Abstract
The ability to selectively target specific biofilm species with antimicrobials would enable control over biofilm consortium composition, with medical applications in treatment of infections on mucosal surfaces that are colonized by a mixture of beneficial and pathogenic microorganisms. We functionalized a genetically engineered multimeric protein with both a targeting moiety (biotin) and either a fluorophore or a photosensitizer (SnCe6). Biofilm microcolonies of Aggregatibacter actinomycetemcomitans, a periodontal pathogen, were targeted with the multifunctional dodecamer. Streptavidin was used to couple biotinylated dodecamer to a biotinylated anti-A. actinomycetemcomitans antibody. This modular targeting approach enabled us to increase the loading of photosensitizer onto the cells by a cycle of amplification. Scanning laser confocal microscopy was used to characterize transport of fluorescently tagged dodecamer into the microcolonies and targeting of the cells with biotin-labeled, fluorescently tagged dodecamer. Light-induced activity of the targeted photosensitizer reduced the viability of A. actinomycetemcomitans biofilm, as indicated by membrane permeability to propidium iodide. The functionalized multimeric protein promises to be a useful tool for controlling periodontal biofilm consortia and offers a modular design whereby moieties that target different species can be readily combined with the functionalized protein construct.
Therapeutic tools that enabled targeted delivery of antimicrobials to specific species in biofilms would expand our capability to treat chronic infections associated with mucosal surfaces where complete sterility is not the natural healthy condition. The oral cavity and the gut are prime examples of mucosal surfaces at which the immune system maintains a healthy microbial presence (3, 50). Mucosal surfaces in the oral cavity are nonsterile environments colonized by biofilm consortia (38). In the gut, some commensals are true symbionts (30, 36), and similarly, it is hypothesized that oral commensals may aid in maintaining a healthy immune homeostasis (35). The implication is that treatment of oral diseases such as periodontal disease can be made more effective by selective elimination of pathogens. By extension, we may also find that tools that enable specific targeting of biofilm pathogens provide effective treatments for chronic infections of the gut (31).
A class of multimeric proteins that self-assemble into highly symmetric quaternary structures (7, 52) shows promise for development into delivery vehicles for therapeutic (15, 16, 44) and imaging agents (41, 51). We selected one of these proteins, a relatively small, spherical, 9-nm-diameter dodecamer, originally isolated from Listeria inocula (LiDps) (21), for targeted delivery of a photosensitizer to Aggregatibacter actinomycetemcomitans biofilm. We genetically added a tetrapeptide to the C terminus of each of the 12 monomeric 18-kDa subunits of LiDps. Cysteines incorporated into this tetrapeptide were labeled with biotin. Since the C terminus is presented on the exterior surface of the assembled dodecamer, these biotins were readily available for binding to streptavidin, a universal coupling protein. An SnCe6 photosensitizer was covalently linked to intrinsic lysines.
Photosensitizers are compounds that produce reactive oxygen species (ROS) upon excitation by light (25). Antimicrobial photodynamic therapy (PDT) has been shown to be effective against oral pathogens (6, 29, 54). SnCe6 has been used previously for targeted antimicrobial PDT (10-12). The primary modes of action of photosensitizers are membrane disruption and, if the photosensitizer is internalized, DNA damage (19). The antimicrobial activity of ROS produced by light-activated photosensitizers is relatively localized. This is especially the case for singlet oxygen (diffusion length of <50 nm), which is thought to be the primary ROS causing cell death (32). The localized action of photosensitizers makes them attractive agents for targeted antimicrobial therapy.
The LiDps genetic construct, dual functionalized with SnCe6 and biotin, is a modular unit that can be combined with any targeting moiety that can be biotinylated. This would accommodate a variety of approaches that have been used to target microbial pathogens, including lectin/carbohydrate interactions (53), antimicrobial peptides (8, 22), and the lysins isolated from bacteriophage (14). We chose to use an antibody as the targeting moiety for these studies since monoclonal antibodies are available for a number of the prevalent periodontal pathogens (5, 9, 17). In addition, antibodies offer a wide range of potential cell surface targets combined with the possibility to engineer high binding affinities (4).
A. actinomycetemcomitans is a Gram-negative periodontal pathogen implicated as the primary etiological agent in localized aggressive periodontitis (13, 37). Rough colony variants isolated from sites of infection form discrete microcolonies when cultured in vitro which exhibit distinctive cohesive as well as adhesive properties (27). We characterized transport of the LiDps dodecamer to the base of A. actinomycetemcomitans microcolonies, demonstrated targeting of the LiDps to the cells in the microcolonies, and showed that SnCe6 targeted to 24-h biofilms produced light-activated membrane disruption. The modular design enabled us to use cycles of alternating exposure to the streptavidin and the LiDps dodecamer to increase the loading of targeted photosensitizer onto the cells.
MATERIALS AND METHODS
Bacterial strain and culture conditions.
Aggregatibacter actinomycetemcomitans (formerly Actinobacillus actinomycetemcomitans) strain D7, a rough colony clinical isolate obtained from the central incisor of an African-American female patient with generalized aggressive periodontitis, was provided by Casey Chen, University of Southern California. The liquid medium was modified tryptic soy broth (MTSB) consisting of (per liter) 30 g tryptic soy broth and 3 g yeast extract. The solid medium was MTSB with 50 ml fetal bovine serum (HyClone) and 15 g Bacto agar added (per liter). Frozen stocks were maintained at −80°C in 20% glycerol and 80% MTSB. Biofilms were cultured in 96-well microtiter plates with glass bottoms (P96G-1.5-5-F; MatTek Corp.) to enable confocal laser scanning microscope (CLSM) images to be acquired. The biofilm inoculum was prepared by looping single colonies, cultured on solid medium at 37°C in 5% CO2 for 72 h, into a 1-ml aliquot of liquid medium and dispersing the suspension of cell aggregates by repeated pipetting followed by vortexing and finally diluting the suspension to obtain approximately 107 CFU/ml. No attempt was made to isolate single cells from cell aggregates as described by Kaplan et al. (27). Biofilms were cultured at 37°C in 5% CO2. Medium was replaced at 24 h for biofilms cultured for 48 h.
Antibody.
