Abstract
Bevirimat (BVM) is the first of a new class of anti-HIV drugs with a novel mode of action known as maturation inhibitors. BVM inhibits the last cleavage of the Gag polyprotein by HIV-1 protease, leading to the accumulation of the p25 capsid-small peptide 1 (SP1) intermediate and resulting in noninfectious HIV-1 virions. Early clinical studies of BVM showed that over 50% of the patients treated with BVM did not respond to treatment. We investigated the impact of prior antiretroviral (ARV) treatment and/or natural genetic diversity on BVM susceptibility by conducting in vitro phenotypic analyses of viruses made from patient samples. We generated 31 recombinant viruses containing the entire gag and protease genes from 31 plasma samples from HIV-1-infected patients with (n = 21) or without (n = 10) prior ARV experience. We found that 58% of the patient isolates tested had a >10-fold reduced susceptibility to BVM, regardless of the patient's ARV experience or the level of isolate resistance to protease inhibitors. Analysis of mutants with site-directed mutations confirmed the role of the V370A SP1 polymorphism (SP1-V7A) in resistance to BVM. Furthermore, we demonstrated for the first time that a capsid polymorphism, V362I (CA protein-V230I), is also a major mutation conferring resistance to BVM. In contrast, none of the previously defined resistance-conferring mutations in Gag selected in vitro (H358Y, L363M, L363F, A364V, A366V, or A366T) were found to occur among the viruses that we analyzed. Our results should be helpful in the design of diagnostics for prediction of the potential benefit of BVM treatment in HIV-1-infected patients.
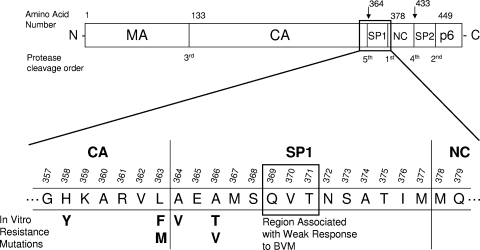
With the continued advancement of antiretroviral therapeutic options, the treatment of HIV-1 infection has greatly improved over the years. However, the development of drug-resistant variants of HIV-1 in infected patients often leads to treatment failure and underscores the need for new therapies for these patients. The naturally occurring medicinal compound betulinic acid and some of its derivatives have been shown to inhibit HIV-1 in novel ways (4-6). Bevirimat (BVM; also known as PA-457, DSB, and MPC-4326) is a derivative of betulinic acid that is currently in phase 2 clinical trials for the treatment of HIV-1 infection (3, 15) and is the first member of a novel class of maturation inhibitors of HIV-1 (8). Evidence has shown that BVM acts by inhibiting the last cleavage of the HIV-1 Gag polyprotein (Fig. 1) (8, 20, 22), preventing the release of the Gag small peptide 1 (SP1) from the Gag capsid (CA or p24) protein. This in turn leads to the formation of an immature p25 CA protein-SP1 instead of the expected p24 CA protein, which cannot condense in a timely fashion to form the capsid core and which gives rise to noninfectious virion progeny. Interestingly, BVM activity in an in vitro cell-free system could be detected only when Gag was able to assemble into an immature particle-like structure, strongly suggesting that the presumed binding of BVM near the CA protein-SP1 cleavage site is highly dependent on the surrounding Gag tertiary structure (8, 13, 21).
FIG. 1.
Schematic of HIV-1 Gag polyprotein showing amino acid residues associated with bevirimat susceptibility. MA, matrix; CA, capsid; SP1, small peptide 1; NC, nucleocapsid; SP2, small peptide 2.
In vitro drug resistance selection experiments with BVM have been conducted and identified several mutations (Fig. 1) in the vicinity of the Gag CA protein-SP1 cleavage site. These included the mutations at the P1′ and P1 residues of the cleavage site, A364V (SP1-A1V) and L363F or L363M (CA protein-L231F or CA protein-L321M), respectively, as well as A366T or A366V (SP1-A3T or SP1-A3V) and H358Y (CA protein-H226Y) (1, 8, 22). Resistance to BVM in these mutants was characterized by their ability to cleave the p25 CA protein-SP1 efficiently in the presence of the drug (up to 3 μM) without significantly affecting their ability to replicate (1). In a phase 2 clinical trial of BVM with antiretroviral (ARV)-experienced patients, about half of the patients treated with BVM did not respond to treatment (mean HIV-1 load reduction, 0.05 log10 copies/ml), while the other half showed a strong response to treatment (mean HIV-1 load reduction, 1.26 log10 copies/ml) after 2 weeks of dosing (7, 12). Genotypic analyses of these patients' HIV-1 isolates have identified three key polymorphic sites in SP1 (Q369, V370, and T371) that are associated with innate resistance to BVM (7, 12), and baseline phenotypic analyses of patient samples from BVM clinical trials have confirmed the presence of >100-fold resistance to BVM among isolates from patients with polymorphisms at these sites (16).
The aim of the present study was to investigate the phenotypic susceptibility to BVM of HIV-1 isolates from HIV-1-infected patients who had not previously been exposed to BVM, in order to shed light on the observed lack of response to BVM in certain patient populations. We hypothesized that prior ARV treatment and, more specifically, treatment with protease inhibitors (PIs) may have led to the presence of adaptive mutations in Gag that could confer resistance to BVM.
