Abstract
Bovine-origin Escherichia coli isolates were tested for resistance phenotypes using a disk diffusion assay and for resistance genotypes using a DNA microarray. An isolate with gentamicin and amikacin resistance but with no corresponding genes detected yielded a 1,056-bp DNA sequence with the closest homologues for its inferred protein sequence among a family of 16S rRNA methyltransferase enzymes. These enzymes confer high-level aminoglycoside resistance and have only recently been described in Gram-negative bacteria.
Aminoglycosides interfere with bacterial 16S rRNA function by binding at the site where codon-anticodon accuracy is assessed (the A site) (14). In Gram-negative pathogens, resistance to aminoglycosides is mediated primarily by enzymes that modify the drug by acetylation, adenylylation, or phosphorylation and less commonly by other methods, including efflux mechanisms (27). Aminoglycoside-producing bacteria (Streptomyces and Micromonospora species) have intrinsic resistance to aminoglycosides through methylation of nucleotides within the A site of 16S rRNA, preventing disruption of translation by the aminoglycoside. The earliest reports of clinical Gram-negative isolates with plasmid-borne rRNA methylase aminoglycoside resistance genes were from Japan (33, 35) and France (14), followed by Taiwan (34), Spain (15, 16), South Korea (19), Belgium (4), and China (7). All were bacteria infecting human patients, except for reports from Spain (16) and China (7) of similar genes in Escherichia coli and Enterobacter cloacae isolates from swine.
As part of a larger study, 81 cattle origin commensal E. coli isolates were assayed for the presence of antibiotic resistance genes using a DNA oligonucleotide microarray (9a). The E. coli isolates used in this study were from different animals on the same farm or, if from the same animal, had different resistance phenotypes. The microarray includes 30 probes for detecting aminoglycoside resistance genes, five of which code for amikacin resistance and 10 for gentamicin resistance (Table 1). The results of the array hybridizations were compared to the resistance phenotypes as measured by a standard disk diffusion assay (2). The panel of antimicrobial disks included four aminoglycosides, amikacin (30 μg), gentamicin (10 μg), kanamycin (30 μg), and streptomycin (10 μg). The resistance breakpoints were those recommended by the Clinical and Laboratory Standards Institute (9). Among the 81 E. coli isolates, 11 isolates were phenotypically resistant to amikacin, gentamicin, kanamycin, and streptomycin, but no amikacin resistance genes were detected by microarray assay. Nine of these isolates were from separate animals, and two were from the same animal but had different resistance profiles (Table 2).
TABLE 1.
Aminoglycoside resistance genes represented by probes on the resistance gene microarray
| Gene | Accession no. | Target(s) | Enzyme encoded | Reference |
|---|---|---|---|---|
| aac(3)-Ia | DQ370505 | Gentamicin | 3-N-Acetyltransferase | 29 |
| aac(3)-Ib | L06157 | Gentamicin | 3-N-Acetyltransferase | 13 |
| aac(3)-Id | AY458224 | Gentamicin | 3-N-Acetyltransferase | 13 |
| aac(3)-III | X13542 | Gentamicin | 3-N-Acetyltransferase | 1 |
| aac(3)-IVa | X01385 | Gentamicin | 3-N-Acetyltransferase | 8 |
| aac(3)-Vb | M97172 | Gentamicin | 3-N-Acetyltransferase | 24 |
| aac(6′)-I30 | AY289608 | Amikacin | 6′-N-Acetyltransferase | 22 |
| aac(6′)-Ib | AY103455 | Amikacin, kanamycin | 6′-N-Acetyltransferase | 13 |
| aac(6′)-IIa | AY123251 | Gentamicin | 6′-N-Acetyltransferase | 22 |
| aac(6′)-Ia | M18967 | Amikacin, kanamycin | 6′-N-Acetyltransferase | 28 |
| aacC1 | U04610 | Gentamicin | 3-N-Acetyltransferase | 13 |
| aacC2 | S68058 | Gentamicin | 3-N-Acetyltransferase | 32 |
| aacCA5 | AY463797 | Gentamicin, kanamycin, amikacin | 3-N-Acetyltransferase | 22 |
| aadA1 | EF422367 | Streptomycin | 3″-Adenylyltransferase | 32 |
| aadA2 | AF071555 | Streptomycin | 3″-Adenylyltransferase | 5 |
| aadA21 | AY171244 | Streptomycin | 3″-Adenylyltransferase | 22 |
| aadA5 | AB126604 | Streptomycin | 3″-Adenylyltransferase | 22 |
| aadA7 | AY458224 | Streptomycin | 3″-Adenylyltransferase | 12 |
| aadB | AY204504 | Gentamicin, kanamycin | 2″-Adenylyltransferase | 13 |
| aadE | AF516335 | Streptomycin | 6-Adenylyltransferase | 13 |
| aph(3)-Ia | V00359 | Kanamycin | 3′-Phosphotransferase | 6 |
| aph(3)-IIa | V00618 | Kanamycin | 3′-Phosphotransferase | 6 |
| aph4 | V01499 | Hygromycin B | Aminocyclitol phosphotransferase | 20 |
| aphA-3 | AF516335 | Kanamycin | 3′-Phosphotransferase | 13 |
| aphA7 | AY509004 | Kanamycin | 3′-Phosphotransferase | 22 |
| aphD | Y00459 | Streptomycin | 6-Phosphotransferase | 10 |
| aphE | X53527 | Streptomycin | 3″-Phosphotransferase | 30 |
| aphIII | V01547 | Kanamycin | 3′-Phosphotransferase | 17 |
| strA | AY055428 | Streptomycin | 6-Phosphotransferase | 3 |
| strB | NC_005014 | Streptomycin | 6-Phosphotransferase | 3 |
TABLE 2.