Monoclonal antibody (Aa-MAb) 225AA2 (18) against A. actinomycetemcomitans, isotype mIgG2b, was purified by affinity chromatography on an AffinityPak protein A column (Thermo Fisher Scientific), following the manufacturer's recommendations with slight modifications. The wash buffer was Dulbecco's phosphate-buffered saline (PBS), pH 8.2, and the elution buffers were 0.1 M sodium citrate at pH 5.0, 4.0, and 3.0. Fractions eluted from the column were concentrated to an optical density at 280 nm (OD280) of approximately 0.2 and dialyzed into 50 mM HEPES (150 mM NaCl, pH 7.0). Purity of Aa-MAb was confirmed by SDS-PAGE under reducing conditions to separate the light and heavy chains. Aa-MAb was biotinylated by reaction for 1 h at room temperature with 0.5 mM succinimidyl ester of biotin incorporating a hydrophilic spacer (product no. 21362, EZ-Link NHS-PEG4-biotin; Thermo Scientific Pierce). Unreacted reagent was separated from the Aa-MAb by using a Bio-Rad spin column (equilibrated in 10 mM Tris, pH 7.4). Immunodot blots were used to confirm the association of the biotin with Aa-MAb and to check the integrity of the epitope binding site. The covalent addition of the biotin reagent to the light chain of the Aa-MAb was further confirmed by liquid chromatography-electrospray mass spectrometry (LC/MS) as described previously (41). Only the light chain (biotinylated or nonbiotinylated) could be detected with LC/MS.
The Aa-MAb 225AA2 epitope has not been characterized at the molecular level. However, its specificity has been characterized (18). The Aa-MAb 225AA2 bound to every A. actinomycetemcomitans strain tested (18 strains with five different serotypes) and was negative for all non-A. actinomycetemcomitans strains tested, consisting of 55 species known to be prominent in the oral cavity (17 genera).
LiDps mutagenesis and expression.
Addition of residues KLFC to the C termini of wild-type LiDps subunits was accomplished by a modification of the QuikChange site-directed mutagenesis (Stratagene) by using primers containing extra nucleotides and pET-30b-based plasmids containing the wild-type LiDps sequence as a template. Primers for mutagenesis contained the nucleotide for the KLFC tetrapeptide (forward primer, 5′-AAGGATCCGAATTCGAGCTCCGTCGAC-3′; reverse primer, 5′-AAGGATCCTTAGCAGAATAGCTTTTCTAAGGAGCTTTTCC-3′). The amplified DNAs were transformed into competent Escherichia coli strain BL21(DE3) (Novagen). For expression and purification of the assembled KLFC LiDps, cultures of the transformed cells were grown overnight at 37°C in 30 mg/ml LB-kanamycin. Protein expression was induced with 1.0 mM isopropyl-β-d-thiogalactopyranoside for 5 h. Cells were pelleted at 3,700 × g and resuspended in 50 mM MES (morpholineethanesulfonic acid, pH 6.5; 100 mM NaCl) with lysozyme (50 mg/ml), RNase (90 mg/ml), and DNase (60 mg/ml). A sequence of sonication and French press treatment was used to lyse cells. Cell fragments were removed by centrifugation at 12,000 × g, and the supernatant was heated to 65°C for 10 min. The suspension was centrifuged at 12,000 × g to remove the aggregated heat-sensitive proteins. Assembled KLFC LiDps was further purified by size exclusion chromatography (SEC) (Superose 6; Amersham Biosciences, Uppsala, Sweden). The identity and purity of the KLFC LiDps protein were confirmed by LC/MS, SDS-PAGE, and transmission electron microscopy (TEM).
Functionalization of the KLFC LiDps.
Purified KLFC LiDps was biotinylated by reaction of 2 mg/ml protein (0.1 mM protein subunit) with 2 mM maleimide polyethylene glycol (PEG) 2 biotin (product no. 21901; EZ-link; Pierce) for 3 h at room temperature in MES buffer (pH 6.5; 100 mM NaCl). KLFC LiDps incorporates no cysteine residues except for those added genetically, and maleimides react specifically with cysteines of proteins (23). Assembled dodecamer was purified from the reaction mixture by SEC. Covalent addition of biotin to the protein subunits was confirmed using LC/MS. For transport studies, fluorescein was added to the cysteines of the KLFC tetrapeptide by using the same protocol but with fluorescein-5 maleimide reagent (Invitrogen). A fluorescent tag (Alexa Fluor 488; Invitrogen) or the photosensitizer (Sn[IV] chlorin e6; Frontier Scientific, Logan, UT) was added to lysines of biotinylated LiDps by reaction with succinimidyl esters. The succinimidyl ester of Alexa Fluor 488 was purchased (A20000; Invitrogen). Carboxylate groups of the succinimidyl ester of the SnCe6 were converted to succinimidyl esters by using a modification of a previously published protocol (10). Sn(IV) chlorin e6 (7 mg), 1-(3-[dimethylamino]propyl)-3-ethylcarbodiimide hydroxychloride (3 mg), and N-hydroxysuccinimide (4 mg) were combined in a flame-dried glass anaerobe jar which was placed in an anaerobe chamber. To this was added 1 ml of dimethylformamide (dry; stored under nitrogen), the jar was sealed, and the mixture was allowed to react for approximately 12 h with mixing. The reaction mixture was aliquoted into 50-μl portions, flash frozen in liquid nitrogen, and lyophilized. The succinimidyl esters of the Alexa Fluor 488 or SnCe6 were added to the biotinylated KLFC LiDps by reaction of 1 mg/ml protein (0.05 mM protein subunit) with 0.5 mM the reagent for 3 h at room temperature in 50 mM phosphate buffer (pH 7.5; 100 mM NaCl). The latter reaction was performed anaerobically in the dark. Prior to addition of the SnCe6 succinimidyl ester, nitrogen gas was bubbled through the protein solution for 10 min. The solution was then transferred immediately into an anaerobe chamber, the reagent was added, and the mixture was covered with foil and mixed. KLFC LiDps fluorescently tagged or labeled with SnCe6 was purified by SEC as described above. According to spectrophotometric determination, the ratios of labeling of the Alexa Fluor 488 and SnCe6 were both approximately four per dodecamer of LiDps. Extensive dialysis did not decrease these ratios. We further measured the extent of nonspecific binding of SnCe6 to the dodecamer. Dodecamer was exposed to SnCe6 under conditions identical to those used for reaction with the succinimidyl ester of SnCe6. In this case, comigration of the absorbance originating from SnCe6 with the dodecamer was negligible.
Transport and targeting of the KLFC LiDps.
