Abstract
Nocardia farcinica strains showing high-level resistance to amikacin were isolated from clinical cases in a Canada-wide bovine mastitis epizootic. Shotgun cloning of the resistance genes in the amikacin-resistant mastitis isolate N. farcinica IFM 10580 (W6220 [Centers for Disease Control and Prevention]) using a multicopy vector system revealed that the 16S rRNA gene with an A-to-G single-point mutation at position 1408 (in Escherichia coli numbering) conferred “moderate” cross-resistance to amikacin and other aminoglycosides to an originally susceptible N. farcinica strain IFM 10152. Subsequent DNA sequence analyses revealed that, in contrast to the susceptible strain, all three chromosomal 16S rRNA genes of IFM 10580, the epizootic clinical strain, contained the same A1408G point mutations. Mutant colonies showing high-level aminoglycoside resistance were obtained when the susceptible strain N. farcinica IFM 10152 was transformed with a multicopy plasmid carrying the A1408G mutant 16S rRNA gene and was cultured in the presence of aminoglycosides for 3 to 5 days. Of these transformants, at least two of the three chromosomal 16S rRNA genes contained A1408G mutations. A triple mutant was easily obtained from a strain carrying the two chromosomal A1408G mutant genes and one wild-type gene, even in the absence of the plasmid. The triple mutant showed the highest level of resistance to aminoglycosides, even in the absence of the plasmid carrying the mutant 16S rRNA gene. These results suggest that the homozygous mutations in the three 16S rRNA genes are responsible for the high-level aminoglycoside resistance found in N. farcinica isolates of the bovine mastitis epizootic.
Members of the genus Nocardia are aerobic Gram-positive soil saprophytes that cause nocardiosis in humans and animals by infecting lung, skin, brain, and other tissues (5). More than 70 species of the genus Nocardia are known (http://www.bacterio.cict.fr/n/nocardia.html), approximately half of which are reported as pathogens. Nocardiosis is on the rise, along with the increase of immunocompromised patients: more than 100 new cases occur annually in Japan (16), with 500 to 1,000 new cases occurring in the United States (26). Of the greatest clinical importance within this genus is Nocardia farcinica: infectious cases attributable to this pathogen are the highest among Nocardia species in Japan and Europe. They often have multidrug resistance (13). Because treatment for nocardiosis relies heavily on chemotherapy, this intrinsic multiple drug resistance presents a serious clinical problem.
Recently, N. farcinica was identified as the causative agent of a Canada-wide epizootic of bovine Nocardia mastitis reported during January 1987 to June 1989, linked to repeated intramammary antibiotic therapy and unsanitary practices during intramammary infusion (8, 18, 22, 28). The results of a recent study suggest that the epizootic resulted from transmission of a unique clone of N. farcinica through intrinsically contaminated dry cow intramammary products (4). Epizootic mastitis isolates of N. farcinica showed high-level resistance to the aminoglycoside amikacin (MIC > 2,048 μg/ml), which has been used for treatment of Nocardia species with drug resistance in dairy farming. Resistance to amikacin has not been observed in Nocardia species isolated from human patients.
Although aminoglycoside resistance in clinical isolates results from chemical modification of drugs in many cases (19), modification of amikacin has not been observed in amikacin-resistant epizootic mastitis strains of N. farcinica (our unpublished data). Moreover, because the level of amikacin resistance in the Canada-wide epizootic is extremely high, a unique resistance mechanism, e.g., an alteration of drug target in these strains, is suggested (4).
In the present study, we identified the resistance determinant that is responsible for the high-level aminoglycoside cross-resistance in N. farcinica isolates obtained in the Canada-wide bovine epizootic mastitis through shotgun cloning of genes from one of these isolates, IFM 10580 (W6220; Centers for Disease Control and Prevention [CDC]) (4). The cloned gene A1408G 16S rRNA conferred broad-spectrum aminoglycoside cross-resistance to an amikacin-susceptible N. farcinica IFM 10152, elucidating the molecular mechanism underlying the high-level aminoglycoside resistance found in the epizootic.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The mastitis epizootic strain N. farcinica IFM 10580, originally named W6220 (4), was provided by the Special Bacteriology Reference Laboratory, CDC. Amikacin-susceptible N. farcinica strain IFM 10152 was maintained at the Medical Mycology Research Center, Chiba University, Chiba, Japan. The strains were cultured in brain heart infusion (BHI; BD Biosciences) broth with or without antibiotics (Table 1) . For cloning experiments, Escherichia coli DH5α was used as a host strain. The E. coli were cultured in Luria-Bertani (LB) medium containing 100 μg of ampicillin or kanamycin/ml. Nocardia-E. coli shuttle vector pNV19, which contains a neomycin resistance marker (6), was obtained from the National Institute of Infectious Diseases, Tokyo, Japan.
