Abstract
Molecular studies have shown that the majority of azole resistance in Aspergillus fumigatus is associated with amino acid substitutions in the cyp51A gene. To obtain insight into azole resistance mutations, the cyp51A gene of 130 resistant and 76 susceptible A. fumigatus isolates was sequenced. Out of 130 azole-resistant isolates, 105 contained a tandem repeat of 34 bp in the promoter region and a leucine-to-histidine substitution in codon 98 (designated TR/L98H). Additionally, in 12 of these TR/L98H resistant isolates, the mutations S297T and F495I were found, and in 1 isolate, the mutation F495I was found. In eight azole-resistant isolates, known azole resistance mutations were detected in codon G54, G138, or M220. In three azole-susceptible isolates, the mutation E130D, L252L, or S400I was found and in 13 azole-susceptible isolates but also in 1 azole-resistant isolate, the mutations F46Y, G98G, M172V, N248T, D255E, L358L, E427K, and C454C were found. All of the nonsynonymous mutations, apart from the mutations in codons G54, G138, and M220 and L98H, were located at the periphery of the protein, as determined by a structural model of the A. fumigatus Cyp51A protein, and were predicted neither to interact with azole compounds nor to affect structural integrity. Therefore, this wide diversity of mutations in the cyp51A gene in azole-susceptible A. fumigatus isolates is not correlated with azole resistance. Based on the Cyp51A protein homology model, the potential correlation of a mutation to azole resistance can be predicted.
Aspergillus fumigatus is the etiological agent of a range of clinical syndromes, most notably invasive aspergillosis (IA), which is associated with significant morbidity and mortality. Advances in the management of IA have been made in recent years by imaging techniques, by the use of non-culture-based diagnostic tools, by improvement in the management of patients with IA, and by the use of more effective antifungal agents, especially azole compounds (2). Azole resistance in clinical A. fumigatus isolates has long been considered to be an uncommon phenomenon, but recently multiazole resistance (MAR) has been reported to be emerging and is increasingly recognized as a cause of treatment failure (7, 18, 23). Resistance significantly complicates patient management, given the limited number of alternative agents with evidence-based efficacy in Aspergillus diseases and the fact that the azoles are the only class of antifungal drugs that can be administered orally. Azoles bind, with one of the nitrogen atoms of the azole ring, to the iron atom of the heme group located in the center of the Cyp51 protein and thereby probably block the access of lanosterol to the active site, where C14 is demethylated (24). This leads to the substitution of methylated sterols and ergosterol depletion in the fungal cell membrane, as well as to the accumulation of toxic sterol intermediates that eventually causes inhibition of fungal cell growth (19). Molecular studies have shown that in the majority of cases azole resistance in A. fumigatus is associated with mutations in the azole drug target enzyme encoded by the cyp51A gene. This has been described for azole-resistant A. fumigatus clinical isolates, as well as for laboratory-induced azole-resistant A. fumigatus mutants (5). Mutations at codon G54 are related to cross-resistance to the azoles itraconazole and posaconazole (4). Also, different mutations in codon M220 have been described in clinical strains that are correlated with itraconazole resistance combined with different patterns of elevated MICs of the other azole drugs (12). Azole-resistant A. fumigatus isolates have been found that showed a glycine-to-cysteine substitution in codon 138 and exhibited a pan-azole resistance phenotype (8). In codon 488, a mutation from glycine to serine was correlated with resistance to voriconazole and itraconazole (1). A recent survey of MAR clinical isolates in the Netherlands showed over 90% dominance of a resistance mechanism consisting of a tandem repeat of 34 bp in the promoter region combined with a leucine-to-histidine substitution at codon 98 (TR/L98H) (11, 17, 18, 20, 23).