Resistance gene microarray hybridization results for E. coli isolates having an aminoglycoside resistance phenotype not explained by any genes detected using the oligonucleotide microarray
| Isolate | Unexplained resistance phenotypea | aac(3)-III | aac(6′)-IIa | aac(3)-IVa | aac(3)-IVa | aacC2 | aadA1 | aadA2 | aadA21 | aadA5 | aph(3′)-Ia | aph4 | aphA7 | strA | strB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1090 | Gentamicin, amikacin | + | + | + | + | + | + | ||||||||
| 2517 | Gentamicin, amikacin | + | + | + | + | ||||||||||
| 2521 | Gentamicin, amikacin | + | + | + | |||||||||||
| 2534 | Amikacin | + | + | + | + | + | + | + | |||||||
| 2538 | Amikacin | + | + | + | + | + | + | + | + | ||||||
| 2545 | Amikacin | + | + | + | + | + | + | + | |||||||
| 2550b | Gentamicin, amikacin | + | + | + | + | + | |||||||||
| 2551b | Amikacin | + | + | + | + | + | + | + | + | ||||||
| 2577 | Gentamicin, amikacin | + | + | + | |||||||||||
| 2612 | Amikacin | + | + | + | + | + | + | + | + | + | |||||
| 2614 | Amikacin | + | + | + | + | + | + | + |
Phenotypic resistance characteristic for which no explanatory gene was detected on the array. All isolates were resistant to the four aminoglycosides, amikacin, gentamicin, kanamycin, and streptomycin, as measured by a standard disk diffusion assay, and all isolates demonstrated a MIC of >32 μg/ml for amikacin and >8 μg/ml for gentamicin. All isolates were PCR positive for the new methyltransferase gene (GenBank accession no. GU201947) described herein.
Isolates 2550 and 2551 were from the same calf fecal sample.
The initial attempts to transform sonicated plasmid DNA fragments from isolates with amikacin resistance but no corresponding gene, as described here, were successful for isolate 2517, which was therefore characterized further. The plasmid profile of E. coli isolate 2517 was performed as previously described (18); it contained two plasmids of approximately 60 and 95 kb. Plasmid DNA was extracted from isolate 2517 (Qiaprep spin miniprep kit; Qiagen, Valencia, CA) and electroporated into competent E. coli cells (GeneHogs; Invitrogen, Carlsbad, CA). Four resulting transformants that grew on gentamicin-supplemented medium (10 μg/ml) were all resistant to amikacin, gentamicin, kanamycin, and streptomycin as measured by disk diffusion assay, as was the donor isolate, 2517. The transformants each had a single plasmid of approximately 95 kb which probably corresponded to the 95-kb plasmid of the donor isolate. Plasmid DNA was extracted from the transformants and sonicated. After blunt-end repair and dephosphorylation, DNA fragments were ligated into a pCRII-Blunt-TOPO vector (Invitrogen) and chemically transformed into One Shot TOP10 cells (Invitrogen). They were then plated onto LB medium with gentamicin (10 μg/ml). The insert DNA was PCR amplified from the resulting transformants using flanking M13 primer binding sites. Two transformants yielded products that were 1,056 and 883 bp in length. Sequencing revealed an 819-bp open reading frame (ORF) (GenBank accession no. GU201947). Within the 1,056-bp fragment, the GC content of the ORF was 37.4%, and the GC content of its flanking sequences was 46.0%.