MATERIALS AND METHODS
Reagents.
The viral plasmid that was used for cloning, pXXLAI, was a gift from John Mellors. That plasmid was derived from the infectious clone pLAI3.2, which was modified to contain an XmaI and an XbaI cloning site within the HIV reverse transcriptase (RT) gene (14). The XmaI restriction site downstream of HIV protease and the naturally occurring SfoI restriction site upstream of HIV gag were used for cloning. The 293T cells used for virus production were purchased from the American Type Culture Collection (ATCC; Manassas, VA). The MT-2 cells used in the phenotypic assays were obtained from the NIH AIDS Research and Reference Reagent Program (Germantown, MD). BVM was obtained from Acme Biosciences, Inc. (Palo Alto, CA). Tenofovir (TFV), atazanavir (ATV), and lopinavir (LPV) were synthesized at Gilead Sciences. The mitochondrial dye used in the phenotypic assays, 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT), was purchased from Sigma-Aldrich (St. Louis, MO).
Patient isolates.
Thirty-one plasma samples from ARV treatment-experienced or -naïve patients were used to generate the virus isolates. None of the subjects chosen had been exposed to BVM. Ten samples from ARV-naïve patients from study GS-01-934 (collected between August 2003 and September 2003) without any resistance mutations in protease or RT were randomly selected. Twenty-one samples from ARV-experienced patients were selected from two different past Gilead studies in order to capture a snapshot of patient susceptibility to bevirimat at two different time periods. Eleven of these samples were from ARV-experienced patients enrolled in study GS-99-907 (collected between February 2000 and July 2000), and the remaining 10 samples were from ARV-experienced patients enrolled in study GS-US-183-0105 (collected between March 2006 and May 2006). The samples from ARV-experienced patients were selected on the basis of the presence of PI resistance-conferring mutations.
Nucleic acid preparation and PCR.
A viral RNA extraction kit (Qiagen, Valencia, CA) was used to obtain HIV-1 RNA from the patient isolates. cDNA was generated with a Ready-To-Go You-Prime First-Strand cDNA kit (GE Healthcare, Buckinghamshire, United Kingdom) with the downstream oligonucleotide RT-21 (5′-CTGTATTTCTGCTATTAAGTCTTTTGATGGG). The cDNA was amplified by two rounds of PCR with Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA). The initial round of PCR (30 cycles) was done with oligonucleotides U563 (5′-GCTTAAGCCTCAATAAAGCTTGCCTTG) and RT155 (5′-GGCCCAATTTTTGAAATTTTCCCTTCC), and the nested 2nd round of PCR (30 cycles) was done with oligonucleotides U613 (5′-GTGACTCTGGTAACTAGAGATCCCTC) and RC-5′-HIV-Xma (5′-CTTTTGGGCCATCCATCCCGGGCTTTAATTTTACTGG). The 2nd round of PCR resulted in the addition of an XmaI restriction site at the 3′ end of the PCR product for further cloning. The approximately 1.8-kb PCR end product containing the entire HIV gag and protease genes from the clinical samples was purified by agarose gel electrophoresis (Qiagen).
Plasmids constructs.
The gel-purified PCR products were subcloned with a Zero Blunt TOPO PCR cloning kit (Invitrogen, Carlsbad, CA). After overnight growth, colonies from the kanamycin plates were pooled in order to maintain the diversity of the HIV quasispecies present in the patient isolates and were grown overnight in 3 ml of LB broth containing kanamycin. Plasmid DNA was extracted with a GenElute HP miniprep kit (Sigma-Aldrich) and digested overnight first with XmaI and then with SfoI (New England Biolabs). The band of interest was gel purified prior to and after digestion with SfoI. Viral plasmid pXXLAI was doubly digested overnight with both XmaI and SfoI and gel purified. The SfoI-XmaI patient-derived HIV DNA fragment (100 ng) was ligated into the pXXLAI doubly digested vector (140 ng) overnight at 16°C with T4 DNA ligase (New England Biolabs). XL-10 Gold ultracompetent cells (Agilent Technologies, Inc., Santa Clara, CA) were transformed with the ligation mixture and were grown overnight in 150 ml LB broth containing ampicillin. After plasmid DNA extraction and purification (Qiagen), the polyclonal plasmid DNA was sequenced (Elim Biopharmaceuticals, Hayward, CA) and used for virus production.
Mutant viruses constructed by site-directed mutagenesis.
DNA fragments containing the mutations of interest were generated in two rounds of PCR. The mutagenesis primers were designed either to change the gag-SP1 gene of specific samples into the wild-type LAI sequence (isolates 1, 3, and 6; see below) or to insert or revert to the wild-type sequence the V362I Gag-capsid mutation (isolates 7, 8, and 12 and LAI laboratory strain; see below) while keeping the remaining sequence intact. The mutagenesis primers were used in the 1st round of PCR, along with the appropriate oligonucleotides (listed above), to generate two DNA fragments containing the mutated sequence either at the 3′ end or at the 5′ end. These fragments were gel purified and used as the template for the 2nd round of PCR by using the same oligonucleotides described above. Viral plasmid DNA (pXXLAI backbone) was generated from the 2nd-round PCR product as described above, except that it was obtained from an individual clone. Mutant plasmids were sequenced as described above.
DNA transfection.