Transport of dodecamer into A. actinomycetemcomitans biofilm microcolonies and targeting of dodecamer to the microcolonies were characterized using SLCM (Leica TCS-SP2-AOBS). Biofilm microcolonies were visualized through the glass coverslip floor of the wells. This was accomplished by filling each well to the brim, covering the row of wells with Parafilm, and inverting the plate. Images were collected with either a 40× 0.8 NA or a 63× 0.9 NA HCX APO L U-V-I water immersion objective. Green and red fluorophores were discriminated by acousto-optical tunable filters. Green fluorophores (fluorescein, Syto 9 [Invitrogen], and Alexa Fluor 488) were excited with a 488-nm laser, and fluorescence was collected from 500 nm to 543 nm. Red fluorophores (Syto 59 [Invitrogen] and propidium iodide [Sigma-Aldrich]) were excited with a 561-nm laser, and fluorescence was collected from 600 nm to 721 nm. Images were taken at 1.0-μm intervals throughout the depth of the biofilms. Stacks were combined in Imaris software (Bitplane AG, Zürich, Switzerland) to yield final images. Transport was evaluated by exposing the biofilm to 1 mg/ml KLFC LiDps labeled with fluorescein and then tracking changes in the fluorescence at the base of the biofilm. Biofilm microcolonies were prestained with Syto 59 for this study so that biofilm microcolonies could be located and brought into approximate focus, which required about 3 min after filling the well with the protein solution. Images of the biofilm were acquired at approximately 4-min intervals by repeating the scan using the same parameters. The fluorescence increase in areas at the base of microcolonies was assessed as mean pixel brightness by using Adobe Photoshop (v7.0). The colonies stained heavily with the Syto 59 nucleic acid stain, resulting in strong red fluorescence and slight bleed-through into the green channel. The corresponding pixel brightness of this bleed-through was treated as background which was subtracted from the raw data values. Targeting was performed by exposing biofilms to a series of solutions in the wells. Biofilms were first washed with 1% bovine serum albumin (BSA) in 10 mM PBS (pH 7.0; 100 mM NaCl) for 5 min. Biofilms were rinsed with PBS after each reaction. Solutions and exposure times were as follows: biotinylated Aa-MAb at approximately 50 μg/ml for 50 min, streptavidin at 50 μg/ml for 30 min, and KLFC LiDps at 100 μg/ml for 50 min. The last two steps were repeated to amplify the level of SnCe6-targeted delivery.
Light exposure.
SnCe6 was excited with a 633-nm HeNe laser (Melles Griot, Carlsbad, CA) defocused with a microscope ocular lens. For light exposure, the plate was inverted as described above. The plate was placed at a specified distance from the objective so that the area of the beam was slightly larger than the area of the floor of the well. Irradiance (W cm−2) was determined using a photometer. The total fluence (J cm−2) was determined by the product of the irradiance and the exposure time. The illumination time was 20 min (1,200 s) to achieve a total fluence of 14 J cm−2, which is in the standard range for antimicrobial PDT (25).
Viability assay.
Membrane integrity of A. actinomycetemcomitans cells in biofilm microcolonies was evaluated using the BacLight Live/Dead bacterial viability assay (Molecular Probes) (28). Saline solutions of Syto 9 and propidium iodide at concentrations recommended by the manufacturer of the kit (L-7012; Molecular Probes) were used. Propidium iodide (red) penetrates cells with compromised membranes, while Syto 9 (green) penetrates all cells. Photomultiplier tube (PMT) sensitivity for the red and green channels was optimized so that signal was well above background while minimizing bleed-through. These optimized settings were used for each experiment. The red/green ratio was quantified as the ratio of pixel intensities of the red and green channels evaluated using MetaMorph software (Molecular Devices Corporation, Downingtown, PA). Microcolonies were selected for analysis, excluding dark regions between the colonies. This was accomplished by “thresholding.” Histograms of number of pixels versus pixel brightness were constructed and then truncated to omit the dark intercolony regions. Mean pixel brightness of the associated truncated histograms was then calculated. The relationship between the red/green ratio and membrane disruption was assessed by using the red/green ratio induced in biofilms exposed to 0.1 mg/ml chlorhexidine digluconate (CHG) for 1 h as an internal standard for each experiment. In preliminary studies, we determined that exposure of cell suspensions of A. actinomycetemcomitans used for biofilm inocula to 0.1 mg/ml CHG for 1 h reduces the number of CFU by 3 orders of magnitude. (The rough colony A. actinomycetemcomitans variant remains primarily in cell aggregates even when dispersed into liquid).
Statistical analysis.
Efficacy of the targeted SnCe6 was evaluated by a paired t test. Three independent replicates were performed on separate days. Each experiment included three or four untreated biofilms with the wells for these biofilms distributed along a row of 10 wells. Two targeted biofilms exposed to light as well as one well with a nontargeted biofilm exposed to light were also included in each experiment. These wells were located toward one side of the row to minimize the effect of scattered laser light on unexposed wells. Red/green ratios were normalized to the red/green ratio obtained for internal standard (the 0.1 mg/ml CHG condition). Means of within-experiment replicates were used for the paired t test.
RESULTS
Transport of LiDps into A. actinomycetemcomitans biofilm microcolonies.
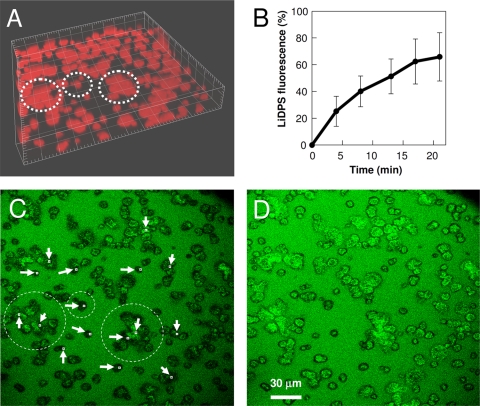
Transport of KLFC LiDps into A. actinomycetemcomitans microcolonies was characterized using SLCM. The absence of targeting allowed transport kinetics to be measured without the additional factor of binding kinetics. The well was filled with a solution of KLFC LiDps, fluorescently tagged via the peptidyl cysteine residues, and the time course of increase in fluorescence was measured at locations at the base of microcolonies (near the substratum) and in the interior of microcolonies with respect to the view in the plane of the substratum. The A. actinomycetemcomitans biofilm was stained with a red nucleic acid stain prior to the transport study (Fig. 1A). Figure 1B shows the mean kinetics of transport of the fluorescently tagged KLFC LiDps to locations at the base of 16 A. actinomycetemcomitans microcolonies. Figure 1B indicates that the mean concentration of dodecamer in the interior regions of A. actinomycetemcomitans microcolonies reached a level of approximately 60% of the fluorescent signal in the bulk (between microcolonies) at 20 min.
FIG. 1.