TABLE 1.
MICs of various aminoglycosides for amikacin-susceptible (IFM 10152/pNV19), amikacin-resistant (IFM 10580), and transformed [IFM 10152/pNV19- 16S(1408G)] N. farcinica strains
| Aminoglycoside | MIC (μg/ml)a for strain: |
|||
|---|---|---|---|---|
| IFM 10152b (day 3) | IFM 10580 (day 3) | IFM 10152/pNV19-16S(1408G) |
||
| Day 3 | Day 5 | |||
| Amikacin | <1 | >1,024 | 64 | >1,024 |
| Kanamycin | 256 | >1,024 | >1,024 | >1,024 |
| Neomycin | 16 | >1,024 | >1,024 | >1,024 |
| Streptomycin | 64 | 128 | 64 | 128 |
| Arbekacin | <1 | >1,024 | 2 | >1,024 |
| Gentamicin | 64 | >1,024 | 256 | >1,024 |
| Tobramycin | 32 | >1,024 | 64 | >1,024 |
| Paromomycin | 8 | >1,024 | >1,024 | >1,024 |
| Apramycin | 32 | >1,024 | 64 | 256 |
| Isepamicin | <1 | >1,024 | 16 | 128 |
| Hygromycin B | 32 | 32 | 32 | 32 |
| Fortimicin | 8 | >1,024 | 16 | >1,024 |
| Ribostamycin | 256 | >1,024 | >1,024 | >1,024 |
| Netilmicin | <1 | >1,024 | 4 | 32 |
All transformed cells were cultured in the presence of each aminoglycosides for 3 or 5 days.
The susceptible strain was transformed with the empty vector.
Cloning of N. farcinica IFM 10580 genes into a plasmid vector.
Genomic DNA of strain IFM 10580 was partially digested with Sau3AI, and 1- to 10-kb fragments were inserted into the unique BamHI site of the Nocardia-E. coli shuttle vector pNV19.
Transformation of the Nocardia strain and recovery of plasmids.
The N. farcinica strain IFM 10152 was transformed using 50 to 100 ng of plasmid DNA by electroporation, as described in a previous report (14). Cells were plated on BHI agar plates containing 50 μg of amikacin/ml and incubated for 2 to 3 days at 37°C. Plasmids were recovered from the amikacin-resistant transformants by using Isoplant II (Nippon Gene Co., Ltd.). The recovered plasmids were reintroduced into the susceptible strain IFM 10152. The resultant transformant was IFM 10152/pNV19-16S(1408G) (see Results and Tables 1 and 2, analysis 2).
TABLE 2.
Relations between the subtypes of genomic and plasmid-encoded 16S rRNA genes and amikacin susceptibility in N. farcinica strains IFM 10152 and IFM 10580
| Analysis | Straina | Nucleotide at position 1408 of genomic 16S rRNA genesb |
pNV19-16S (1408G)c | Amikacin susceptibility (MIC, μg/ml)d | ||
|---|---|---|---|---|---|---|
| rrnA | rrnB | rrnC | ||||
| 1 | IFM 10152 (amikacin susceptible) | A | A | A | - | Susceptible (<1) |
| 2 | IFM 10152/pNV19-16S(1048G) | A | A | A | + | Moderately resistant (∼64) |
| 3 | IFM 10152-M2/pNV19-16S(1048G)* | G | A | G | + | Highly resistant (>1,024) |
| 4 | IFM 10152-M2* | G | A | G | - | Susceptible (<1) |
| 5 | IFM 10152-M3* | G | G | G | - | Highly resistant (>1,024) |
| 6 | IFM 10580 (amikacin resistant) | G | G | G | - | Highly resistant (>1,024) |
*, Strains IFM 10152-M2 and IFM 10152-M3 were generated as described in the text.
Nucleotides at position 1408 are either wild-type (A) or mutated (G).
The plasmid carrying 16S rRNA gene with an A1408G mutation is either present (+) or absent (-).