The cyp51A sequence of Mycobacterium tuberculosis shows 28% sequence identity over 464 residues with the cyp51A sequence of A. fumigatus. Therefore, it was possible to derive the structure of the wild-type CYP51A protein of A. fumigatus from the crystal structure of M. tuberculosis by homology modeling (6, 26). The carefully optimized and checked structure of the protein was used to characterize the mutations in the CYP51A homology model. Two ligand access channels were previously identified in the model which probably not only accommodate the natural substrates but are also used by the azole compounds for docking (6, 26). The aims of our study were to obtain insight into the distribution of mutations in the cyp51A gene and to correlate the presence of cyp51A mutations with the phenotype regarding their susceptibility to azole compounds by using the Cyp51A protein homology model.
MATERIALS AND METHODS
The Department of Medical Microbiology of the Radboud University Nijmegen Medical Center has a policy of routinely storing all Aspergillus isolates cultured from clinical specimens, irrespective of their clinical significance. In addition, Aspergillus species cultured from the indoor or outdoor hospital environment or as part of research collaborations are stored.
For several research studies performed at our center, 2,925 A. fumigatus isolates were screened for resistance to itraconazole by subculturing on Sabouraud agar slants that contained 4 or 8 mg/liter itraconazole. For isolates that grew on itraconazole-containing agar (ITZ+), the in vitro activities of amphotericin B, itraconazole, voriconazole, posaconazole, and caspofungin were determined by using the Clinical and Laboratory Standards Institute M38-A reference method (13). For classification of the MICs, the recently proposed interpretative breakpoints were used (22). Isolates resistant to more than one azole compound but not all of them were designated MAR, while isolates with MICs in the resistant range for all clinically licensed mold-active azoles were designated pan-azole resistant (22).
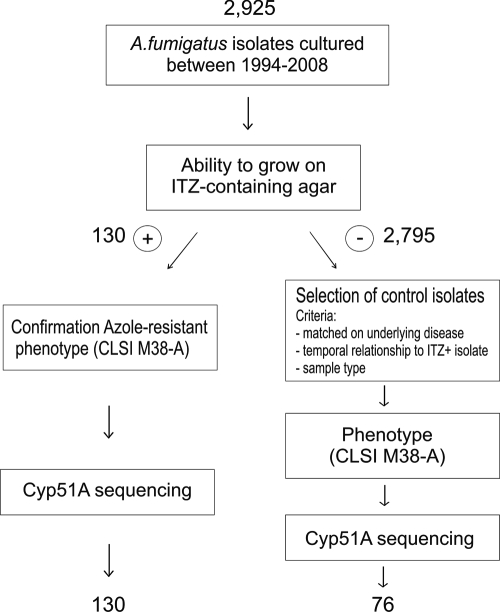
Of a total of 2,925 A. fumigatus isolates, 130 were able to grow on itraconazole-containing agar. If resistance to itraconazole was confirmed, the cyp51A gene was fully sequenced. From the isolates that failed to grow on itraconazole-containing agar (ITZ−), controls were selected. In total, 76 ITZ− isolates were randomly selected and matched on the basis of (i) the underlying disease of the patient, (ii) the temporal relationship to an ITZ+ isolate, or (iii) the sample type (Fig. 1).
FIG. 1.
Procedure for selection of ITZ-resistant and -susceptible A. fumigatus isolates for sequencing of the cyp51A gene.