Transport of fluorescently tagged KLFC LiDps into A. actinomycetemcomitans biofilm microcolonies. (A) Biofilm microcolonies stained with Syto 59 (4.167-μm tick spacing). Microcolonies circled with the dashed lines are the same ones circled in panel C. (B) Transport kinetics of fluorescently tagged KLFC Dps into regions at the base of biofilm microcolonies. The solid line connects data which are the means of measurements at 16 locations; error bars represent the standard deviations. (C) Section acquired near the substratum 4 min after exposure of the biofilm to the fluorescently tagged KLFC LiDps; regions used to acquire the data presented in panel B are indicated. (D) Section acquired near the substratum 21 min after exposure of the biofilm to the fluorescently tagged KLFC LiDps.
Figures 1C and D are SLCM images taken at a section proximal to the substratum interface 5 and 20 min, respectively, after exposure of the biofilm to the fluorescently tagged KLFC LiDps. Locations where data were acquired to construct Fig. 1B are indicated. There was a 20% increase in the fluorescence signal from the bulk solution (between the microcolonies) between the first and second time points (4 and 8 min, respectively), which then remained constant. This is likely due to diffusion of the multimeric protein into a static boundary layer near the substratum, accompanied, perhaps, by nonspecific adsorption onto the substratum. The 100% value for fluorescence was taken as the value for the later times points in order to construct the data presented in Fig. 1B. One possible reason for the exclusion of fluorescence from peripheral regions of the microcolonies may be that there is a greater density of extracellular matrix in these regions. However, we have no direct evidence for this.
Dual-functionalized LiDps for targeting.
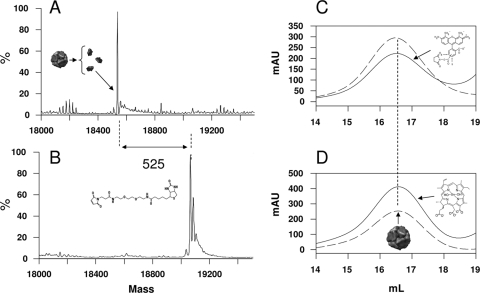
The 12 terminating cysteines (C) on the tetrapeptides of the KLFC LiDps construct provide ideal attachment sites for targeting moieties since they are displaced from the exterior dodecamer shell (21) by the peptide spacer (KLFC). The biotinylation reagent provides a further extension of the spacer length (3 nm) between the biotin and the dodecamer external surface. The covalent addition of one biotin maleimide functional group to each of the monomeric subunits was confirmed using LC/MS (Fig. 2A and B). Some oxidation of the biotinylated product is indicated by the appearance of small higher-molecular-mass bands spaced 16 Da apart (26).
FIG. 2.
Dual-functionalized KLFC LiDps. (A and B) LC/MS deconvolutions of mass spectra of monomeric subunits of the dodecamer. (A) Mass of nonfunctionalized KLFC LiDps monomeric subunits (18,540 Da). (B) Mass of biotinylated KLFC LiDps (KLFC-LiDps-B) monomeric subunits (19,065 Da). The difference in mass between the subunits in panels A and B (525 Da) is expected for addition of the biotinylation reagent to the cysteines. The deconvolved spectrum in panel B indicates that all the 12 subunits of KLFC LiDps were biotinylated. (C and D) Size exclusion chromatograms of biotinylated KLFC LiDps tagged with Alexa Fluor 488 (KLFC-LiDps-B-AF) (C) or functionalized with the SnCe6 (KLFC-LiDps-B-photosensitizer) (D). The solid lines represent the absorbance at 280 nm (protein). The dashed lines indicate the absorbances of the Alexa Fluor 488 (488 nm) or photosensitizer (410 nm) (C and D, respectively). The vertical straight line indicates the position at which the dodecamer elutes.
Biotinylated KLFC LiDps (KLFC-LiDps-B) was further functionalized with either Alexa Fluor 488 (KLFC-LiDps-B-AF) or SnCe6 (KLFC-LiDps-B-SnCe6). KLFC-LiDps-B-AF is an analog of the KLFC-LiDps-B-SnCe6 with Alexa Fluor 488 substituted for SnCe6. Succinimidyl esters of these compounds were added to intrinsic lysines of the dodecamer. Association of these functionalities with the dodecamer was confirmed by comigration of a distinguishing absorbance band of the Alexa Fluor 488 or SnCe6 with the dodecamer on SEC (Fig. 2C and D). Extensive dialysis did not reduce the ratio of the functional group to protein (determined spectrophotometrically), indicating that the functional groups were covalently bound to the lysines.
Targeting of A. actinomycetemcomitans biofilm microcolonies with LiDps.
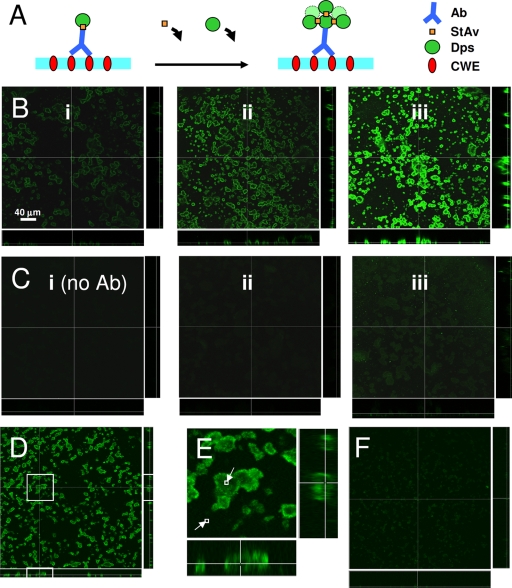
KLFC-LiDps-B-AF was used to characterize targeting of A. actinomycetemcomitans biofilm since SnCe6 fluorescence was not in an appropriate range for SLCM detection. The targeting approach used for both KLFC-LiDps-B-AF and KLFC-LiDps-B-SnCe6 is illustrated in Fig. 3A. Streptavidin was used to couple KLFC-LiDps-B-AF to biotinylated Aa-MAb bound to the biofilm cells. The loading of the fluorophore per Aa-MAb epitope binding site was subsequently increased by repeating cycles of exposure to streptavidin and biotinylated dodecamer, with a consequent amplification in fluorescence signal. The first row of SLCM images in Fig. 3B (panels i, ii, and iii) are 48-h A. actinomycetemcomitans biofilms targeted with KLFC-LiDps-B-AF. The mean fluorescence within 48-h A. actinomycetemcomitans biofilm microcolonies was increased by factors of 3.4 and 6.3 for one and two repeated cycles of exposure to streptavidin and KLFC-LiDps-B-AF, respectively (Fig. 3B, panels ii and iii). Binding to A. actinomycetemcomitans microcolonies was primarily mediated through the specific Aa-MAb/epitope interaction, as indicated by the much lower fluorescent signal acquired when the Aa-MAb was omitted from the series of reactions (Fig. 3C, panels i, ii, and iii). There was a slight amount of nonspecific binding of the dodecamer to the A. actinomycetemcomitans biofilm microcolonies, indicated by a fluorescent signal in the green channel which was above background and which was also amplified by cycles of exposure to streptavidin and KLFC-LiDps-B-AF. The KLFC-LiDps-B-AF fluorescence within microcolonies was lower by factors of 248, 802, and 8 (Fig. 3B and C, from left to right, respectively) when Aa-MAb was omitted from the reaction.