A BLASTx (NCBI) query of the deduced protein indicated that its amino acid sequence was most similar to that of 16S rRNA methyltransferases that confer high-level aminoglycoside resistance (11). The newly identified gene was amplified in each of the remaining 10 isolates identified with the same phenotype-genotype discordance (Table 2), using PCR primers GM1-Forward (5′-ATGAATATTGATGAAATGGTTGC) and GM1-Reverse (5′-TGATTGATTTCCTCCGTTTTTG). To confirm the aminoglycoside resistance phenotype in the 11 isolates that were PCR positive for the newly discovered gene, those isolates were tested for MICs using the Trek Diagnostics (Cleveland, OH) Sensititre plate COMEQ3F. The MICs for amikacin and gentamicin were greater than 32 μg/ml and 8 μg/ml, respectively.
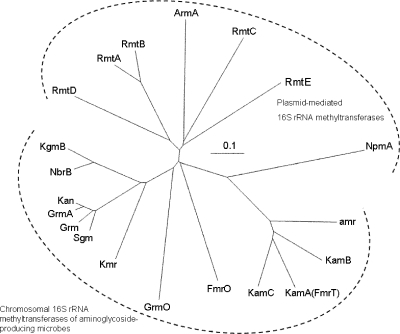
To assess the phylogenetic relationships between the newly identified gene and other 16S rRNA methyltransferase genes, its inferred amino acid sequence and those of previously identified 16S rRNA methyltransferases were analyzed using CLUSTALW (http://www.ddbj.nig.ac.jp/search/top-e.html). The results were illustrated with the TreeViewX program, version 0.5.0 for Macintosh OS X (Fig. 1). It is clear from this analysis and from its low percent identity (33%) to the closest match in the NCBI protein database that the deduced product of the aminoglycoside resistance gene identified in this study is distinct from other recognized homologues. This gene product can be designated RmtE (11). Its proximity on the tree to other enzymes for which the G1405 methylation site has been confirmed (ArmA [21] and RmtB [23]) suggests that the newly discovered RmtE probably also methylates that site (Fig. 1).
FIG. 1.
Phylogenetic relationships among 16S rRNA methyltransferases. Twenty amino acid sequences of known 16S rRNA methyltransferases were compared in CLUSTALW, and the results were illustrated using the TreeViewX program, version 0.5.0 for Macintosh OS X. GenBank or EMBL accession numbers associated with each gene are as follows: RmtA, AB120321; RmtB, AB117036; ArmA, AF550415; RmtC, AB194779; RmtD, DQ914960; RmtE, GU201947; NpmA, AB261016; FmrO, D13171; GrmO, AY524043; Grm, M55521; Sgm, A45282; GrmA, AY524043; Kmr, AB164642; Kan, AJ414669; NbrB, AF038408; KgmB, AAB20100; KamA (FmrT), D13170; KamC, AAA26499; KamB, CAF33037; and Amr, AAB08.
This is the first report of cattle-associated field isolates of E. coli bearing a plasmid-mediated aminoglycoside resistance gene of this type. The isolates in which the newly discovered RmtE gene was detected were collected from calves in a type of facility associated with intense antimicrobial use (25, 26, 31). While it seems biologically plausible that antibiotic selection pressure may promote the acquisition of novel resistance mechanisms by commensal enteric bacteria, we lack data on antimicrobial use associated with the specific animals from which these isolates originated. The difference between the GC content of the ORF itself and the GC content of the flanking regions is high, supporting the idea of a gene transfer event between species or genera of bacteria. As we have not characterized the plasmid or mobile elements associated with the gene, we cannot make further inferences about its origins. The array hybridization results for the isolates that were PCR positive for the novel gene indicate variations in the presence or absence of specific aminoglycoside resistance genes in this collection of isolates (Table 2). Because the E. coli isolates described here were from a single farm, these results cannot be generalized with regard to occurrence or prevalence on other premises.
Our purpose here is to report the finding of a previously unreported aminoglycoside resistance gene in the family of 16S rRNA methyltransferases in an E. coli isolate from an unusual setting and location. Further characterization, including protein expression, determination of the methylation site, and description of the plasmid and mobile elements associated with the new gene, is to follow.
Acknowledgments
This project was funded with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract numbers N01-A1-30054 and N01-A1-30055.