All isolates were identified to the A. fumigatus species level by macroscopic and microscopic morphology, the ability to grow at 48°C, and sequencing of the β-tubulin gene. PCR amplification and subsequent sequencing of the β-tubulin gene were performed by using forward primer 5′-AATTGGTGCCGCTTTCTG-3′ and reverse primer 5′-AGTTGTCGGGACGGAATAG-3′. Cycling and sequencing conditions were the same as those described for the amplification of the cyp51A gene. Sequences were compared with those of A. fumigatus, Aspergillus lentulus, Aspergillus viridinutans, Aspergillus brevipes, Aspergillus novofumigatus, Aspergillus fumigatiaffinis, and Neosartorya species, all obtained from the CBS Fungal Biodiversity Center, Utrecht, Netherlands, by using the neighbor-joining method for phylogenetic analysis (18). For selected isolates, the full sequence of the cyp51A gene was determined. For this, conidia from each strain were inoculated into 15 ml of GYEP medium (2% glucose, 0.3% yeast extract, 1% peptone) and grown for 48 h at 37°C. Mycelial mats were recovered, dried, and subjected to a DNA isolation protocol as described previously (18). PCR amplification and subsequent sequencing of the cyp51A gene were performed by using forward primer 5′-ATGGTGCCGATGCTATGG-3′ and reverse primer 5′-CTGTCTCACTTGGATGTG-3′. Cycling conditions were as follows: 95°C for 5 min and 40 cycles of 94°C for 30 s, 58°C for 45 s, and 72°C for 2 min, followed by a final extension at 72°C for 7 min. All DNA sequences were determined using a BigDye Terminator version 3.1 cycle sequencing kit (ABI) and an ABI 3100 DNA sequencer. Sequences were aligned with a reference cyp51A sequence (GenBank accession no. AF338659) to identify mutations.
A homology model of the A. fumigatus cyp51A gene was built using the crystal structure of M. tuberculosis Cyp51A as a template (PDB code 1e9x). The two proteins show 28% sequence identity over 464 residues. The WHAT IF web server (http://swift.cmbi.ru.nl) was used for model building, and Yasara was used for energy minimization and analysis. The structure was initially optimized in a vacuum with all backbone atoms fixed. The locations of the mutations were pinpointed, and they were introduced into the Cyp51A homology model by using the VMD software (9). Furthermore, the mutations were investigated for their potential interactions with azole compounds by considering the ligand binding residues, the reported substrate access channel residues, and known mutation hot spots (10, 26).
RESULTS AND DISCUSSION
A total of 206 A. fumigatus isolates, 130 ITZ+ and 76 ITZ−, were analyzed for mutations in the cyp51A gene. A wide variety of mutations was found in the cyp51A gene of ITZ+ and ITZ− isolates, both known and not known from the literature to be correlated with azole resistance (Table 1). In 113 ITZ+ isolates, cyp51A mutations known to be correlated with azole resistance were found. In 105 ITZ+ isolates, a tandem repeat of 34 bp in the promoter region and a leucine-to-histidine substitution at codon 98 (TR/L98H) were found, and in 8 ITZ+ isolates, other known mutations were found (11).