The transfection reagent TransIT-LT1 (Mirus Bio Corporation, Madison, WI) was used to transfect 7 μg of the viral plasmid DNA (polyclonal or clonal) in 2 million 293T cells seeded in T-25 cell culture flasks in a 6-ml volume 1 day before transfection. The cell culture supernatant containing the virus stock was harvested 2 days after transfection and was monitored by p24 and infectivity assays.
Phenotypic assays.
The susceptibilities of the mutant viruses to bevirimat, tenofovir, lopinavir, and atazanavir were measured in a 5-day cell-killing assay with MT-2 cells and by use of the XTT assay, as described previously (11, 18). In short, infection of 2.4 million MT-2 cells with either mutant or wild-type HIV-1 LAI was carried out in 1-ml tubes at 37°C with gentle rocking. The amount of virus used for the infections was determined by titration of each viral construct's infectivity and was set as the amount of virus that gave a signal-to-noise ratio in the range of 2.5 to 4.0 in the XTT assay. That amount corresponds to a multiplicity of infection of approximately 0.001 when commercially available HIV-1 strain IIIb (Applied Biotechnologies, Inc., Columbia, MD) is used. The cells were plated in 96-well plates containing compound dilutions or no-drug controls with ≈17,000 cells/well in a 200-μl volume. After a 5-day incubation at 37°C in 5% CO2, 100 μl of medium from each well was removed and replaced with 100 μl of 1.67 mg/ml XTT. The plates were incubated at 37°C for 1 h, and the absorbance (450 nm minus 650 nm) was measured with a Vmax microplate reader (Molecular Devices, Inc., Sunnyvale, CA) to measure the cytopathic effect generated by the viral infection.
Data analysis.
For the phenotypic assays, percent cell death in the drug-containing wells in comparison to that in the uninfected cell control was calculated by using the Excel program (Microsoft, Redmond, WA), and the data were plotted by using the SigmaPlot program (SPSS, Chicago, IL). The effective concentration required to inhibit viral replication by 50% (EC50) was also determined by using the SigmaPlot program. Sequencing data were analyzed by use of the SeqMan and MegAlign programs (DNAStar, Madison, WI).
Nucleotide sequence accession number.
The sequences described here have been submitted to GenBank under reference numbers HM116810 to HM116840.
RESULTS
Phenotypic susceptibility of patient isolates.
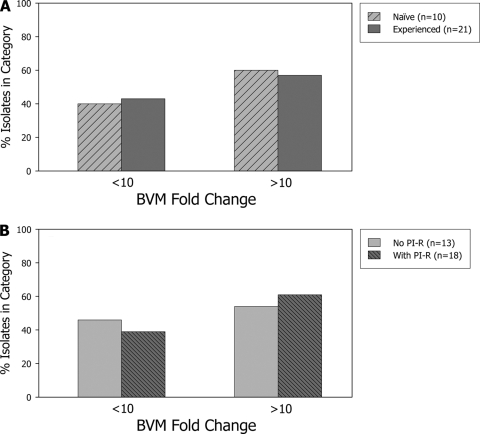
Recombinant pLAI viruses containing the entire gag-protease region of HIV-1 from 21 ARV-experienced patients without prior BVM exposure and from 10 ARV-naïve patients were analyzed phenotypically. Phenotypic susceptibility to BVM, as well as to two protease inhibitors (lopinavir and atazanavir) and TFV, was determined by the XTT assay in MT-2 cells. These analyses revealed that over half of the patient isolates tested (18/31; 58%) (Table 1) showed a highly significant >10-fold reduced susceptibility to BVM compared to that of wild-type HIV-1 strain LAI, and the majority of isolates with reduced susceptibility (16/18) showed >100-fold resistance to BVM. The remaining 13 isolates had BVM susceptibilities ranging from 0.7- to 5.2-fold the susceptibility of the wild-type control. Six of these 13 isolates showed significantly reduced susceptibility to BVM compared to that of the wild type (range, 1.8- to 5.2-fold), likely reflecting the range of nearly wild-type susceptibility to BVM in these complex patient isolates. Among the 18 isolates showing >10-fold reduced susceptibility to BVM, 12 were from ARV-experienced patients (12/21; 57%), while the remaining 6 isolates were from ARV-naïve patients (6/10; 60%). Conversely, among the 13 isolates with BVM susceptibility within 10-fold of that of the wild type, 9 isolates were from ARV-experienced patients (9/21; 43%) and 4 isolates were from ARV-naïve patients (4/10; 40%). These results are depicted graphically in Fig. 2A and demonstrate no association between ARV treatment experience and BVM resistance (P = 1.00, Fisher's exact test).
TABLE 1.