MICs were determined on day 3 of incubation.
Establishment of strains highly resistant to aminoglycosides.
By incubating IFM 10152/pNV19-16S(1408G) cells in the presence of 1,024 μg of amikacin/ml at 37°C for 5 days, N. farcinica strains that were highly resistant to aminoglycosides [IFM 10152-M2/pNV19-16S(1408G); see Table 2, analysis 3] were obtained. Strain IFM 10152-M2 (see Table 2, analysis 4) was obtained by eliminating the plasmid pNV19-16S(1408G) from the host cells by incubating the cells in BHI medium without amikacin at 37°C for 2 days and then isolating colonies on BHI agar plates. Elimination of the plasmid was confirmed by examining whether the host cells regained neomycin sensitivity. Genomic 16S rRNA gene sequences of the plasmid-eliminated cells were analyzed by using PCR amplification with specific primers for the three genes (see below), followed by sequencing.
Strain IFM 10152-M3 was obtained spontaneously by incubating IFM 10152-M2 in the presence of 1,024 μg of amikacin/ml for 1 day.
Susceptibility test.
Susceptibilities to amikacin and other aminoglycosides were determined by using the broth microdilution method in BHI medium with a final bacterial concentration of 104 CFU/ml. Cells were incubated in the presence of various aminoglycosides at 37°C for 3 or 5 days. The optical densities at 600 nm (OD600) were then measured to determine the MICs.
The terms “susceptible” and “resistant” were used based on CLSI standards (21).
DNA techniques.
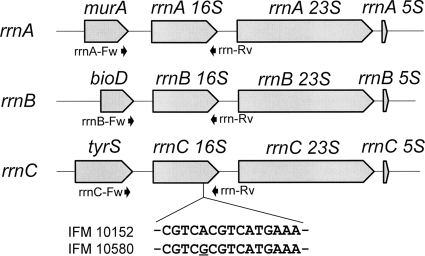
Genomic 16S rRNA genes rrnA, rrnB, and rrnC of N. farcinica strains (15) were amplified by using the following forward primers rrnA-Fw (5′-TACCCGAACTTCGTCGAG-3′), rrnB-Fw (5′-ACACCTCGCAATCGGGTACG-3′), and rrnC-Fw (5′-GTGAACAACCAGAAGATCGC-3′), respectively (Fig. 1). The reverse primer rrn-Rv (5′-GTACGGCTACCTTGTTAC-3′) was used for all three genes. For nucleotide sequencing, BigDye Terminator v3.1 cycle sequencing kits and a 3130 Genetic Analyzer (Applied Biosystems) were used in accordance with the manufacturer's instructions.
FIG. 1.
N. farcinica rRNA operons, rrnA, rrnB, rrnC, and upstream genes. Positions of forward and reverse PCR primers used in the present study are indicated by arrows. Partial sequences of the A-sites for wild-type (IFM 10152) and the epizootic aminoglycoside resistant (IFM 10580) strains are also shown.
The 16S rRNA sequence of IFM 10580 (W6220) was deposited to GenBank (GenBank/EMBL/DDBJ accession no. EF452728) (4).
RESULTS
To identify molecular determinants responsible for the high-level amikacin resistance observed in N. farcinica isolate as a causative pathogen of the Canada-wide bovine mastitis epizootic, we performed shotgun cloning for the resistant genes of the mastitis isolate N. farcinica IFM 10580, using the Nocardia-E. coli shuttle vector pNV19 (6) and the amikacin-susceptible N. farcinica strain IFM 10152 as a host strain. Transformants were selected on BHI agar plates containing amikacin (50 μg/ml). Of 24 plasmids isolated independently from the amikacin-resistant colonies, 10 showed moderate amikacin resistance (MIC ≈ 64 μg/ml) when reintroduced to the susceptible strain IFM 10152 (MIC < 1 μg/ml). Sequencing of insert DNAs (2 to 6 kb) of the isolated plasmids that conferred amikacin resistance revealed that all 10 inserts contained the entire region of N. farcinica 16S rRNA gene. In particular, some inserts contained the coding region of the 16S rRNA gene and their short flanking sequences. The adjacent sequences to respective 16S rRNA genes in the isolated plasmids indicated that these 16S rRNA genes were derived either from the rrnA or the rrnC rRNA operon, which are two of the three rRNA operons present in N. farcinica genome (15). Comparison of the nucleotide sequences of 16S rRNA genes in the resistant strain with those of amikacin-susceptible N. farcinica strain IFM 10152 revealed that 16S rRNA gene sequences had guanine-for-adenine substitutions at the conserved 16S rRNA position 1408 (in E. coli numbering), which constitutes the aminoacyl site (A-site) of the ribosomal decoding region (helix 44 of 16S rRNA), and corresponds to the binding target of 2-deoxystreptamine (2-DOS) aminoglycoside antibiotics, as shown previously in E. coli (8). No other substitution was observed in the isolated 16S rRNA genes compared to rrnA and rrnB genes of the susceptible IFM 10152 strain.