TABLE 1.
Mutations found in the cyp51A gene of A. fumigatus and azole susceptibility phenotypes
| Phenotype and no. of strains | MIC (range)a (μg/ml) |
Mutation(s) in cyp51A gene | Source(s) (no.) of isolate(s) | ||
|---|---|---|---|---|---|
| ITZ | VRC | POS | |||
| ITZ+ (total n = 130) | |||||
| 1 | >16 | 0.25 | 0.25 | G54Eb | Clinical |
| 1 | >16 | 0.25 | >16 | G54Wb | Clinical |
| 1 | >16 | 8 | >16 | G138Cb | Clinical |
| 1 | >16 | 2 | 2 | M220Rb | Clinical |
| 2 | >16 | 1 | 0.5 | M220Ib | Clinical |
| 1 | >16 | 1 | 0.5 | M220Vb | Clinical |
| 1 | >16 | 2 | >16 | M220Kb | Clinical |
| 1 | >16 | >16 | 1 | F46Y, G89G, M172V, N248T, D255E, L358L, E427K, C454C | Clinical |
| 12 | 16->16 | 1-4 | 0.063-0.25 | TR,c L98H, S297T, F495I | Clinical (11), environmental (1) |
| 1 | >16 | 2 | 0.125 | TR, L98H, F495I | Clinical |
| 1 | >16 | 4 | 0.5 | TR, L98H, S52T | Environmental |
| 1 | >16 | 8 | 0.25 | TR, L98H, Q88H | Clinical |
| 1 | >16 | 8 | 0.5 | TR, L98H, N125I | Clinical |
| 1 | >16 | 4 | 0.5 | TR, L98H, Q141H | Environmental |
| 1 | >16 | 4 | 0.5 | TR, L98H, A284A | Clinical |
| 1 | >16 | 8 | 1 | TR, L98H, L339L | Clinical |
| 86 | 4->16 | 2-16 | 0.25-1 | TR, L98H | Clinical |
| 16 | 8->16 | 0.25-4 | 0.125->16 | None | Clinical (13), environmental (3) |
| ITZ− (total n = 76) | |||||
| 1 | 0.125 | 0.5 | 0.016 | F46Y, G89G | Clinical |
| 1 | 0.25 | 0.25 | 0.063 | F46Y, G89G, E427K, C454C | Clinical |
| 1 | 0.25 | 2 | 0.063 | F46Y, G89G, L358L, E427K, C454C | Clinical |
| 1 | 0.25 | 1 | 0.125 | F46Y, G89G, M172V, L358L, C454C | Environmental |
| 1 | 0.125 | 1 | 0.063 | F46Y, M172V, L358L, E427K, C454C | Clinical |
| 5 | 0.125-0.5 | 0.5-1 | 0.063-0.125 | F46Y, G89G, M172V, L358L, E427K, C454C | Clinical (4), environmental (1) |
| 1 | 0.25 | 1 | 0.063 | F46Y, G89G, M172V, N248T, D255E, E427K | Clinical |
| 2 | 0.25 | 0.25 | 0.063 | F46Y, G89G, M172V, N248T, D255E, L358L, E427K, C454C | Clinical |
| 1 | 0.5 | 0.5 | 0.125 | E130D | Clinical |
| 1 | 0.25 | 1 | 0.5 | L252L | Clinical |
| 1 | 1 | 2 | 0.25 | S400I | Clinical |
| 60 | 0.125 | 0.5 | 0.031 | None | Clinical (47), environmental (13) |
Each isolate was tested once. When multiple isolates contain the same mutation, the MIC range is given. VRC, voriconazole; POS, posaconazole.
Mutation known from the literature to be correlated with azole resistance.
TR; tandem repeat of 34 bp in the promoter region of the cyp51A gene.
In 19 isolates that contained the TR/L98H mutations, additional mutations in the cyp51A gene (S297T, F495I, S52T, Q88H, N125I, Q141H, A284A, and L339L) were found. In one ITZ+ isolate, the mutations F46Y, G98G, M172V, N248T, D255E, L358L, E427K, and C454C were found, and in 16 ITZ+ isolates, no mutations were found in the cyp51A gene. In 13 ITZ− isolates, combinations of the mutations F46Y, G98G, M172V, N248T, D255E, L358L, E427K, and C454C were found, and in three ITZ− the mutations, E130D, L252L, and S400I were found. In 60 ITZ− isolates, no mutations were found in the cyp51A gene.
ITZ+ isolates. (i) Cyp51A mutations known to be correlated with azole resistance.