FIG. 3.
Targeting of A. actinomycetemcomitans biofilm microcolonies with KLFC-LiDps-B-AF. (A) Illustration of presumed cluster formation produced by cycles of exposure to streptavidin followed by KLFC-LiDps-B-AF, resulting in an amplification of the fluorescent signal at sites of Aa-MAb binding. Ab, Aa-MAb; StAv, streptavidin; Dps, KLFC Dps; CWE, cell wall epitope. (B) Forty-eight-hour biofilm microcolonies targeted with biotinylated KLFC Dps tagged with Alexa Fluor 488 for one to three cycles of exposure to streptavidin followed by KLFC-LiDps-B-AF. One cycle means exposure to Aa-MAb followed by streptavidin followed by KLFC Dps. (i) One cycle. (ii) Two cycles. (iii) Three cycles. (C) Biofilms treated the same as described for panel B except that the Aa-MAb was omitted from the first step. (D) Twenty-four-hour biofilm targeted with biotinylated KLFC Dps tagged with Alexa Fluor 488. (E) Magnified view of a selected portion of panel D, showing the relatively even distribution of fluorescence from the targeted dodecamer; pixel brightness computed for two small regions indicated that even the darkest region of the microcolonies was twice as fluorescent as the background (small squares). (F) Biofilm was treated the same as described for panel D but the Aa-MAb was omitted.
Images of sections in the plane of the interface and sections taken perpendicular to the interface both indicated that the fluorescence distribution in the microcolonies was nonuniform for 48-h biofilms targeted with dodecamer, with a denser concentration of fluorescence near the peripheries of microcolonies. In contrast, the fluorescence originating from nonspecific interactions (Fig. 3C) was uniformly distributed throughout the A. actinomycetemcomitans colonies. This is more evident if the images are contrast enhanced (data not shown). This result suggests that the distribution of fluorescence in targeted A. actinomycetemcomitans microcolonies reflects the Aa-MAb epitope distribution in 48-h A. actinomycetemcomitans microcolonies, rather than originating from hindered transport (consistent with the transport study).
In contrast with 48-h targeted biofilms, 24-h targeted biofilms show a more even uniform distribution of KLFC-LiDps-B-AF (Fig. 3D, E, and F). The brightness/contrast of these images was optimized (identically) to show the pattern of fluorescence. Figure 3E shows a magnified view of a microcolony. A comparison of the pixel brightness levels within the two squares showed that even fluorescence within the darkest region of a large microcolony was twice that of the background. Figure 3F shows the level of nonspecific binding of the KLFC-LiDps-B-AF to the A. actinomycetemcomitans microcolonies when the Aa-MAb was omitted.
Light-induced membrane disruption of SnCe6-targeted A. actinomycetemcomitans biofilm.
The red/green ratio determined from the viability assay upon exposure of biofilms to 0.1 mg/ml CHG for 1 h was used as an internal standard for assessing the efficacy of light-induced membrane disruption of SnCe6-targeted cells. The rationale for this was that CHG has been shown previously to be efficacious against A. actinomycetemcomitans biofilms (48), and the primary mode of action of CHG is membrane disruption (24). According to the viability assay, there was a clear breakpoint between 0.1 mg/ml CHG and 0.01 mg/ml CHG (see Fig. S1 in the supplemental material). A. actinomycetemcomitans biofilms exposed to these two CHG concentrations for 1 h were included in each experiment as internal standards.
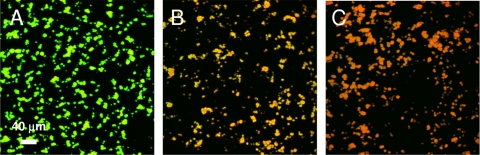
A. actinomycetemcomitans biofilms were targeted with KLFC-LiDps-B-SnCe6 by using the same protocol as that for KLFC-LiDps-B-AF (Fig. 3A). In a preliminary experiment, we obtained no evidence of membrane disruption of 48-h biofilms. We proceeded to test 24-h biofilms and obtained evidence of membrane disruption for biofilms targeted using two cycles of exposure to streptavidin followed by KLFC-LiDps-B-SnCe6. We attempted to gather data demonstrating a correlation between loss of viability and cycles of amplification by using the KLFC-LiDps-B-SnCe6 construct, but the variability was too great to make any strong claims. As indicated in Fig. 4, membrane disruption induced by exposure of SnCe6-targeted A. actinomycetemcomitans microcolonies to light (Fig. 4B) was intermediate between that of untreated microcolonies (Fig. 4A) and that of microcolonies exposed to 0.1 mg/ml CHG (Fig. 4C). The mean red/green ratio for SnCe6-targeted microcolonies exposed to light compared to those exposed to 0.1 mg/ml CHG was 0.62 (standard deviation [SD] of 0.21) for three independent experiments. The red/green ratio for the SnCe6-targeted microcolonies exposed to light was higher than those for both untreated microcolonies and SnCe6-targeted microcolonies not exposed to light at the 5% level of confidence (Table 1). The red/green ratio for the SnCe6-targeted microcolonies exposed to light was higher than those for both microcolonies exposed to light but not targeted with SnCe6 and microcolonies exposed to 0.01 mg/ml CHG at the 10% level of confidence (Table 1). Data for all three experiments are shown in the supplemental material (see Fig. S2).
FIG. 4.
Membrane disruption of A. actinomycetemcomitans biofilm microcolonies targeted with the photosensitizer assessed by the BacLight Live/Dead bacterial viability assay (images of biofilm microcolonies). (A) Not treated. (B) Targeted with photosensitizer and exposed to light. (C) Exposed to 0.1 mg/ml CHG.
TABLE 1.
