data not shown). Figure 5 shows fermentor data for one of the five ΔvanY clones compared to those of the wild-type strain. Growth, glucose consumption, pH, dissolved oxygen, and A40926 production profiles were similar in both strains.
FIG. 4.
Population analysis profile of Nonomuraea sp. ATCC 39727 ΔvanY mutant. Colony-forming units in the presence of A40926 (♦), teicoplanin (▪), and vancomycin (▴) are shown. Results are the average of three independent experiments in which the SD was less than 5%.
FIG. 5.
Comparison of A40926 production in 2-liter-batch fermentations of Nonomuraea sp. ATCC 39727 (A and B) and the ΔvanY mutant (C and D). Panels A and C: time course of pH, ▴, solid line; pO2, ▴, dashed line; glucose, ▪, dotted line; growth curve measured as dry weight, •, dashed line. Panels B and D, production of A40926 measured by HPLC analysis expressed as μg/ml, filled bars.
Heterologous expression of vanY in Streptomyces venezuelae and analysis of the resulting resistance phenotype.
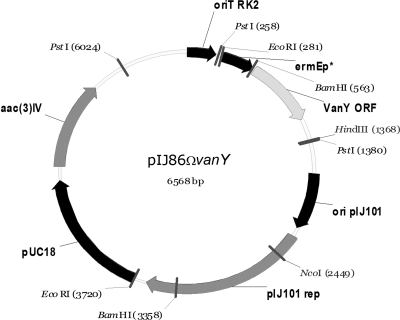
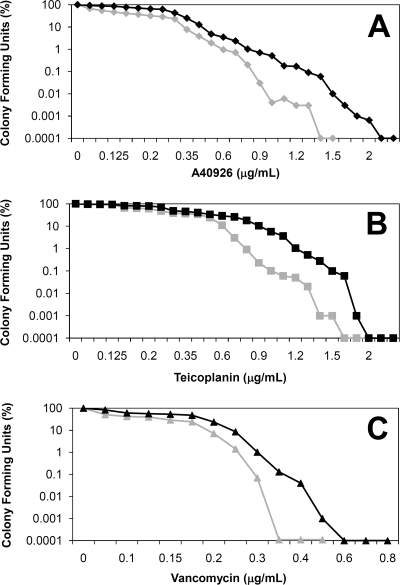
vanY was cloned in the multicopy vector pIJ86, which contains the constitutive ermE* promoter to drive the expression of the inserted gene. pIJ86ΩvanY (Fig. 6) was then introduced into Streptomyces venezuelae by conjugation from E. coli. S. venezuelae was selected as host for heterologous expression since, unlike S. coelicolor, it does not possess vanHAX genes and is very sensitive to glycopeptides. PAPs of the pIJ86ΩvanY transformants and of the parental strain indicated that expression of vanY increased the level of resistance to A40926, teicoplanin, and vancomycin (Fig. 7). These data confirmed the role of vanY in conferring glycopeptide resistance in the absence of vanHAX genes.
FIG. 6.
Map of pIJ86ΩvanY plasmid.
FIG. 7.
Population analysis profile of Streptomyces venezuelae containing pIJ86 (light lines) or pIJ86ΩvanY (dark lines). Colony-forming units in the presence of A40926 (A), teicoplanin (B), and vancomycin (C) are shown. Results are the average of three independent experiments in which the SD was less than 5%.
Conclusions.
vanHAX gene clusters similar to those first described for enterococci were found in several glycopeptide producing actinomycetes (26, 27, 30, 32). Recently, we described constitutive glycopeptide resistance in the teicoplanin producer A. teichomyceticus ATCC 31121 (4), which also contains a vanHAX gene cluster directing the synthesis of a peptidoglycan precursor terminating in d-Ala-d-Lac (24, 32, 33). The glycopeptide A40926 is produced by Nonomuraea sp. ATCC 39727 and belongs to the same lipoglycopeptide family as teicoplanin. Despite the similarities between the two antibiotics, neither a typical vanHAX gene cluster nor a similar glycopeptide resistance phenotype was apparent in Nonomuraea sp. ATCC 39727. In this work, we have demonstrated the involvement of vanY, usually considered an ancillary but not essential resistance gene in enterococci, in determining the resistance of Nonomuraea sp. ATCC 39727 to glycopeptides. This mechanism of glycopeptide resistance is based on the cleavage of the terminal d-alanine residue from the peptidoglycan precursor. The tetrapeptide produced by the action of the VanY d,d-carboxypeptidase is a poor substrate for glycopeptide binding, thus reducing the level of susceptibility to A40926, teicoplanin, and vancomycin.
This resistance mechanism plays an important role in the initial phases of A490926 production when cells of the producer organism are actively growing, and its induction appears to be coregulated with the onset of antibiotic production. vanY is located in the dbv cluster for A40926 biosynthesis (34), but its deletion does not affect glycopeptide biosynthesis. This is consistent with the observation that vanY is transcribed from its own promoter and in the direction opposite to most of the other contiguous genes in the dbv biosynthetic cluster. Specific induction of resistance by A40926 itself and during antibiotic production suggests induction of vanY transcription by increasing endogenous levels of A40926. During stationary phase, tolerance, rather than resistance, accommodates the high concentrations of glycopeptides achieved, as demonstrated in other glycopeptide producers with similarly complex life cycles (4).