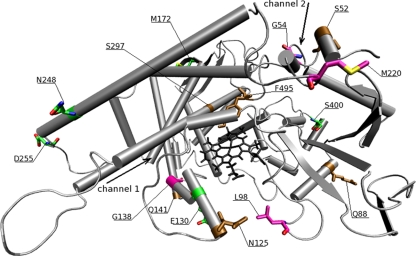
In eight ITZ+ isolates, mutations changing amino acid G54, G138, or M220 in the cyp51A-encoded protein were detected (Table 1). These mutations were all known to be correlated with azole resistance (4, 8, 12). The amino acids in codons G54, G138, and M220 were all located in close proximity to the opening of one of the two ligand access channels of the protein (Fig. 2). The ligand access channels are thought to be used by the azole compounds to enter the active site of the protein, and alterations of these openings of these channels due to amino acid changes probably disturb the docking of azole molecules. The G138C mutation was located in a helix of channel 1 close to the heme cofactor, and isolates harboring this mutation show a pan-azole resistance phenotype (Fig. 2). This was also described previously for G138R, in which the arginine was predicted to clash with one of the heme's side chains and with the side chains of the neighboring residues (26). Amino acids G54 and M220 are located in loops in close proximity to the opening of channel 2. The M220R, -I, and -V mutations produced an itraconazole resistance phenotype combined with an increased MIC of voriconazole, while M220K produced an itraconazole and posaconazole resistance phenotype. The mutation G54W produced an itraconazole and posaconazole resistance yet voriconazole susceptibility phenotype. As glycine is replaced with the larger and hydrophobic tryptophan residue, it might sufficiently close the entrance of access channel 2 to disturb the docking of large molecules such as those of itraconazole and posaconazole. The long tails of itraconazole and posaconazole need to make interactions along the opening and within the channel for stable docking toward the heme center. The voriconazole molecule, however, is much smaller than those of itraconazole and posaconazole since it lacks a long tail. It does not need to make interactions at the entrance surface of the channel. Therefore, changes in residues in codon 54, which is located at the entrance of the channel, do not affect voriconazole but do have a dramatic effect on the docking of itraconazole and posaconazole. Furthermore, the G54E mutation produced only an itraconazole resistance phenotype. The main difference in structure between posaconazole and itraconazole is the presence of the hydroxyl group at the end of the long tail of posaconazole that may create a hydrogen bond interaction on the surface of the protein. Moreover, it is not clear whether an additional oxygen atom in the core scaffold of itraconazole plays any role in preventing the drug from accessing the active site in the presence of the G54E mutation. Therefore, not only the specific location of an amino acid change is important but the amino acid replacement is of at least the same importance for the azole susceptibility phenotype. Although there are two ligand access channels present, the change of an amino acid in the opening in one channel is already sufficient for certain azole resistance phenotypes. It seems that the structural changes in one channel affect the docking of certain azole compounds for the whole protein.
FIG. 2.
Mapping of mutations found in the A. fumigatus cyp51A gene in the CYP51A homology model. The two ligand access channels are indicated by arrows. The heme cofactor is black. Mutations correlated with azole resistance are purple, and mutations correlated with an azole-susceptible profile are green. Mutations additionally found in azole-resistant strains are brown.
Out of 130 azole-resistant isolates, 105 contained the TR/L98H mutations. It was shown that leucine 98 is not located close to either the active heme center or any of the two ligand access channels but is located on a loop that partly forms an arch-like structure that is highly conserved among the members of the CYP51 family of proteins (Fig. 2). In a preliminary study, we have used molecular dynamic simulations changing the leucine to histidine in codon 98. In these studies, we observed that the flexibility of this arch-like structure increased considerably, thereby narrowing the diameter of the entry of the ligand access channels (data not shown). These changes may result in the MAR phenotype by affecting the ligand access channels important for the docking of azole compounds.
(ii) ITZ+ isolates with other cyp51A mutations.
In each of six isolates with the TR/L98H mutations, an additional mutation was found, four nonsynonymous mutations, namely, S52T, Q88H, N125I, and Q141H, and two synonymous mutations, L339L and A284A (Table 1). Introduction of these mutations into the CYP51A homology model showed that these mutations were not located in conserved regions of the protein but were all distributed on the periphery of the protein, except for two, S52T and N125I (Fig. 2). S52 was located at the entrance of channel 2; however, the serine is replaced with an only slightly larger threonine that is similar to serine, an alcoholic residue with intermediate hydrophobicity. The N125I mutation represents a change from electrophilic asparagine to a large nonpolar isoleucine and may have a larger impact on resistance. However, as the isolates all contain the TR/L98H mutations and exhibit the MAR phenotype, these additional nonsynonymous mutations probably play only a minor or no role at all in the azole resistance phenotype and no differences in MIC values were observed between these isolates and those containing only the TR/L98H mutations. It is important to correlate changes in the structural integrity of the protein with susceptibility to azole compounds because structural changes could affect the docking of the azole molecules, with azole resistance as a possible consequence. The amino acid residues on the periphery of the protein generally have a lower conservation level and are not essential for the functionality of the protein; therefore, amino acid changes in these residues will not influence structural integrity. The structural integrity of the Cyp51A protein is not changed by the two synonymous mutations L339L and A284A, since the amino acids are not changed. However, the possibility of changes at the RNA structural level due to silent mutations cannot be excluded, as synonymous mutations in RNA regulatory elements might alter transcription, splicing, and other regulatory processes, although there is no evidence that they may actually play a role in A. fumigatus (21, 25).