Statistical level of significance for targeted light-induced membrane damage
| Paira | Relative damageb | P valuec |
|---|---|---|
| PSL, NT | 3.78 | 0.0184 |
| PSL, PS | 2.77 | 0.0286 |
| PSL, L | 4.94 | 0.0995 |
| PSL, CHG (0.01) | 4.29 | 0.0904 |
PSL, targeted with photosensitizer and exposed to light; NT, not treated; PS, targeted with photosensitizer, no light exposure; L, exposed to light, not targeted; CHG (0.01), exposed to 0.01 mg/ml CHG.
Ratio of mean red/green ratios (PSL/paired condition) for three independent experiments.
Determined by a paired t test for three independent experiments.
DISCUSSION
Our results indicate that the KLFC LiDps can be transported into A. actinomycetemcomitans microcolonies during a reasonable time period, that cells within the microcolonies can be targeted with the dodecamer, and that this multimeric protein can be used as a vehicle for targeted delivery of a photosensitizer to A. actinomycetemcomitans microcolonies. The susceptibility of A. actinomycetemcomitans biofilms to antimicrobials has been shown to decrease as the biofilm matures (45). Consistent with these results, we observed no light-induced decrease in the viability of 48-h biofilms targeted with photosensitizer, while there was clear evidence of light-induced membrane disruption of 24-h biofilms targeted with a photosensitizer. The more uniform Aa-MAb epitope distribution in 24-h A. actinomycetemcomitans microcolonies may have been partially responsible for this result (Fig. 3). In general, mechanical plaque removal is not sufficient to completely eliminate oral biofilm (2), and this has been shown to be the case for A. actinomycetemcomitans (34). Selective killing of A. actinomycetemcomitans early-stage biofilms would presumably enhance the ability of commensals to compete with A. actinomycetemcomitans during recolonization (47), thus conferring a measure of colonization resistance to periodontal tissues (33). In addition, the recolonization by a consortium in which commensals were predominant might aid in establishing a healthy immune balance at diseased sites, allowing periodontal tissues more leverage to eliminate intracellular (invasive) A. actinomycetemcomitans.
Transport of macromolecules through a polymicrobial oral biofilm was found previously to be significantly hindered (49). The shape of the data curves presented in Fig. 1B suggests that the LiDps dodecamer attained about 90% saturation in central regions at the base of the A. actinomycetemcomitans biofilm microcolonies at 20 min. The fluorescence in these regions at 20 min was about 35 to 95% of the fluorescence in the bulk, with a mean of about 60%. An extensive review of the literature indicated that the diffusion coefficient of macromolecules the size of the LiDps (molecular mass, 222 kDa) could be reduced in biofilms by as much as a factor of about 0.02 compared to the diffusion coefficient in water (39). This translates to about 17 min to reach 90% saturation in a 30-μm-thick planar slab, indicating that the transport kinetics of the LiDps dodecamer that we measured are consistent with previous studies. In terms of eventual clinical application, the mean fluorescence at 5 min in locations that were relatively inaccessible was approximately 25% of the bulk fluorescence. Estimating the volume excluded by the cells to be 50%, the intercellular concentration at 5 min in the interior portions of the biofilm was about 50% of the bulk concentration. This may be sufficient to effectively target A. actinomycetemcomitans biofilms during a 5-min exposure period if the affinity coefficient for binding of the targeting moieties is sufficiently high.
Advantages of the targeting approach we used in this study include modularity and ability to amplify the loading. The modularity of the system allows different targeting moieties to be combined easily with a single functionalized multimeric protein. We have shown previously that loading of a small molecule onto a 12-nm, 24-subunit small heat shock protein from Methanococcus jannaschii can be increased substantially by incorporating a polymer confined to the interior cavity delineated by the protein shell (1). Although the nucleation and stepwise growth required to synthesize the polymer are fairly sophisticated, the process can be streamlined to produce large quantities of the biotinylated product. Recently, we synthesized an asymmetrically biotinylated LiDps dodecamer that enables one-step targeting via an antibody while preserving the modular design (42). Another advantage of our targeting approach is the possibility of increasing the loading with a sequence of applications. Previously, we characterized signal amplification that can be obtained by exploiting the streptavidin/biotin interaction to obtain clusters of the cowpea chlorotic mottle virus (CCMV) capsule bound at nucleation sites (40). We acquired atomic force microscopy (AFM) images to confirm formation of CCMV clusters at a planar interface (43). Here, we used a similar approach to increase the photosensitizer loading at targeted sites. Although the amplification in the fluorescence signal shown in Fig. 3B suggests that aggregated clusters of the dodecamer are forming at sites of Aa-MAb binding as depicted in Fig. 3A, we have no direct evidence to support this interpretation. With respect to clinical relevance, topical application of a sequence of agents may be feasible if the application time is within 5 min. However, for some applications, it will obviously be desirable to have a one-step administration.
The antimicrobial action of photosensitizers against Gram-negative bacteria is known to be less than that against Gram-positive bacteria (19, 25). One method to enhance photosensitizer action against Gram-negative bacteria is to couple the photosensitizer to a cationic peptide or polymer (20, 46). This enhancement may originate from the localized concentration of photosensitizer near the cell membrane or, alternatively, from disruption of the membrane by the cationic moieties. The former explanation is consistent with the loss of viability we observed for cells targeted with photosensitizer and exposed to light. We cannot claim that cells in biofilms were in fact killed, since we have no direct evidence for this. However, our results indicate that membranes of photosensitizer-targeted cells exposed to light were more permeable than were controls, and thus, these cells may be sufficiently damaged to substantially decrease their competitiveness when growing in a developing oral consortium.
Supplementary Material
Acknowledgments
This work was supported by NIH grant 1R21DE019237-01A1 to M.Y.
We thank Casey Chen for the Aa strain and for advice on culturing the biofilms. We thank Homa Zadeh for useful discussions and Helga Lüthi-Schaller and Betsy Pitts for technical assistance.