BVM phenotypic susceptibility and genotypic data for the 31 patient-derived HIV isolates
| BVM FCa | Prior ARVb | Isolate no.c | Amino acid change at the indicated position in the following protein: |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Capsidd |
SP1e |
||||||||||||||||||||||
| 357 (G)f | 358 (H) | 359 (K) | 360 (A) | 361 (R) | 362 (V) | 363 (L) | 364 (A) | 365 (E) | 366 (A) | 367 (M) | 368 (S) | 369 (Q) | 370 (V) | 371 (T) | 372 (N) | 373 (S) | 374 (A) | 375 (T) | 376 (I) | 377 (M) | |||
| 0.7 | Exp. | 22 | —j | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 0.9 | Naïve | 32 | — | — | — | — | — | — | — | — | — | — | — | — | — | I/V | — | G | — | — | — | — | — |
| 1.0 | Exp. | 7 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 1.2 | Exp. | 8 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 1.2 | Exp. | 21 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | V | — |
| 1.3 | Exp. | 26 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | S | P | — | N | — | — |
| 1.3 | Exp. | 28 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 1.8** | Naïve | 36 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | Q | — | — | N | A | — | — |
| 2.1* | Exp. | 20 | S | — | — | — | — | — | — | — | — | — | — | — | H | — | — | — | — | — | — | V | — |
| 2.2** | Exp. | 5 | — | — | — | — | — | — | — | — | — | — | — | — | H/Q | — | — | — | — | — | N | — | — |
| 4.6* | Naïve | 37 | S | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | V | — |
| 4.7** | Exp. | 9 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | N | V | — |
| 5.2** | Naïve | 30 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 11** | Exp. | 25 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | Delg | — | P | — | — | — | — |
| 36** | Exp. | 27 | S | — | — | — | — | — | — | — | — | — | — | — | — | A | — | — | — | — | — | — | — |
| >100** | Exp. | 1 | — | — | — | — | — | — | — | — | — | — | — | C | — | A | Del | — | — | N | — | — | — |
| >100** | Exp. | 3 | S | — | — | — | — | — | — | — | — | — | — | — | — | A | Del | H/N | — | — | — | — | — |
| >100** | Exp. | 6 | — | — | — | — | — | — | — | — | — | — | — | — | — | A | — | — | A | T | — | — | — |
| >100** | Exp. | 10 | — | — | — | — | — | — | — | — | — | — | — | — | — | I | Del | — | P/S | — | — | V | — |
| >100** | Naïve | 31 | — | — | — | — | — | — | — | — | — | — | — | — | — | A | S | A/T | — | N/T | — | V/I | — |
| >100** | Naïve | 33 | — | — | — | — | — | — | — | — | — | — | — | G | — | M | — | — | T | — | — | — | — |
| >100** | Naïve | 34 | S | — | — | — | — | — | — | — | — | — | — | — | — | A | — | T | P | — | A | — | — |
| >100** | Naïve | 35 | S | — | — | — | — | — | — | — | — | — | — | — | — | A | Q | S | — | — | N | M | — |
| >100** | Naïve | 38 | — | — | — | — | — | — | — | — | — | — | — | — | — | A/V | — | S/G | Xh | Xi | A/T | M/I | — |
| >100** | Exp. | 23 | S | — | — | — | — | I | — | — | — | — | — | — | — | A | Del | — | — | — | A | — | — |
| >100** | Exp. | 24 | — | — | — | — | — | I | — | — | — | — | — | — | — | A | Del | T | — | — | A | — | — |
| >100** | Exp. | 12 | S | — | — | — | — | I | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| >100** | Naïve | 39 | S | — | — | — | — | I | — | — | — | — | — | — | — | — | — | — | — | — | A | — | — |
| >100** | Exp. | 2 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | A | G | A | — | — |
| >100** | Exp. | 4 | S | — | — | — | — | — | — | — | — | — | — | — | — | — | — | A | — | T | N | — | — |
| >100** | Exp. | 29 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | A | — | — |
Fold change (FC) in the bevirimat EC50 compared to the wild-type EC50 of 34 nM against HIV-1 LAI. The statistical significance of the fold changes in the EC50s compared to the wild-type EC50 is expressed as P < 0.01 (**) and P < 0.05 (*). The results are the averages of at least three independent experiments.
ARV experienced (Exp.) or naïve.
Isolates designated in boldface had both genotypic and phenotypic PI resistance. Data are shown in Table 2.
The last 7 amino acids of the Gag-capsid protein sequences are shown.
The complete amino acid sequences of Gag-SP1 are shown (14 residues).
The letters in parentheses represent the wild-type sequence.
Del, deletion.
A complex S373P/T/Q/K amino acid mixture was found at that site.
A complex A374A/G/T/S amino acid mixture was found at that site.
—, same as wild type.
FIG. 2.
Fold change in the BVM EC50 for the 31 patient-derived isolates compared to the EC50 for the wild-type control stratified according to antiretroviral therapy status (A) or the presence of protease inhibitor resistance (PI-R) (B).
PI resistance was detected in 18 of the 31 isolates and was characterized by the presence of at least three PI resistance-conferring mutations (among the PI resistance-conferring mutations D30N, V32I, L33F, M46I/L, I47V, G48V, I50V, I54V/M/L, L76V, V82A/F/L/T, I84V, and L90M) and at least 4-fold reduced susceptibility to either LPV or ATV (Table 2). Among the 18 isolates showing >10-fold reduced susceptibility to BVM, 11 isolates had PI resistance (11/18; 61%), while the other 7 isolates did not (7/13; 54%); and among the 13 isolates with BVM susceptibility within 10-fold of that of the wild type, 7 isolates had PI resistance (7/18; 39%), while the other 6 isolates did not (6/13; 46%). These results (shown in Fig. 2B) fail to demonstrate an association between PI resistance and BVM resistance in these patient isolates (P = 0.73, Fisher's exact test). The susceptibility to TFV was within 1.9-fold of that of the wild type (range, 0.3- to 1.9-fold; data not shown), consistent with the fact that all 31 recombinant viruses carried the wild-type RT sequence.