Susceptibilities to several aminoglycosides in the amikacin-susceptible clinical strain IFM 10152 carrying pNV19 (empty vector) (IFM 10152/pNV19), amikacin-resistant clinical strain IFM 10580, and IFM 10152 carrying pNV19 containing the 16S rRNA gene with A1408G point mutation [IFM 10152/pNV19-16S(1408G)] are compared in Table 1. The susceptible strain IFM 10152 had susceptibility to arbekacin, isepamicin, netilmicin, and amikacin (MICs < 1 μg/ml). On the other hand, the amikacin-resistant clinical strain IFM 10580 was highly resistant to most of 2-DOS aminoglycosides (MICs > 1,024 μg/ml), although it demonstrated MICs to streptomycin and hygromycin B that were comparable to those of the wild-type strain (128 and 32 μg/ml, respectively). Streptomycin and hygromycin B are known to target the ribosome at a site that is distinct from the A-site (20). On the other hand, IFM 10152/pNV19-16S(1408G) demonstrated MICs that were approximately 1 (paromomycin) to >512 (arbekacin) doubling dilutions lower than those of IFM 10580, but the MICs were 2- to >128-fold larger for all aminoglycosides (except streptomycin and hygromycin B) compared to IFM 10152/pNV19 on day 3 after the start of incubation (Table 1). The MICs of IFM 10152/pNV19-16S(1408G) for many aminoglycosides were increased further on day 5 of incubation (Table 1, last column), although those for streptomycin and hygromycin B were only slightly affected or unaffected, even after day 5 of incubation. Although the levels of resistance to some aminoglycoside were low in IFM 10152/pNV19-16S(1408G) compared to the resistant strain IFM 10580, the spectrum of the resistance was similar between the clinically isolated resistant strain IFM 10580 and IFM 10152/pNV19-16S(1408G) (for a few exceptions, e.g., netilmicin, see the Discussion). These results suggest that the A1408G mutation of the 16S rRNA gene is responsible for resistance to amikacin and many other 2-DOS aminoglycosides observed in N. farcinica IFM 10580.
The genome sequencing project has shown that N. farcinica IFM 10152 has three rRNA operons, rrnA, rrnB, and rrnC, each containing 16S rRNA genes, where the nucleotide at position 1408 is adenine (1408A, see Table 2, analysis 1) (15). Therefore, we investigated three 16S rRNA gene sequences in the amikacin-resistant clinical isolate IFM 10580 after specific amplification of the respective genes. The result showed that all three 16S rRNA genes in IFM 10580 harbored identical A1408G mutations, suggesting that homogeneity for mutated 16S rRNA might contribute to its high-level resistance to amikacin and other 2-DOS aminoglycosides (see Table 2, analysis 6).
The IFM 10152 strain carrying pNV19-16S(1408G) showed only moderate resistance to amikacin (MIC ≈ 64 μg/ml) and other aminoglycosides on day 3 of incubation (Table 1, second column from the right). Based on a previous report that rRNA-mediated mutational resistance to aminoglycoside is recessive in Mycobacterium smegmatis (24), we considered that it is likely that IFM 10152/pNV19-16S(1408G) expresses both wild-type (1408A) and mutated (1408G) 16S rRNA genes, which were derived from chromosomal and plasmid-encoded 16S rRNA genes, respectively. On day 5 of the incubation, however, we obtained cells that had apparently acquired higher levels of resistance to many aminoglycosides including amikacin (Table 1, last column). We considered it possible that these cells might have acquired chromosomal mutations in 16S rRNA genes in the presence of the multicopy plasmid carrying the mutated 16S rRNA gene under the stress of the high aminoglycoside concentration.