Footnotes
Published ahead of print on 12 April 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Abedin, M. J., L. Liepold, P. Suci, M. Young, and T. Douglas. 2009. Synthesis of a cross-linked branched polymer network in the interior of a protein cage. J. Am. Chem. Soc. 131:4346-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adriaens, P. A., and L. M. Adriaens. 2004. Effects of nonsurgical periodontal therapy on hard and soft tissues. Periodontol. 2000 36:121-145. [DOI] [PubMed] [Google Scholar]
- 3.Artis, D. 2008. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 8:411-420. [DOI] [PubMed] [Google Scholar]
- 4.Barderas, R., J. Desmet, P. Timmerman, R. Meloen, and J. I. Casal. 2008. Affinity maturation of antibodies assisted by in silico modeling. Proc. Natl. Acad. Sci. U. S. A. 105:9029-9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cridland, J. C., V. Booth, F. P. Ashley, M. A. Curtis, R. F. Wilson, and P. Shepherd. 1994. Preliminary characterization of antigens recognized by monoclonal-antibodies raised to Porphyromonas gingivalis and by sera from patients with periodontitis. J. Periodont. Res. 29:339-347. [DOI] [PubMed] [Google Scholar]
- 6.de Oliveira, R. R., H. O. Schwartz-Filho, A. B. Novaes, and M. Taba. 2007. Antimicrobial photodynamic therapy in the non-surgical treatment of aggressive periodontits: a preliminary randomized controlled clinical study. J. Periodontol. 78:965-973. [DOI] [PubMed] [Google Scholar]
- 7.Douglas, T., and M. Young. 2006. Viruses: making friends with old foes. Science 312:873-875. [DOI] [PubMed] [Google Scholar]
- 8.Eckert, R., J. He, D. K. Yarbrough, F. X. Qi, M. H. Anderson, and W. Y. Shi. 2006. Targeted killing of Streptococcus mutans by a pheromone-guided “smart” antimicrobial peptide. Antimicrob. Agents Chemother. 50:3651-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggert, F. M., M. H. McLeod, and G. Flowerdew. 2001. Performance of a commercial immunoassay for detection and differentiation of periodontal marker bacteria: analysis of immunochemical performance with clinical samples. J. Periodontol. 72:1201-1209. [DOI] [PubMed] [Google Scholar]
- 10.Embleton, M. L., S. P. Nair, B. D. Cookson, and M. Wilson. 2004. Antibody-directed photodynamic therapy of methicillin resistant Staphylococcus aureus. Microb. Drug Resist. 10:92-97. [DOI] [PubMed] [Google Scholar]
- 11.Embleton, M. L., S. P. Nair, B. D. Cookson, and M. Wilson. 2002. Selective lethal photosensitization of methicillin-resistant Staphylococcus aureus using an IgG-tin(IV) chlorin e6 conjugate. J. Antimicrob. Chemother. 50:857-864. [DOI] [PubMed] [Google Scholar]
- 12.Embleton, M. L., S. P. Nair, W. Heywood, D. C. Menon, B. D. Cookson, and M. Wilson. 2005. Development of a novel targeting system for lethal photo sensitization of antibiotic-resistant strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3690-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fine, D. H., K. Markowitz, D. Furgang, K. Fairlie, J. Ferrandiz, C. Nasri, M. McKiernan, and J. Gunsolley. 2007. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: longitudinal cohort study of initially healthy adolescents. J. Clin. Microbiol. 45:3859-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischetti, V. A. 2008. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 11:393-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flenniken, M. L., L. O. Liepold, B. E. Crowley, D. A. Willits, M. J. Young, and T. Douglas. 2005. Selective attachment and release of a chemotherapeutic agent from the interior of a protein cage architecture. Chem. Commun. (Camb.) 4:447-449. [DOI] [PubMed] [Google Scholar]
- 16.Flenniken, M. L., D. A. Willits, A. L. Harmsen, L. O. Liepold, A. G. Harmsen, M. J. Young, and T. Douglas. 2006. Melanoma and lymphocyte cell-specific targeting incorporated into a heat shock protein cage architecture. Chem. Biol. 13:161-170. [DOI] [PubMed] [Google Scholar]
- 17.Gmür, R., B. Guggenheim, E. Giertsen, and T. Thurnheer. 2000. Automated immunofluorescence for enumeration of selected taxa in supragingival dental plaque. Eur. J. Oral Sci. 108:393-402. [DOI] [PubMed] [Google Scholar]
- 18.Gmür, R., and T. Thurnheer. 1996. Monoclonal antibodies for the rapid identification in clinical samples of Peptostreptococcus micros and Actinobacillus actinomycetemcomitans serotypes, a, d, and e. Med. Microbiol. Lett. 5:335-349. [Google Scholar]
- 19.Hamblin, M. R., and T. Hasan. 2004. Photodynamic therapy: a new antimicrobial approach to infectious disease? Photochem. Photobiol. Sci. 3:436-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamblin, M. R., D. A. O'Donnell, N. Murthy, K. Rajagopalan, N. Michaud, M. E. Sherwood, and T. Hasan. 2002. Polycationic photosensitizer conjugates: effects of chain length and Gram classification on the photodynamic inactivation of bacteria. J. Antimicrob. Chemother. 49:941-951. [DOI] [PubMed] [Google Scholar]
- 21.Ilari, A., S. Stefanini, E. Chiancone, and D. Tsernoglou. 2000. The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat. Struct. Biol. 7:38-43. [DOI] [PubMed] [Google Scholar]
- 22.He, J., M. H. Anderson, W. Y. Shi, and R. Eckert. 2009. Design and activity of a ‘dual-targeted’ antimicrobial peptide. Int. J. Antimicrob. Agents 33:532-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermanson, G. T. 1996. Bioconjugate techniques. Academic Press, New York, NY.
- 24.Jang, H. O., G. S. Geen, S. H. Shin, I. K. Chung, and I. Yun. 2004. Chlorhexidine digluconate alters structural parameters of outer membranes isolated from cultured Actinobacillus actinomycetemcomitans, poster B10, p. 196a. Biophys. Soc. 48th Annu. Meet. Biophysical Society, Bethesda, MD.