In 12 isolates with the TR/L98H mutations, the nonsynonymous mutations S297T and F495I were found, and in 1 isolate with the TR/L98H mutations, only the F495I mutation was found (Table 1). Interestingly, although both mutations are located close to the active site of the protein, they do not seem to affect resistance, as the TR/L98H mutations already produce the MAR phenotype and no differences in MIC values were observed between these isolates and those containing only the TR/L98H mutations. Apart from that, although threonine is slightly larger than serine and both residues contain a hydroxyl group, in the case of the second observed mutation, F495I, the hydrophobic phenylalanine is replaced with a hydrophobic though not aromatic isoleucine. The slight differences are not expected to influence the binding of the external ligands, and therefore these additional mutations do not seem to affect protein function either (brown, Fig. 2).
(iii) ITZ+ isolates without cyp51A mutations.
In 16 azole-resistant isolates, no mutations were found in the cyp51A gene. Azole-resistant A. fumigatus isolates without mutations in the cyp51A gene have been reported previously in the literature, which suggests that alternative mechanisms of azole resistance exist (7). The upregulation of efflux pumps has been described as a mechanism of azole resistance in Candida albicans previously; however, for clinical A. fumigatus strains, this possibility has only been described once (3, 16). A selection of two of these ITZ+ isolates did not show any significant increase in the mRNA expression levels of the ABC transporter AtrF or the multidrug resistance protein MDR3 or MDR4 compared to ITZ− isolates (unpublished observations). Therefore, in these isolates, an as-yet-unknown mechanism of azole resistance must be present. Possibly, the cyp51A gene could be upregulated, although no changes in the promoter region of the cyp51A gene were present. Further investigation is warranted to characterize the alternative mechanisms of resistance in these isolates.
ITZ− isolates.
In 60 of 76 azole-susceptible isolates, no mutations were found in the cyp51A gene. However, in 3 susceptible isolates, either the nonsynonymous mutation E130D or S400I or the synonymous mutation L252L was found. E130D and S400I are both located on the periphery of the protein and, indeed, do not seem to interact with azole compounds. Also, in different combinations, five nonsynonymous mutations, F46Y, M172V, N248T, D255E, and E427K, and three synonymous mutations, G98G, L358L, and C454C, were found in 13 azole-susceptible isolates. These mutations were also found in one azole-resistant isolate without any other known mutation in the cyp51A gene related to azole resistance. These exact same mutations have been described previously in the literature to be found in both azole-resistant and -susceptible isolates (7, 14). Therefore, amino acid changes found in the protein encoded by the cyp51A gene are not exclusively correlated with the development of azole resistance. Introducing the nonsynonymous amino acid alterations in the CYP51A homology model shows that these amino acid changes are all distributed at the periphery of the protein, not close to any of the two ligand access channels (Fig. 2), and are not located in any of the conserved regions of the CYP51A protein. Therefore, no effect on the biological activity of the CYP51A protein or the docking of the azole compounds in the ligand access channels is expected.