To examine this possibility, we eliminated (cured) the plasmid pNV19-16S(1408G) from the highly resistant mutants obtained by cultivating IFM 10152/pNV19-16S(1408G) in the presence of high concentrations (1,024 μg/ml) of amikacin for 5 days and determined the chromosomal 16S rRNA gene sequences. The results showed that in those mutants, two of the three 16S rRNA genes (rrnA 16S and rrnC 16S) had been altered to mutated alleles harboring 1408G (Table 2, analysis 4). These mutant strains containing one wild-type and two mutated chromosomal rRNA genes were susceptible to amikacin (MIC < 1 μg/ml) in the absence of the plasmid carrying mutated 16S rRNA gene (Table 2, analysis 4) but were highly resistant (MIC > 1,024 μg/ml) when transformed with the plasmid pNV19-16S(1408G) (Table 2, analysis 3).
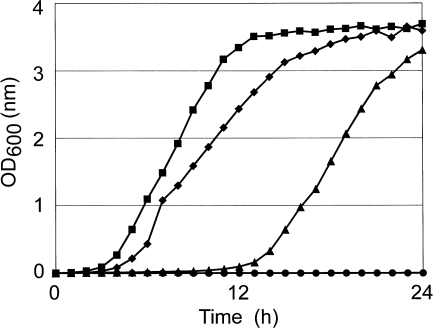
When the amikacin-susceptible mutant strain IFM 10152-M2 was incubated in the presence of a high concentration (1,024 μg/ml) of amikacin, cell growth was observed after 24 h of incubation (Fig. 2). The strain growing thereafter was highly resistant to amikacin (Table 2, analysis 6, MIC > 1,024 μg/ml). Examination of the 16S rRNA gene sequences of the resistant isolate revealed that 1408A of the third 16S rRNA gene, rrnB, was also mutated to 1408G in addition to rrnA and rrnC 16S rRNA genes. Such highly resistant mutants that are homozygously mutated in the three chromosomal 16S rRNA genes (IFM 10152-M3) were obtained from IFM 10152-M2 at a high rate (2.3 × 10−3). In contrast to IFM 10152-M2, IFM 10152-M3 and the clinical resistant strain IFM 10580 grew significantly faster in the presence of 1,024 μg of amikacin/ml without a lag time of 12 h (Fig. 2). Together, these results suggest that high-level aminoglycoside resistance mediated by 16S rRNA mutation is recessive and that the cells become resistant only when 1408G mutant 16S rRNAs are expressed homozygously or significantly abundant in comparison to the wild-type 16S rRNA.
FIG. 2.
Growth curves of N. farcinica strains: IFM 10152 (wild type) (•), the clinical isolate IFM 10580 (⧫), IFM 10152-M2 (▴), and IFM 10152-M3 (▪) in the presence of 1,024 μg of amikacin/ml.
We also investigated the 16S rRNA sequences of four N. farcinica isolates obtained from the bovine mastitis epizootic in Canada other than strain IFM 10580. These were strains W6218 (IFM 10850) from Alberta, W6222 (IFM 10851) from Newfoundland, W6224 (IFM 10849) from Nova Scotia, and W6226 (IFM 14848) from Ontario, Canada (4). The results showed that all three copies of 16S rRNA genes in all four isolates had the same A1408G mutations. We encountered no substitution other than the A1408G mutation of 16S rRNA genes in the four amikacin-resistant clinical isolates examined. Together with the restriction fragment length polymorphism of chromosomal DNA and ribotyping study of the isolates performed previously (4), the results suggest that these Nocardia mastitis isolates might have derived from a single origin.
DISCUSSION
This study demonstrated that the homozygous triplicate point mutations that occurred at the position 1408 (in E. coli numbering) of each chromosomal 16S rRNA gene were responsible for the high-level 2-DOS aminoglycoside cross-resistance observed in the N. farcinica isolates as a causative pathogen of Canada-wide bovine mastitis epizootic.