In addition to revealing new fundamental insights, a thorough understanding of A40926 resistance in the producing microorganism may prove useful in the future surveillance of emerging mechanisms of resistance to clinically used glycopeptide antibiotics. Moreover, it may reveal new understanding of the molecular mechanisms linking antibiotic production to self-resistance, thus enabling knowledge-based approaches to strain improvement for glycopeptide-producing actinomycetes.
Acknowledgments
This work was supported by FAR 2007-2008-2009 to F.M. and by the MIUR fellowship to G.L.M. We also thank the Consorzio Interuniversitario per le Biotecnologie (CIB) and Progetto Cariplo, Promuovere Capitale Umano d'Eccellenza, for their support and for the fellowships to G.L.M. and E.B. L.F., A.H., and M.B. were funded by grants from the Biotechnology and Biological Sciences Research Council, United Kingdom.
Footnotes
Published ahead of print on 22 March 2010.
REFERENCES
- 1.Arthur, M., F. Depardieu, L. Cabanie, P. Reynolds, and P. Courvalin. 1998. Requirement of the VanY and VanX d,d-peptidases for glycopeptide resistance in enterococci. Mol. Microbiol. 30:819-830. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, M., F. Depardieu, H. A. Snaith, P. E. Reynolds, and P. Courvalin. 1994. Contribution of VanY d,d-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob. Agents Chemother. 38:1899-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltrametti, F., A. Lazzarini, C. Brunati, E. Selva, and F. Marinelli. 2003. Production of demannosyl-A40926 by a Nonomuraea sp. ATCC 39727 mutant strain. J. Antibiot. (Tokyo) 56:310-313. [DOI] [PubMed] [Google Scholar]
- 4.Beltrametti, F., A. Consolandi, L. Carrano, F. Bagatin, R. Rossi, L. Leoni, E. Zennaro, E. Selva, and F. Marinelli. 2007. Resistance to glycopeptide antibiotics in the teicoplanin producer is mediated by van gene homologue expression directing the synthesis of a modified cell wall peptidoglycan. Antimicrob. Agents Chemother. 51:1135-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltrametti, F., S. Jovetic, M. Feroggio, L. Gastaldo, E. Selva, and F. Marinelli. 2004. Valine influences production and complex composition of glycopeptide antibiotic A40926 in fermentations of Nonomuraea sp. ATCC 39727. J. Antibiot. (Tokyo). 57:37-44. [DOI] [PubMed] [Google Scholar]
- 6.Beltrametti, F., A. Lazzarini, C. Brunati, A. Marazzi, S. Jovetic, E. Selva, and F. Marinelli. 2003. Production and characterization of monochlorinated and dechlorinated A40926 derivatives. J. Antibiot. (Tokyo). 56:773-782. [DOI] [PubMed] [Google Scholar]
- 7.Cornaglia, G., and G. M. Rossolini. 2009. Forthcoming therapeutic perspectives for infections due to multidrug-resistant Gram-positive pathogens. Clin. Microbiol. Infect. 15:218-223. [DOI] [PubMed] [Google Scholar]
- 8.Cundliffe, E. 1992. Self-protection mechanisms in antibiotic producers. Ciba Found. Symp. 171:199-214. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donadio, S., M. Sosio, E. Stegmann, T. Weber, and W. Wohlleben. 2005. Comparative analysis and insights into the evolution of gene clusters for glycopeptide antibiotic biosynthesis. Mol. Genet. Genomics 274:40-50. [DOI] [PubMed] [Google Scholar]
- 11.Evers, S., R. Quintiliani, Jr., and P. Courvalin. 1996. Genetics of glycopeptide resistance in enterococci. Microb. Drug Resist 2:219-223. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein, B. P., E. Selva, L. Gastaldo, M. Berti, R. Pallanza, F. Ripamonti, P. Ferrari, M. Denaro, V. Arioli, and G. Cassani. 1987. A40926, a new glycopeptide antibiotic with anti-Neisseria activity. Antimicrob. Agents Chemother. 31:1961-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 15.Hiramatsu, K. 1999. Mechanisms of methicillin and vancomycin resistance in Staphylococcus aureus. Clin. Infect. Dis. 5:221-237. [Google Scholar]
- 16.Hiramatsu, K. 1998. Vancomycin resistance in Staphylococcus aureus. Drug Resist. Updat. 1:135-150. [DOI] [PubMed] [Google Scholar]
- 17.Hong, H. J., M. I. Hutchings, and M. J. Buttner. 2008. Vancomycin resistance VanS/VanR two-component systems. Adv. Exp. Med. Biol. 631:200-213. [DOI] [PubMed] [Google Scholar]
- 18.Hong, H. J., M. I. Hutchings, L. M. Hill, and M. J. Buttner. 2005. The role of the novel Fem protein VanK in vancomycin resistance in Streptomyces coelicolor. J. Biol. Chem. 280:13055-13061. [DOI] [PubMed] [Google Scholar]
- 19.Hong, H. J., M. I. Hutchings, J. M. Neu, G. D. Wright, M. S. Paget, and M. J. Buttner. 2004. Characterization of an inducible vancomycin resistance system in Streptomyces coelicolor reveals a novel gene (vanK) required for drug resistance. Mol. Microbiol. 52:1107-1121. [DOI] [PubMed] [Google Scholar]
- 20.Hu, Y., J. S. Helm, L. Chen, X. Y. Ye, and S. Walker. 2003. Ramoplanin inhibits bacterial transglycosylases by binding as a dimer to lipid II. J. Am. Chem. Soc. 125:8736-8737. [DOI] [PubMed] [Google Scholar]
- 21.Kahne, D., C. Leimkuhler, W. Lu, and C. Walsh. 2005. Glycopeptide and lipoglycopeptide antibiotics. Chem. Rev. 105:425-448. [DOI] [PubMed] [Google Scholar]
- 22.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 23.Lancini, G., and B. Cavalleri. 1990. Glycopeptide antibiotics of the vancomycin group, p. 159-178. In H. Kleinkauf and H. V. Dohren (ed.), Biochemistry of peptide antibiotics. Walter de Gruyter, Berlin, Germany.