We gratefully acknowledge the contribution of the phylogenetic analysis by Yoshichika Arakawa, Department of Bacterial Pathogenesis and Infection Control, National Institute of Infectious Diseases, Tokyo 208-0011, Japan.
Footnotes
Published ahead of print on 5 April 2010.
REFERENCES
- 1.Allmansberger, R., B. Brau, and W. Piepersberg. 1985. Genes for gentamicin-(3)-N-acetyl-transferases III and IV. II. Nucleotide sequences of three AAC(3)-III genes and evolutionary aspects. Mol. Gen. Genet. 198:514-520. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, A. W., M. M. W. Kirby, J. C. Sherris, and M. Turck. 1966. Antibiotic susceptibility testing by a standard single disk method. Am. J. Clin. Pathol. 45:493-496. [PubMed] [Google Scholar]
- 3.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogaerts, P., M. Galimand, C. Bauraing, A. Deplano, R. Vanhoof, R. De Mendonca, H. Rodriguez-Villalobos, M. Struelens, and Y. Glupczynski. 2007. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J. Antimicrob. Chemother. 59:459-464. [DOI] [PubMed] [Google Scholar]
- 5.Briggs, C. E., and P. M. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruant, G., C. Maynard, S. Bekal, I. Gaucher, L. Masson, R. Brousseau, and J. Harel. 2006. Development and validation of an oligonucleotide microarray for detection of multiple virulence and antimicrobial resistance genes in Escherichia coli. Appl. Environ. Microbiol. 72:3780-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, L., Z. L. Chen, J. H. Liu, Z. L. Zeng, J. Y. Ma, and H. X. Jiang. 2007. Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J. Antimicrob. Chemother. 59:880-885. [DOI] [PubMed] [Google Scholar]
- 8.Chen, S., S. Zhao, P. F. McDermott, C. M. Schroeder, D. G. White, and J. Meng. 2005. A DNA microarray for identification of virulence and antimicrobial resistance genes in Salmonella serovars and Escherichia coli. Mol. Cell. Probes 19:195-201. [DOI] [PubMed] [Google Scholar]
- 9.CLSI. 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 3rd ed. Approved Standard. CLSI document M31-A3. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9a.Davis, M. A., J. Y. Lim, Y. Soyer, H. Harbottle, Y. F. Chang, D. New, L. H. Orfe, T. E. Besser, and D. R. Call. Development and validation of a resistance and virulence gene microarray targeting Escherichia coli and Salmonella enterica. J. Microbiol. Methods, in press. [DOI] [PMC free article] [PubMed]
- 10.Distler, J., A. Ebert, K. Mansouri, K. Pissowotzki, M. Stockmann, and W. Piepersberg. 1987. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 15:8041-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doi, Y., J. Wachino, and Y. Arakawa. 2008. Nomenclature of plasmid-mediated 16S rRNA methylases responsible for panaminoglycoside resistance. Antimicrob. Agents Chemother. 52:2287-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doublet, B., F. X. Weill, L. Fabre, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Variant Salmonella genomic island 1 antibiotic resistance gene cluster containing a novel 3′-N-aminoglycoside acetyltransferase gene cassette, aac(3)-Id, in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 48:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frye, J. G., T. Jesse, F. Long, G. Rondeau, S. Porwollik, M. McClelland, C. R. Jackson, M. Englen, and P. J. Fedorka-Cray. 2006. DNA microarray detection of antimicrobial resistance genes in diverse bacteria. Int. J. Antimicrob. Agents 27:138-151. [DOI] [PubMed] [Google Scholar]
- 14.Galimand, M., P. Courvalin, and T. Lambert. 2003. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Zorn, B., A. Catalan, J. A. Escudero, L. Dominguez, T. Teshager, C. Porrero, and M. A. Moreno. 2005. Genetic basis for dissemination of armA. J. Antimicrob. Chemother. 56:583-585. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez-Zorn, B., T. Teshager, M. Casas, M. C. Porrero, M. A. Moreno, P. Courvalin, and L. Dominguez. 2005. armA and aminoglycoside resistance in Escherichia coli. Emerg. Infect. Dis. 11:954-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 18.