- 25.Jori, G. 2006. Photodynamic therapy of microbial infections: state of the art and perspectives. J. Environ. Pathol. Toxicol. Oncol. 25:505-519. [DOI] [PubMed] [Google Scholar]
- 26.Kang, S., L. Y. Mou, J. Lanman, S. Velu, W. J. Brouillette, and P. E. Prevelige. 2009. Synthesis of biotin-tagged chemical cross-linkers and their applications for mass spectrometry. Rapid Commun. Mass Spectrom. 23:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan, J. B., M. F. Meyenhofer, and D. H. Fine. 2003. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J. Bacteriol. 185:1399-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J., B. Pitts, P. S. Stewart, A. Camper, and J. Yoon. 2008. Comparison of the antimicrobial effects of chlorine, silver ion, and tobramycin on biofilm. Antimicrob. Agents Chemother. 52:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komerik, N., and A. J. MacRobert. 2006. Photodynamic therapy as an alternative antimicrobial modality for oral infections. J. Environ. Pathol. Toxicol. Oncol. 25:487-504. [DOI] [PubMed] [Google Scholar]
- 30.Martens, E. C., N. M. Koropatkin, T. J. Smith, and J. I. Gordon. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 284:24673-24677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Medina, M., P. Naves, J. Blanco, X. Aldeguer, J. E. Blanco, M. Blanco, C. Ponte, F. Soriano, A. Darfeuille-Michaud, and L. J. Garcia-Gil. 2009. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC). BMC Microbiol. 9:202-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochsner, M. 1997. Photophysical and photobiological processes in the photodynamic therapy of tumours. J. Photochem. Photobiol. B 39:1-18. [DOI] [PubMed] [Google Scholar]
- 33.Raj, P. A., and A. R. Dentino. 2005. Intraoral delivery of antimicrobials. Drug News Perspect. 18:362-374. [DOI] [PubMed] [Google Scholar]
- 34.Renvert, S., M. Wikström, G. Dahlén, J. Slots, and J. Egelberg. 1990. On the inability of root debridement and periodontal surgery to eliminate Actinobacillus actinomycetemcomitans from periodontal pockets. J. Clin. Periodontol. 17:351-355. [DOI] [PubMed] [Google Scholar]
- 35.Roberts, F. A., and R. P. Darveau. 2002. Beneficial bacteria of the periodontium. Periodontol. 2000 30:40-50. [DOI] [PubMed] [Google Scholar]
- 36.Round, J. L., and S. K. Mazmanian. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9:313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schacher, B., F. Baron, M. Rossberg, M. Wohlfeil, R. Arndt, and P. Eickholz. 2007. Aggregatibacter actinomycetemcomitans as indicator for aggressive periodontitis by two analysing strategies. J. Clin. Periodontol. 34:566-573. [DOI] [PubMed] [Google Scholar]
- 38.Socransky, S. S., and A. D. Haffajee. 2002. Dental biofilms: difficult therapeutic targets. Periodontol. 2000 28:12-55. [DOI] [PubMed] [Google Scholar]
- 39.Stewart, P. S. 1998. A review of experimental measurements of effective diffusive permeabilities and effective diffusion coefficients in biofilms. Biotechnol. Bioeng. 59:261-272. [DOI] [PubMed] [Google Scholar]
- 40.Suci, P., M. T. Klem, M. Young, and T. Douglas. 2008. Signal ampflication using nanoplatform cluster formation. Soft Matter 4:2519-2523. [Google Scholar]
- 41.Suci, P. A., D. L. Berglund, L. Liepold, S. Brumfield, B. Pitts, W. Davison, L. Oltrogge, K. O. Hoyt, S. Codd, P. S. Stewart, M. Young, and T. Douglas. 2007. High-density targeting of a viral multifunctional nanoplatform to a pathogenic, biofilm-forming bacterium. Chem. Biol. 14:387-398. [DOI] [PubMed] [Google Scholar]
- 42.Suci, P. A., S. Kang, M. Young, and T. Douglas. 2009. A streptavidin-protein cage Janus particle for polarized targeting and modular functionalization. J. Am. Chem. Soc. 131:9164-9165. [DOI] [PubMed] [Google Scholar]
- 43.Suci, P. A., M. T. Klem, F. T. Arce, T. Douglas, and M. Young. 2006. Assembly of multilayer films incorporating a viral protein cage architecture. Langmuir 22:8891-8896. [DOI] [PubMed] [Google Scholar]
- 44.Suci, P. A., Z. Varpness, E. Gillitzer, T. Douglas, and M. Young. 2007. Targeting and photodynamic killing of a microbial pathogen using protein cage architectures functionalized with a photosensitizer. Langmuir 23:12280-12286. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi, N., K. Ishihara, T. Kato, and K. Okuda. 2007. Susceptibility of Actinobacillus actinomycetemcomitans to six antibiotics decreases as biofilm matures. J. Antimicrob. Chemother. 59:59-65. [DOI] [PubMed] [Google Scholar]
- 46.Tegos, G. P., M. Anbe, C. M. Yang, T. N. Demidova, M. Satti, P. Mroz, S. Janjua, F. Gad, and M. R. Hamblin. 2006. Protease-stable polycationic photosensitizer conjugates between polyethyleneimine and chlorin(e6) for broad-spectrum antimicrobial photoinactivation. Antimicrob. Agents Chemother. 50:1402-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teughels, W., S. K. Haake, I. Sliepen, M. Pauwels, J. Van Eldere, J. J. Cassiman, and M. Quirynen. 2007. Bacteria interfere with A. actinomycetemcomitans colonization. J. Dent. Res. 86:611-617. [DOI] [PubMed] [Google Scholar]
- 48.Thrower, Y., R. J. Pinney, and M. Wilson. 1997. Susceptibilities of Actinobacillus actinomycetemcomitans biofilms to oral antiseptics. J. Med. Microbiol. 46:425-429. [DOI] [PubMed] [Google Scholar]
- 49.Thurnheer, T., R. Gmür, S. Shapiro, and B. Guggenheim. 2003. Mass transport of macromolecules within an in vitro model of supragingival plaque. Appl. Environ. Microbiol. 69:1702-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tlaskalova-Hogenova, H., R. Stepankova, T. Hudcovic, L. Tuckova, B. Cukrowska, R. Lodinova-Zadnikova, H. Kozakova, P. Rossmann, J. Bartova, D. Sokol, D. P. Funda, D. Borovska, Z. Rehakova, J. Sinkora, J. Hofman, P. Drastich, and A. Kokesova. 2004. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol. Lett. 93:97-108. [DOI] [PubMed] [Google Scholar]
- 51.Uchida, M., M. L. Flenniken, M. Allen, D. A. Willits, B. E. Crowley, S. Brumfield, A. F. Willis, L. Jackiw, M. Jutila, M. J. Young, and T. Douglas. 2006. Targeting of cancer cells with ferrimagnetic ferritin cage nanoparticles. J. Am. Chem. Soc. 128:16626-16633. [DOI] [PubMed] [Google Scholar]
- 52.Uchida, M., M. T. Klem, M. Allen, P. Suci, M. Flenniken, E. Gillitzer, Z. Varpness, L. O. Liepold, M. Young, and T. Douglas. 2007. Biological containers: protein cages as multifunctional nanoplatforms. Adv. Mater. 19:1025-1042. [Google Scholar]
- 53.Umamaheshwari, R. B., and N. K. Jain. 2003. Receptor mediated targeting of lectin conjugated gliadin nanoparticles in the treatment of Helicobacter pylori. J. Drug Target. 11:415-424. [DOI] [PubMed] [Google Scholar]
- 54.Wilson, M. 2004. Lethal photosensitisation of oral bacteria and its potential application in the photodynamic therapy of oral infections. Photochem. Photobiol. Sci. 3:412-418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.