TABLE 2.
Protease inhibitor phenotypic susceptibilities and HIV-1 Gag-capsid and protease amino acid sequences for all 31 patient-derived isolates
| BVM FCa | Isolate no. | LPV FCb | ATV FCb | HIV-1 Gag capsid amino acid sequencec | HIV-1 protease amino acid sequenced |
|---|---|---|---|---|---|
| 0.7 | 22 | 0.9 | 1.3 | V215L H219Q I223V N252S P255A L268M A340G | L19I E35D M36I S37N L63P V77I I93L |
| 0.9 | 32 | 0.5 | 1.2 | I138L A146P I147L V159I V215L I223A N252S/N T280V S310T | R41K/R I62V L63P V77I I93L |
| 1.0 | 7 | 4.1** | 1.0 | I138L I147L S173A V215L I223V T242N G248A N252A I256T T280V E312D | K20R M36I R41K M46I L63Q I64V P79S V82F L90M |
| 1.2 | 8 | 28.7** | 29** | V215T | L10I L24I L33F S37N R41K I54V D60E I62V L63P A71V V77I V82A I84V |
| 1.2 | 21 | 0.6 | 0.8 | I138L A146P I147L/I V159I/V E211D V215L N252H/N R286K/R K331R/K | E35D M36I S37N I62V L63C |
| 1.3 | 26 | 46.5** | 571** | I138L A146S I147L E312D A326S L337M A340G | T4S L10F V11L I13V I15V K20A V32I L33F M36I S37D K43T M46I I54L I62V L63P A71I I72K G73T I84V I85V L89V L90M |
| 1.3 | 28 | 99.3** | 14.4** | I138M | L10I I13V L23I L33F S37N M46I I54V K55R R57K Q58E D60E I62V L63P A71I I72V L76V V77I V82A I84V I85V L89M L90M I93L |
| 1.8** | 36 | 1.1 | 1.3 | I138L A146P S165N/S E207D V215L E312D | K14R/K I15V E35D S37D R57K/R L63P |
| 2.1* | 20 | 13.3** | 51.9** | I138L V215L V218P H219Q M228I E230D G248T T280A E312D A340G G357S | L10F A22V D30N S37N K45R I54L I62V L63P A71V V77I I84V N88D Q92K |
| 2.2** | 5 | 12.1** | 63.2** | I138M V159I T280V S310T | I13V D30N L33I E35D M36I S37N M46I/L I54V I62V L63P I66M/I A71N/T I72T/I V82L/V N88D L90M I93L/I |
| 4.6* | 37 | 0.8 | 0.9 | V135I I138P V143I Q182H V215L/V V218P/A H219Q I223V E230D N252S T280A R286K K302R G357S | S37D R41K L63P I93L |
| 4.7** | 9 | 16.5** | 44.4** | I138L A146S I147L V215L N252H T280A A340G E345D | L10I I13V K20M L33V M36I S37N I54V R57K I62V L63P I66L A71V G73S P79A I84V L90M |
| 5.2** | 30 | 0.7 | 0.9 | I138L V215L R286K Q311A | I13V E35D S37N L63P |
| 11** | 25 | 58.9** | 184** | I138M S173A V215L N252S I256T S310T/S A340G E345D | L10F V11I I13V K20I V32I L33F E34Q E35D M36I S37N K43T M46I F53Y I54L Q61N I62V L63T I64V A71L T74S L76V I84V L90M I93L |
| 36** | 27 | 29.2** | 52.1** | I138L S173T/S V215L L268M R275K/R T280V S310T E319D A341del G357S | L10I I15V L19V V32I L33F M46I I54V K55R R57K L63P C67F H69Q A71V G73C/G V82A L90M Q92K |
| >100** | 1 | 22.1** | 62.9** | I138L V159I E203D V215L I223V T280V E312D V323I | L10I K20R M36I S37N G48V I54M I62V L63P A71V I72V V82A I84V L90M I93L/I |
| >100** | 3 | 13** | 15.1** | I138L A146P I147L V159I S176A V215L H219Q/H T242N G248A T280V A340G G357S | L10I T31S/T V32I M36I/M S37C R41K M46I F53L I54V R57K L63P I64V A71V V82A L90M |
| >100** | 6 | 3.4** | 29.2** | I138L V215L | L10I S37N M46L G48V R57K L63P I66F A71V V82T I84V |
| >100** | 10 | 15.3** | 8.5** | I138L I147L V159I V215L G248A I256V T280V R286K N315H | L10I K20R E35D M36I S37N R41K K45R F53L I54V L63P I64V A71T V82A L90M I93L |
| >100** | 31 | 1.1 | 1.9* | I138L S173T V215L I223N M228L G248Q N252H T280V E312D | I13V K14R/K M36I S37N I64V H69K I72V |
| >100** | 33 | 1.6* | 1.8** | I138L V215L V218P H219Q I223V M228I G248T N252S N253T I256M E312D A326S | T12S/T I13V/I I15V L19I S37T L63V/A/P/L I72V/I |
| >100** | 34 | 1.4 | 1.6* | I138L I147L V159I V215L T280V A309C S310T E312D G357S | L10I I13V E35D S37N I62V L63P I64M |
| >100** | 35 | 1.0 | 0.9 | V159I I223T N252S A326S/A A340G G357S | I15V G16E E35D S37N L63H I72V |
| >100** | 38 | 0.9 | 1.2 | I138L I147L V215L H219Q/H I223V N252H P292S A336S/A A340G | E35D S37A L63P |
| >100** | 23 | 203** | 521** | I138L V215L E319D A340G G357S V362I | L10V V11L V32I L33F E34Q E35D M36L S37T M46I I47V G48V I54M I62V L63P I64V I72V T74S V77I V82A I84V T91S |
| >100** | 24 | 69** | 590** | I138L V159I V215L I223A T239S N252S T280V R286K S310T V362I | L10V I13V I15V K20R D30N V32I L33F E35D M36I S37N K43T M46I I47V F53L I54L I62V L63P I66F A71V T74S V82A N88D L89V L90M I93V |
| >100** | 12 | 1.1 | 1.2 | I138L E207D V215L V218P H219Q M228I G248T T280S E312D A340G G357S V362I | S37N I62V/I L63P I64L V82I |