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang, H. Y., K. Y. Kim, J. Kim, J. C. Lee, Y. C. Lee, D. T. Cho, and S. Y. Seol. 2008. Distribution of conjugative-plasmid-mediated 16S rRNA methylase genes among amikacin-resistant Enterobacteriaceae isolates collected in 1995 to 1998 and 2001 to 2006 at a university hospital in South Korea and identification of conjugative plasmids mediating dissemination of 16S rRNA methylase. J. Clin. Microbiol. 46:700-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaster, K. R., S. G. Burgett, R. N. Rao, and T. D. Ingolia. 1983. Analysis of a bacterial hygromycin B resistance gene by transcriptional and translational fusions and by DNA sequencing. Nucleic Acids Res. 11:6895-6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liou, G. F., S. Yoshizawa, P. Courvalin, and M. Galimand. 2006. Aminoglycoside resistance by ArmA-mediated ribosomal 16S methylation in human bacterial pathogens. J. Mol. Biol. 359:358-364. [DOI] [PubMed] [Google Scholar]
- 22.Michael, G. B., P. Butaye, A. Cloeckaert, and S. Schwarz. 2006. Genes and mutations conferring antimicrobial resistance in Salmonella: an update. Microbes Infect. 8:1898-1914. [DOI] [PubMed] [Google Scholar]
- 23.Perichon, B., P. Courvalin, and M. Galimand. 2007. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob. Agents Chemother. 51:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rather, P. N., R. Mierzwa, R. S. Hare, G. H. Miller, and K. J. Shaw. 1992. Cloning and DNA sequence analysis of an aac(3)-Vb gene from Serratia marcescens. Antimicrob. Agents Chemother. 36:2222-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raymond, M. J., R. D. Wohrle, and D. R. Call. 2006. Assessment and promotion of judicious antibiotic use on dairy farms in Washington State. J. Dairy Sci. 89:3228-3240. [DOI] [PubMed] [Google Scholar]
- 26.Sawant, A. A., L. M. Sordillo, and B. M. Jayarao. 2005. A survey on antibiotic usage in dairy herds in Pennsylvania. J. Dairy Sci. 88:2991-2999. [DOI] [PubMed] [Google Scholar]
- 27.Shakil, S., R. Khan, R. Zarrilli, and A. U. Khan. 2008. Aminoglycosides versus bacteria—a description of the action, resistance mechanism, and nosocomial battleground. J. Biomed. Sci. 15:5-14. [DOI] [PubMed] [Google Scholar]
- 28.Tenover, F. C., D. Filpula, K. L. Phillips, and J. J. Plorde. 1988. Cloning and sequencing of a gene encoding a 6′-N-acetyltransferase from an R factor of Citrobacter diversus. J. Bacteriol. 170:471-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenover, F. C., K. L. Phillips, T. Gilbert, P. Lockhart, P. J. O'Hara, and J. J. Plorde. 1989. Development of a DNA probe from the deoxyribonucleotide sequence of a 3-N-aminoglycoside acetyltransferase [AAC(3)-I] resistance gene. Antimicrob. Agents Chemother. 33:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trower, M. K., and K. G. Clark. 1990. PCR cloning of a streptomycin phosphotransferase (aphE) gene from Streptomyces griseus ATCC 12475. Nucleic Acids Res. 18:4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.USDA. 2010. Dairy 2007, Heifer Calf Health and Management Practices on U.S. Dairy Operations, 2007. No. 550.0110. Animal and Plant Health Inspection Service, U.S. Department of Agriculture, Fort Collins, CO. http://www.aphis.usda.gov/vs/ceah/ncahs/nahms/dairy/dairy07/Dairy07_ir_CalfHealth.pdf.
- 32.van Hoek, H. A. M., I. M. J. Scholtens, A. Cloeckaert, and H. J. M. Aarts. 2005. Detection of antibiotic resistance genes in different Salmonella serovars by oligonucleotide microarray analysis. J. Microbiol. Methods 62:13-23. [DOI] [PubMed] [Google Scholar]
- 33.Yamane, K., Y. Doi, K. Yokoyama, T. Yagi, H. Kurokawa, N. Shibata, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Genetic environments of the rmtA gene in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 48:2069-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan, J. J., J. J. Wu, W. C. Ko, S. H. Tsai, C. L. Chuang, H. M. Wu, Y. J. Lu, and J. D. Li. 2004. Plasmid-mediated 16S rRNA methylases conferring high-level aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J. Antimicrob. Chemother. 54:1007-1012. [DOI] [PubMed] [Google Scholar]
- 35.Yokoyama, K., Y. Doi, K. Yamane, H. Kurokawa, N. Shibata, K. Shibayama, T. Yagi, H. Kato, and Y. Arakawa. 2003. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 362:1888-1893. [DOI] [PubMed] [Google Scholar]