Due to the importance of CYP51 in antifungal drug studies, many CYP51 homology models have been made, e.g., C. albicans, Cryptococcus neoformans, Penicillium digitatum, and Saccharomyces cerevisiae, by using the crystal structure of M. tuberculosis CYP51 as a template (10, 15, 26, 27). Although this approach has several limitations, functionally important regions are conserved among the members of this fungal CYP51 family. A wide diversity of amino acid changes probably not correlated with resistance has been described by others (7, 14). In this study, we showed that the use of a CYP51A homology model can be very informative in predicting the effects of these amino acid changes. Specific residues in the CYP51A protein are of importance for azole docking, and identifying the location of a mutation in the CYP51A protein can help to predict whether a mutation can be considered a polymorphism or whether it has a potential correlation with azole resistance. Taking into account the diversity of cyp51A mutations, new mutations found in the cyp51A gene in azole-resistant isolates should be interpreted with care. By using a CYP51A protein homology model, a mutation can be investigated for a correlation with azole resistance. Subsequently, by placing the mutation into a wild-type strain by recombinant analysis, the impact of a mutation can be studied at the protein level to make a conclusive correlation with azole resistance.
Footnotes
Published ahead of print on 12 April 2010.
REFERENCES
- 1.Bellete, B., H. Raberin, J. Morel, P. Flori, J. Hafid, and R. T. Sung. 2010. Acquired resistance to voriconazole and itraconazole in a patient with pulmonary aspergilloma. Med. Mycol. 48:197-200. [DOI] [PubMed] [Google Scholar]
- 2.Boudewijns, M., P. E. Verweij, and W. J. Melchers. 2006. Molecular diagnosis of invasive aspergillosis: the long and winding road. Future Microbiol. 1:283-293. [DOI] [PubMed] [Google Scholar]
- 3.Cannon, R. D., E. Lamping, A. R. Holmes, K. Niimi, P. V. Baret, M. V. Keniya, K. Tanabe, M. Niimi, A. Goffeau, and B. C. Monk. 2009. Efflux-mediated antifungal drug resistance. Clin. Microbiol. Rev. 22:291-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz-Guerra, T. M., E. Mellado, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2003. A point mutation in the 14alpha-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira, M. E., A. L. Colombo, I. Paulsen, Q. Ren, J. Wortman, J. Huang, M. H. Goldman, and G. H. Goldman. 2005. The ergosterol biosynthesis pathway, transporter genes, and azole resistance in Aspergillus fumigatus. Med. Mycol. 43(Suppl. 1):S313-S319. [DOI] [PubMed] [Google Scholar]
- 6.Gollapudy, R., S. Ajmani, and S. A. Kulkarni. 2004. Modeling and interactions of Aspergillus fumigatus lanosterol 14-alpha demethylase ‘A’ with azole antifungals. Bioorg. Med. Chem. 12:2937-2950. [DOI] [PubMed] [Google Scholar]
- 7.Howard, S. J., D. Cerar, M. J. Anderson, A. Albarrag, M. C. Fisher, A. C. Pasqualotto, M. Laverdiere, M. C. Arendrup, D. S. Perlin, and D. W. Denning. 2009. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 15:1068-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howard, S. J., I. Webster, C. B. Moore, R. E. Gardiner, S. Park, D. S. Perlin, and D. W. Denning. 2006. Multi-azole resistance in Aspergillus fumigatus. Int. J. Antimicrob. Agents 28:450-453. [DOI] [PubMed] [Google Scholar]
- 9.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33-38. [DOI] [PubMed] [Google Scholar]
- 10.Lewis, D. F., A. Wiseman, and M. H. Tarbit. 1999. Molecular modelling of lanosterol 14 alpha-demethylase (CYP51) from Saccharomyces cerevisiae via homology with CYP102, a unique bacterial cytochrome P450 isoform: quantitative structure-activity relationships (QSARs) within two related series of antifungal azole derivatives. J. Enzyme Inhib. 14:175-192. [DOI] [PubMed] [Google Scholar]