| >100** | 39 | 0.8 | 1.4 | A146P I147L V215L M228I G248A R286K A326S G357S V362I | K14R E35D M36I S37N R41K K45R D60E I62V I64V |
| >100** | 2 | 15.5** | 10.7** | A146P S173A V215L H219P E230D N252S R264K L268M A340G | L10I I15V K20R M36I S37N M46I L63P T80A V82A N83H I84V I85T L90M |
| >100** | 4 | 15.2** | 13.3** | A146P V159I K162RQ182ST186I V215L I223G/V N252G P255A E260D E312D G357S | L10R S37N M46L I62V L63P A71V T74S V82T L90M I93L |
| >100** | 29 | 89.3** | 3.2* | I138M A146P R286K T303V A326S | L10F I13V V32I M36L S37N L38W P39Q M46I I47V I50V I62V L63P A71I I72V V82A |
Fold change (FC) in the bevirimat EC50 compared to the wild-type EC50 of 34 nM against HIV-1 LAI. The statistical significance of the fold changes compared to the wild-type EC50 is expressed as P < 0.01 (**) and P < 0.05 (*). The results are the averages of at least three independent experiments.
Fold change (FC) in the LPV and ATV EC50s compared to the wild-type EC50s of 24.4 nM and 4.8 nM, respectively. The statistical significance of the fold changes in the EC50s compared to the wild-type EC50 is expressed as P < 0.01; (**) and P < 0.05 (*). The results are the averages of at least two independent experiments.
Amino acid numbers are based on the whole HIV-1 Gag polyprotein. The capsid protein starts at amino acid 133 of Gag. Amino acid changes compared to the consensus sequence of HIV-1 strain LAI are shown. Mutations shown in boldface were found uniquely in isolates 2, 4, and 29.
Amino acid changes compared to the consensus sequence of HIV-1 strain LAI are shown. Mutations shown in boldface for isolates 2, 4, and 29 were not found in BVM-sensitive isolates.
Genotypic analysis of patient isolates.
The HIV-1 gag gene from the 31 patient-derived isolates was sequenced entirely in order to study the relationship between the observed susceptibility to BVM and the presence or the absence of the previously defined polymorphisms in Gag-SP1 at amino acid positions 369, 370, and 371 that were associated with a reduced clinical response to BVM (7, 12), and other polymorphisms near the capsid-SP1 cleavage site or elsewhere in the capsid protein. In all cases in which the V370A or V370M polymorphism or a T371 deletion in Gag-SP1 was present, reduced susceptibility to BVM was observed (reduced 11-fold to >100-fold compared with the susceptibility of the wild type). A Q369H polymorphism was found in two isolates with nearly wild-type susceptibility and therefore did not appear to play a role in resistance in our sample set.
Five patient isolates (isolates 12, 39, 2, 4, and 29) showing >100-fold reduced susceptibility to BVM compared with the susceptibility of the wild type had no natural polymorphisms at position 369, 370, or 371 of Gag-SP1, suggesting that natural genetic variations conferring resistance to BVM exist beyond these three residues. Two of these isolates (isolates 12 and 39) carried a V362I mutation in the capsid protein that could be involved in natural resistance to BVM due to its presence near the CA protein-SP1 cleavage site where BVM is thought to bind. In addition, the V362I mutation was not found in any of the isolates showing wild-type or nearly wild-type susceptibility to BVM. Phenotypic analyses of mutant viruses with the V362I mutation constructed by site-directed mutagenesis are described below. For the remaining three isolates (isolates 2, 4, and 29), resistance to BVM could be associated with the presence of polymorphisms at Gag-SP1 residue 372, 373, 374, or 375 and/or could possibly occur in conjunction with other polymorphisms in the capsid protein (Table 2). Common polymorphisms, such as I138L/M, A146P/S, I147L, V159I, V215L, H219Q, I223V/A, G248A/T, N252S/A/H, T280V/A, R286K, S310T, E312D, A340G, and G357S (each of which was found in at least six isolates), were similarly partitioned between BVM-sensitive and BVM-resistant isolates and are therefore unlikely to play a role in resistance to BVM. However, other polymorphisms, such as K162R, Q182S, T186I, H219P, N252G, E260D, R264K, and T303V, were found uniquely in these three resistant isolates and may play a role in explaining the resistance to BVM.