Aminoglycosides have been divided into two major classes: those containing the streptamine moiety, such as streptomycin, and those containing the 2-DOS moiety, such as the neomycins. The 2-DOS-containing aminoglycosides bind to the end of helix 44 in 16S rRNA near the location of the A-site, which binds tRNA (20). The nucleotide at position 1408 resides in the A-site and 2-DOS aminoglycosides block bacterial growth through interference with protein synthesis. High-level aminoglycoside resistance attributable to alterations of antibiotic-binding targets might involve methylation of conserved regions within 16S rRNA molecules, e.g., an adenine at either position 1405 or position 1408 of 16S rRNA. Modifications of these types have been found in aminoglycoside-producing organisms such as Streptomyces and Micromonospora spp. (2) and recently in clinical strains of Gram-negative bacteria such as Pseudomonas aeruginosa (29), Klebsiella pneumoniae (10), and Serratia marcescens (7). On the other hand, mutational alterations of 16S rRNA at position 1408 have been found to mediate acquired high-level cross-resistance to 2-DOS aminoglycosides in E. coli and M. smegmatis in vivo (9, 27). However, clinical high-level resistance to aminoglycosides attributable to 1408G mutation in the 16S rRNA has been found only in Mycobacterium species carrying a single chromosomal rRNA operon, such as M. tuberculosis, M. abscessus, and M. chelonae (1, 23); it has not been found in organisms that carry more than two rRNA operons, such as members of the enterobacteria family, staphylococci, and streptococci. It has been considered that acquisition of aminoglycoside resistance by rRNA mutation might not occur or that it might be rare because most bacteria harbor multiple rRNA operons and because the mutant allele is usually recessive in strains that are heterozygous for wild-type and mutated rRNAs, as shown in the M. smegmatis strain, which carries two rRNA operons (24), although partial dominance of aminoglycoside resistance attributable to 16S rRNA mutation has also been reported in E. coli (25). The strains of N. farcinica have three chromosomal rRNA operons. The bovine mastitis epizootic clinical strains of N. farcinica, including IFM 10580, represent the first clinical case of high-level aminoglycoside resistance attributable to rRNA alteration in bacterial species carrying more than two copies of chromosomal rRNA operons.
The results of these analyses of transformants carrying 1408G mutated rRNA and spontaneous mutants that acquired the 1408G point mutation in the chromosomal 16S rRNA genes revealed that aminoglycoside resistance mediated by the rRNA mutation was recessive in N. farcinica strains. Introduction of mutated 16S rRNA gene by extrachromosomal replication of the multicopy plasmid [pNV19-16S (1408G)] conferred moderate levels of cross-resistance to 2-DOS aminoglycosides (e.g., MIC ≈ 64 μg/ml for amikacin) to susceptible strain IFM 10152. On the other hand, highly resistant (MIC > 1,024 for amikacin) mutants were obtained spontaneously when moderately resistant cells were cultured in the presence of 1,024 μg of amikacin/ml for a few days (Table 1, last column). The results show that the strains which are highly resistant to aminoglycosides had two 1408G point-mutated copies (rrnA 16S and rrnC 16S) and one wild-type copy in chromosomal 16S rRNA genes [Table 2, analysis3, IFM 10152-M2/pNV19-16S(1408G)]. The heterozygous mutants were amikacin susceptible (MIC < 1 μg/ml) in the absence of the plasmid carrying mutated rRNA (Table 2, analysis 4, IFM 10152-M2) but were highly resistant to amikacin (MIC > 1,024 μg/ml) in the presence of the plasmid (Table 2, analysis 3). The cells became highly resistant when the third chromosomal 16S rRNA gene, rrnB 16S, obtained A1408G mutation (Table 2, analysis 5, IFM 10152-M3). These results suggest rRNA mutation-mediated aminoglycoside resistance is recessive, supporting a previous report in M. smegmatis, where A1408G point mutations were artificially introduced into one or two of 16S rRNA genes (24).
At present, it remains unknown how the clinical amikacin-resistant strains acquired multiple homozygous A1408G point mutations in the three chromosomal 16S rRNA genes. We think it is unlikely that the highly amikacin-resistant clinical strains arose through recombination between chromosomal 16S rRNA genes and plasmids, as observed in experimental conditions (24, present work, Table 1). We assume in the epizootic a single A1408G mutation occurred at either one of the three 16S rRNA genes, followed by successive homozygous mutations in the other two rrn 16S genes in response to the continued used of aminoglycosides at a high concentration, resulting in the amikacin-resistant clinical strains. Indeed, the highly resistant chromosomal mutants (IFM 10152-M3) were easily obtained from the susceptible chromosomal mutant without carrying the plasmids (IFM 10152-M2) in the presence of amikacin (at a frequency of one mutant from 2.3 × 103 cells) (Table 2). The underlying mechanisms for the homozygous mutation might be gene conversions among 16S rRNAs, as have been reported for E. coli, Vibrio parahaemolyticus, and other strains (11, 12, 17), or other novel but unknown mechanisms. Similar homologous recombination has also been hypothesized in Staphylococcus aureus 23S rRNA genes, where mutant gene dose-dependent increase in MIC to linezolid was observed (3). It is of great importance to reveal the mechanisms involved in the homozygous mutations for purposes of clinical treatment and antibiotic therapy, as well as for biological interests.