- 11.Mellado, E., G. Garcia-Effron, L. Alcazar-Fuoli, W. J. Melchers, P. E. Verweij, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51:1897-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellado, E., G. Garcia-Effron, L. Cazar-Fuoli, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2004. Substitutions at methionine 220 in the 14alpha-sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob. Agents Chemother. 48:2747-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi. Approved standard M38-A. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 14.Rodriguez-Tudela, J. L., L. Alcazar-Fuoli, E. Mellado, A. Alastruey-Izquierdo, A. Monzon, and M. Cuenca-Estrella. 2008. Epidemiological cutoffs and cross-resistance to azole drugs in Aspergillus fumigatus. Antimicrob. Agents Chemother. 52:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheng, C., Z. Miao, H. Ji, J. Yao, W. Wang, X. Che, G. Dong, J. Lu, W. Guo, and W. Zhang. 2009. Three-dimensional model of lanosterol 14 alpha-demethylase from Cryptococcus neoformans: active-site characterization and insights into azole binding. Antimicrob. Agents Chemother. 53:3487-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slaven, J. W., M. J. Anderson, D. Sanglard, G. K. Dixon, J. Bille, I. S. Roberts, and D. W. Denning. 2002. Increased expression of a novel Aspergillus fumigatus ABC transporter gene, atrF, in the presence of itraconazole in an itraconazole resistant clinical isolate. Fungal Genet. Biol. 36:199-206. [DOI] [PubMed] [Google Scholar]
- 17.Snelders, E., R. A. Huis In 't Veld, A. J. Rijs, G. H. Kema, W. J. Melchers, and P. E. Verweij. 2009. Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Appl. Environ. Microbiol. 75:4053-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snelders, E., H. A. van der Lee, J. Kuijpers, A. J. Rijs, J. Varga, R. A. Samson, E. Mellado, A. R. Donders, W. J. Melchers, and P. E. Verweij. 2008. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 5:e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanden Bossche, H., L. Koymans, and H. Moereels. 1995. P450 inhibitors of use in medical treatment: focus on mechanisms of action. Pharmacol. Ther. 67:79-100. [DOI] [PubMed] [Google Scholar]
- 20.van Leer-Buter, C., R. P. Takes, K. M. Hebeda, W. J. Melchers, and P. E. Verweij. 2007. Aspergillosis—and a misleading sensitivity result. Lancet 370:102. [DOI] [PubMed] [Google Scholar]
- 21.van Ooij, M. J., D. A. Vogt, A. Paul, C. Castro, J. Kuijpers, F. J. van Kuppeveld, C. E. Cameron, E. Wimmer, R. Andino, and W. J. Melchers. 2006. Structural and functional characterization of the coxsackievirus B3 CRE(2C): role of CRE(2C) in negative- and positive-strand RNA synthesis. J. Gen. Virol. 87:103-113. [DOI] [PubMed] [Google Scholar]
- 22.Verweij, P. E., S. J. Howard, W. J. Melchers, and D. W. Denning. 2009. Azole-resistance in Aspergillus: proposed nomenclature and breakpoints. Drug Resist. Updat. 12:141-147. [DOI] [PubMed] [Google Scholar]
- 23.Verweij, P. E., E. Mellado, and W. Melchers. 2007. Multiple-triazole-resistant aspergillosis. N. Engl. J. Med. 356:1481-1483. [DOI] [PubMed] [Google Scholar]
- 24.Waterman, M. R., and G. I. Lepesheva. 2005. Sterol 14 alpha-demethylase, an abundant and essential mixed-function oxidase. Biochem. Biophys. Res. Commun. 338:418-422. [DOI] [PubMed] [Google Scholar]
- 25.Wray, G. A. 2007. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8:206-216. [DOI] [PubMed] [Google Scholar]
- 26.Xiao, L., V. Madison, A. S. Chau, D. Loebenberg, R. E. Palermo, and P. M. McNicholas. 2004. Three-dimensional models of wild-type and mutated forms of cytochrome P450 14alpha-sterol demethylases from Aspergillus fumigatus and Candida albicans provide insights into posaconazole binding. Antimicrob. Agents Chemother. 48:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao, L., D. Liu, Q. Zhang, S. Zhang, J. Wan, and W. Xiao. 2007. Expression and homology modeling of sterol 14alpha-demethylase from Penicillium digitatum. FEMS Microbiol. Lett. 277:37-43. [DOI] [PubMed] [Google Scholar]