Phenotypic analysis of mutants with site-directed mutations (SDMs).
In order to gain insight into the suspected role of the SP1 polymorphisms as well as the hypothesized role of the V362I capsid mutation in resistance to BVM, nine mutant viruses were constructed by site-directed mutagenesis of patient isolates or wild-type laboratory strain HIV-1 LAI. The SP1 polymorphisms from isolates 1 and 3 were mutated back to the wild-type sequence; the SP1 polymorphisms from isolate 6 were reverted back to the wild-type sequence in a stepwise fashion, generating two additional intermediate viruses carrying either the V370A mutation alone or both the S373A and A374T mutations; and the V362I capsid mutation was added to isolates 7 and 8 and the LAI laboratory strain and was mutated back to the wild-type sequence for patient isolate 12 (Table 3). In all three cases of reversion of the SP1 polymorphisms to the wild-type sequence, full susceptibility to BVM was restored, going from >100-fold BVM resistance before mutagenesis down to 0.6-, 0.7-, and 0.4-fold of that for the wild type for isolates SDM-1, SDM-3, and SDM-6a, respectively, after mutagenesis (Table 3). Furthermore, the analysis of the intermediate mutants showed that the V370A mutation in SP1 by itself is entirely responsible for the strong reduction in susceptibility to BVM observed for isolates SDM-6c and 6 (both of which had >100-fold BVM resistance), while the SP1 polymorphisms present in mutant SDM-6b and isolate 6 (S373A and A374T) played no role in resistance to BVM on their own. Additionally, these data indicate that none of the capsid polymorphisms present in isolates 1, 3, and 6 play any role in resistance to BVM. For isolate SDM-12, full susceptibility to BVM was restored by reversion of the V362I mutation to the wild-type sequence, suggesting that no other polymorphisms in this isolate alter susceptibility to BVM. In agreement with this observation, susceptibility to BVM was entirely lost in all three cases in which the V362I capsid mutation was added to viruses that were previously fully susceptible to BVM (mutants SDM-7, SDM-8, and SDM-LAI). Altogether, these results clearly indicate that the observed reduced susceptibility to BVM was strongly associated with the presence of polymorphisms in SP1—most notably, V370A—as well as the presence of the previously undescribed V362I capsid mutation.
TABLE 3.
Phenotypic susceptibility to BVM and genotypic data for the mutants with site-directed mutations
| Isolate no.a | BVM FCb | Amino acid change at the indicated position in the following protein: |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Capsidc |
SP1d |
|||||||||||||||||||||
| 357 (G)e | 358 (H) | 359 (K) | 360 (A) | 361 (R) | 362 (V) | 363 (L) | 364 (A) | 365 (E) | 366 (A) | 367 (M) | 368 (S) | 369 (Q) | 370 (V) | 371 (T) | 372 (N) | 373 (S) | 374 (A) | 375 (T) | 376 (I) | 377 (M) | ||
| 1 | >100** | —g | — | — | — | — | — | — | — | — | — | — | C | — | A | Delf | — | — | — | — | — | — |
| SDM-1 (wt SP1) | 0.6 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 3 | >100** | S | — | — | — | — | — | — | — | — | — | — | — | — | A | Del | H/N | — | — | — | — | — |
| SDM-3 (wt SP1) | 0.7 | S | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 6 | >100** | — | — | — | — | — | — | — | — | — | — | — | — | — | A | — | — | A | T | — | — | — |
| SDM-6a (wt SP1) | 0.4* | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| SDM-6b (wt V370) | 0.7 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | A | T | — | — | — |
| SDM-6c (wt S373/A374) | >100** | — | — | — | — | — | — | — | — | — | — | — | — | — | A | — | — | — | — | — | — | — |
| 7 | 1 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| SDM-7 (V362I) | >100** | — | — | — | — | — | I | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 8 | 1.2 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| SDM-8 (V362I) | >100* | — | — | — | — | — | I | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| LAI | 1 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| SDM-LAI (V362I) | >100** | — | — | — | — | — | I | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 12 | >100** | S | — | — | — | — | I | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| SDM-12 (wt V362) | 0.7 | S | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
Mutants constructed by site-directed mutagenesis (SDM) were created by PCR from patient-derived isolates (isolates 1, 3, 6, 7, 8, and 12) or the LAI laboratory strain, with the specific changes in SP1 (isolates 1, 3, and 6) or the capsid protein (V362I; isolates 7, 8, 12 and LAI laboratory strain) being as shown. All other protease and Gag sequence were identical to those in the patient isolate or laboratory strain. wt, wild type.
Fold change (FC) in the bevirimat EC50 compared to wild-type EC50 of 34 nM against HIV-1 LAI. The statistical significance of the fold changes in EC50s compared to the wild-type EC50 is expressed as follows P < 0.01 (**) and P < 0.05 (*). The results are the averages of at least three independent experiments.
The last 7 amino acids of the Gag-capsid protein sequences are shown.
The complete amino acid sequences of the Gag-SP1 protein are shown (14 residues).
The letters in parentheses represent the wild-type sequence.
Del, deletion.
—, same as wild type.