Although the transformants of the susceptible strain IFM 10152 carrying plasmid-encoded mutated rRNA gene [IFM 10152/pNV19-16S(1408G)] acquired high-level resistance to 2-DOS aminoglycosides upon prolonged incubation in the presence of amikacin (Table 1, day 5), the spectrum of the aminoglycoside sensitivities differed slightly from that of the clinical resistant strain IFM 10580 (Table 1, e.g., netilmicin). Such a difference might be explained by a ratio or amounts of the wild-type (1408A) and the mutant (1408G) 16S rRNA molecules in the respective strains (only the mutant 16S rRNA is present in the clinical resistant strains, but both wild-type and the mutant rRNA are present in the transformants), and also by a difference in affinities of aminoglycosides to the two different types of the rRNA molecular complexes. Alternatively, some aminoglycosides, e.g., netilmicin, might have some mode of action in addition to binding to the A-site of the 16S rRNA complex.
The results of the present work generalized the previous finding that the mutant allele of 16S rRNA, which confers aminoglycoside resistance, is recessive in microbes having more than two 16S rRNA operons. Nevertheless, the present evidence conflicts with the idea that aminoglycoside resistance might seldom be obtained by multiple mutations of 16S rRNA genes in microbes having multiple 16S rRNA operons. We have not encountered clinical cases of “human” infection with aminoglycoside-resistant microbe harboring the serial 16S rRNA mutations, but we must remain aware of such cases for the future.
Acknowledgments
This study was supported by a Grant-in-Aid for Scientific Research (C) and Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
We are grateful to Eric Dabbs for critical comments related to the manuscript.
Footnotes
Published ahead of print on 22 March 2010.
REFERENCES
- 1.Alangaden, G. J., B. N. Kreiswirth, A. Aouad, M. Khetarpal, F. R. Igno, S. L. Moghazeh, E. K. Manavathu, and S. A. Lerner. 1998. Mechanism of resistance to amikacin and kanamycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:1295-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beauclerk, A. A., and E. Cundliffe. 1987. Sites of action of two rRNA methylases responsible for resistance to aminoglycosides. J. Mol. Biol. 193:661-671. [DOI] [PubMed] [Google Scholar]
- 3.Besier, S., A. Ludwig, J. Zander, V. Brade, and T. A. Wichelhaus. 2008. Linezolid resistance in Staphylococcus aureus: gene dosage effect, stability, fitness costs, and cross-resistances. Antimicrob. Agents Chemother. 52:1570-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, J. M., K. D. Cowley, K. I. Manninen, and M. M. McNeil. 2007. Phenotypic and molecular epidemiologic evaluation of a Nocardia farcinica mastitis epizootic. Vet. Microbiol. 125:66-72. [DOI] [PubMed] [Google Scholar]
- 5.Brown-Elliott, B. A., J. M. Brown, P. S. Conville, and R. J. Wallace, Jr. 2006. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin. Microbiol. Rev. 19:259-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiba, K., Y. Hoshino, K. Ishino, T. Kogure, Y. Mikami, Y. Uehara, and J. Ishikawa. 2007. Construction of a pair of practical Nocardia-Escherichia coli shuttle vectors. Jpn. J. Infect. Dis. 60:45-47. [PubMed] [Google Scholar]
- 7.Doi, Y., K. Yokoyama, K. Yamane, J. Wachino, N. Shibata, T. Yagi, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob. Agents Chemother. 48:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferns, L., I. Dohoo, and A. Donald. 1991. A case-control study of Nocardia mastitis in Nova Scotia dairy herds. Can. Vet. J. 32:673-677. [PMC free article] [PubMed] [Google Scholar]
- 9.Fourmy, D., M. I. Recht, S. C. Blanchard, and J. D. Puglisi. 1996. Structure of the A site of Escherichia coli 16S rRNA complexed with an aminoglycoside antibiotic. Science 274:1367-1371. [DOI] [PubMed] [Google Scholar]