- 24.Li, T. L., F. Huang, S. F. Haydock, T. Mironenko, P. F. Leadlay, and J. B. Spencer. 2004. Biosynthetic gene cluster of the glycopeptide antibiotic teicoplanin: characterization of two glycosyltransferases and the key acyltransferase. Chem. Biol. 11:107-119. [DOI] [PubMed] [Google Scholar]
- 25.Marcone, G. L., L. Carrano, F. Marinelli, and F. Beltrametti. 2010. Protoplast preparation and reversion to the normal filamentous growth in antibiotic-producing uncommon actinomycetes. J. Antibiot. (Tokyo) 63:83-88. [DOI] [PubMed] [Google Scholar]
- 26.Marshall, C. G., G. Broadhead, B. K. Leskiw, and G. D. Wright. 1997. d-Ala-d-Ala ligases from glycopeptide antibiotic-producing organisms are highly homologous to the enterococcal vancomycin-resistance ligases VanA and VanB. Proc. Natl. Acad. Sci. U. S. A. 94:6480-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall, C. G., I. A. Lessard, I. Park, and G. D. Wright. 1998. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 42:2215-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olano, C., F. Lombo, C. Mendez, and J. A. Salas. 2008. Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab. Eng. 10:281-292. [DOI] [PubMed] [Google Scholar]
- 29.Pootoolal, J., J. Neu, and G. D. Wright. 2002. Glycopeptide antibiotic resistance. Annu. Rev. Pharmacol. Toxicol. 42:381-408. [DOI] [PubMed] [Google Scholar]
- 30.Pootoolal, J., M. G. Thomas, C. G. Marshall, J. M. Neu, B. K. Hubbard, C. T. Walsh, and G. D. Wright. 2002. Assembling the glycopeptide antibiotic scaffold: the biosynthesis of A47934 from Streptomyces toyocaensis NRRL15009. Proc. Natl. Acad. Sci. U. S. A. 99:8962-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Serina, S., F. Radice, S. Maffioli, S. Donadio, and M. Sosio. 2004. Glycopeptide resistance determinants from the teicoplanin producer Actinoplanes teichomyceticus. FEMS Microbiol. Lett. 240:69-74. [DOI] [PubMed] [Google Scholar]
- 33.Sosio, M., H. Kloosterman, A. Bianchi, P. de Vreugd, L. Dijkhuizen, and S. Donadio. 2004. Organization of the teicoplanin gene cluster in Actinoplanes teichomyceticus. Microbiology 150:95-102. [DOI] [PubMed] [Google Scholar]
- 34.Sosio, M., S. Stinchi, F. Beltrametti, A. Lazzarini, and S. Donadio. 2003. The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by Nonomuraea species. Chem. Biol. 10:541-549. [DOI] [PubMed] [Google Scholar]
- 35.Stinchi, S., S. Azimonti, S. Donadio, and M. Sosio. 2003. A gene transfer system for the glycopeptide producer Nonomuraea sp. ATCC 39727. FEMS Microbiol. Lett. 225:53-57. [DOI] [PubMed] [Google Scholar]
- 36.Stinchi, S., L. Carrano, A. Lazzarini, M. Feroggio, A. Grigoletto, M. Sosio, and S. Donadio. 2006. A derivative of the glycopeptide A40926 produced by inactivation of the beta-hydroxylase gene in Nonomuraea sp. ATCC 39727. FEMS Microbiol. Lett. 256:229-235. [DOI] [PubMed] [Google Scholar]
- 37.Stuttard, C. 1982. Temperate phages of Streptomyces venezuelae: lysogeny and host specificity shown by SV1 and SV2. J. Gen. Microbiol. 128:115-121. [Google Scholar]