- 10.Galimand, M., P. Courvalin, and T. Lambert. 2003. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Escalona, N., J. Romero, and R. T. Espejo. 2005. Polymorphism and gene conversion of the 16S rRNA genes in the multiple rRNA operons of Vibrio parahaemolyticus. FEMS. Microbiol. Lett. 246:213-219. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto, J. G., B. S. Stevenson, and T. M. Schmidt. 2003. Rates and consequences of recombination between rRNA operons. J. Bacteriol. 185:966-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hitti, W., and M. Wolff. 2005. Two cases of multidrug-resistant Nocardia farcinica infection in immunosuppressed patients and implications for empiric therapy. Eur. J. Clin. Microbiol. Infect. Dis. 24:142-144. [DOI] [PubMed] [Google Scholar]
- 14.Ishikawa, J., K. Chiba, H. Kurita, and H. Satoh. 2006. Contribution of rpoB2 RNA polymerase beta subunit gene to rifampin resistance in Nocardia species. Antimicrob. Agents Chemother. 50:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa, J., A. Yamashita, Y. Mikami, Y. Hoshino, H. Kurita, K. Hotta, T. Shiba, and M. Hattori. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. U. S. A. 101:14925-14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kageyama, A., K. Yazawa, J. Ishikawa, K. Hotta, K. Nishimura, and Y. Mikami. 2004. Nocardial infections in Japan from 1992 to 2001, including the first report of infection by Nocardia transvalensis. Eur. J. Epidemiol. 19:383-389. [DOI] [PubMed] [Google Scholar]
- 17.Liao, D. 2000. Gene conversion drives within genic sequences: concerted evolution of rRNA genes in bacteria and archaea. J. Mol. Evol. 51:305-317. [DOI] [PubMed] [Google Scholar]
- 18.Manninen, K. I., R. A. Smith, and L. O. Kim. 1993. Highly presumptive identification of bacterial isolates associated with the recent Canada-wide mastitis epizootic as Nocardia farcinica. Can. J. Microbiol. 39:635-641. [DOI] [PubMed] [Google Scholar]
- 19.Mingeot-Leclercq, M. P., Y. Glupczynski, and P. M. Tulkens. 1999. Aminoglycosides: activity and resistance. Antimicrob. Agents Chemother. 43:727-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S rRNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. Approved standard M24-A. National Committee for Clinical Laboratory Standards, Wayne, PA. [PubMed]
- 22.Ollis, G. W., M. Schoonderwoerd, and C. Schipper. 1991. An investigation of risk factors for nocardial mastitis in central Alberta dairy herds. Can. Vet. J. 32:227-231. [PMC free article] [PubMed] [Google Scholar]
- 23.Prammananan, T., P. Sander, B. A. Brown, K. Frischkorn, G. O. Onyi, Y. Zhang, E. C. Bottger, and R. J. Wallace, Jr. 1998. A single 16S rRNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 177:1573-1581. [DOI] [PubMed] [Google Scholar]
- 24.Prammananan, T., P. Sander, B. Springer, and E. C. Bottger. 1999. RecA-Mediated gene conversion and aminoglycoside resistance in strains heterozygous for rRNA. Antimicrob. Agents Chemother. 43:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recht, M. I., S. Douthwaite, and J. D. Puglisi. 1999. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 18:3133-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russo, T. A. 2005. Agents of actinomycosis, p. 2924-2934. In G. L. Mandell, Bennette, J. E., Dolin, R (ed.), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 6th ed., vol. 2. Elsevier, New York, NY. [Google Scholar]
- 27.Sander, P., T. Prammananan, and E. C. Bottger. 1996. Introducing mutations into a chromosomal rRNA gene using a genetically modified eubacterial host with a single rRNA operon. Mol. Microbiol. 22:841-848. [DOI] [PubMed] [Google Scholar]
- 28.Stark, D. A., and N. G. Anderson. 1990. A case-control study of Nocardia mastitis in Ontario dairy herds. Can. Vet. J. 31:197-201. [PMC free article] [PubMed] [Google Scholar]
- 29.Yokoyama, K., Y. Doi, K. Yamane, H. Kurokawa, N. Shibata, K. Shibayama, T. Yagi, H. Kato, and Y. Arakawa. 2003. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 362:1888-1893. [DOI] [PubMed] [Google Scholar]