DISCUSSION
We conducted in vitro phenotypic analyses of 31 recombinant HIV-1 isolates containing the entire gag and protease genes extracted from 31 patient samples in an otherwise wild-type HIV-1 background (HIV-1 LAI). The patients in the study were either ARV naïve (n = 10) or ARV experienced (n = 21) and included patients with extensive protease inhibitor experience and PI resistance. In contrast to our original hypothesis that prior ARV treatment and, in particular, protease inhibitor resistance may have induced the selection of mutations in Gag that could have impaired susceptibility or the response to BVM, we found that in the 31 patient isolates that we analyzed, similar proportions of viruses showed resistance to BVM, regardless of the patient's ARV experience or the presence of PI resistance. Given the limited number of samples that we have tested, our results cannot rule out the possibility that PI resistance contributes to BVM resistance, as has been observed by others (17). However, our results clearly show that resistance to BVM is found independently of both PI resistance and treatment experience and is observed in the presence of natural polymorphisms in HIV-1. As a consequence of this genetic diversity, we found that nearly 60% (18/31; 58%) of the samples tested in this study showed innate reduced susceptibility to BVM, consistent with the clinical trial response data, which indicated that 52% of patients did not respond to BVM treatment (7, 12).
In the reported clinical trial, researchers had determined that a poor response to BVM was associated with the presence of polymorphisms at positions Q369, V370, and T371 in SP1 (7, 12). In our study, we have observed an association between reduced susceptibility to BVM and the presence of polymorphisms at position V370. The important role of the V370A polymorphism in the resistance to BVM was confirmed through the phenotypic analysis of a recombinant mutant that was constructed by site-directed mutagenesis and that contained the V370A polymorphism by itself, which was able to confer >100-fold resistance to BVM compared with that of the wild type. These results are in agreement with data for patient isolates and phenotypic data for mutants constructed by site-directed mutagenesis published earlier (16). It is noteworthy that the natural polymorphism V370I (SP1-V7I), which conferred reduced susceptibility to BVM in association with a T371 deletion in our study (isolate 10), was identified in several in vitro resistance selection experiments performed with a low BVM concentration (85 nM, or 50 ng/ml); in those experiments, it was thought to play a compensatory role in replication (1, 2). Interestingly, one of our isolates (isolate 32) with a V370I-V370V mixture showed wild-type BVM susceptibility, while another isolate (isolate 38) with a V370A-V370V mixture showed >100-fold decreased susceptibility to BVM, suggesting that these two substitutions at V370 differentially affect BVM susceptibility.
The presence of the T371 deletion often coincided with the presence of a polymorphism at V370, making the role of the T371 deletion in resistance more difficult to assess. In our data set, one isolate (isolate 25) had the T371 deletion alone and showed an 11-fold reduced susceptibility to BVM. This is consistent with earlier data that showed that a mutant constructed by site-directed mutagenesis with the T371 deletion alone had a 52.2-fold reduced susceptibility to BVM, while a V370A mutant had >151.4-fold reduced susceptibility to BVM (16), suggesting that the T371 deletion plays a less critical role than V370A in resistance to BVM. Interestingly, the T371 deletion was found only in PI-resistant isolates in our data set, suggesting the possibility that that deletion was coselected by treatment with protease inhibitors. However, the T371 deletion was not reported among 82 PI-experienced patients in another study (10). Of note, the T371Q polymorphism observed in isolate 36 was associated with wild-type BVM susceptibility, and a T371I mutation (SP1-T8I) was reported to arise in a selection experiment without use of the drug (1).
In our data set, two isolates had a Q369H polymorphism, with both viruses showing nearly wild-type susceptibility to BVM. Similarly, mutant viruses constructed by site-directed mutagenesis with either Q369A or Q369H alone have been reported by others to have nearly wild-type BVM susceptibility (16), suggesting that these Q369 polymorphisms play no role in resistance to BVM when they are found independently of the V370 or T371 polymorphism. Furthermore, we were able to show that the S373A and A374T SP1 polymorphisms on their own had no effect on resistance to BVM.
Two patient isolates (isolates 12 and 39) with no BVM resistance-conferring polymorphisms in SP1 were found to have >100-fold reduced susceptibility to BVM. These isolates carried the V362I polymorphism in the capsid protein (CA protein-V230I), which, we suspected, could be involved in resistance to BVM because of its location at the P2 position of the CA protein-SP1 cleavage site. We constructed by site-directed mutagenesis mutant viruses in which we either reverted the V362I mutation from isolate 12 to the wild-type sequence or added the V362I mutation to isolates with wild-type BVM susceptibility (isolates 7 and 8 and laboratory HIV strain LAI). These analyses have shown that reversion of the isoleucine back to a valine at position 362 in isolate 12 completely restored susceptibility to BVM, while mutation of the valine to isoleucine at position 362 in the three wild-type isolates led to the complete loss of BVM susceptibility. These data clearly demonstrate that the previously undescribed V362I natural polymorphism in the CA protein is a mutation that confers resistance to BVM. Notably, this mutation was reported to arise in the background of a mutant virus containing the L363M mutation during in vitro resistance selection with a high BVM concentration and was thought to be a secondary mutation that compensated for the replication defect (2). The effect of the V362I mutation on the replication capacity of the viruses was not formerly explored in this study; however, in our assays viruses carrying that mutation did not show any replication defect. The V362I mutation was found in 13% (4/31) of the patients in our study, and other estimates of the prevalence of this mutation in the patient population have been shown to vary from 7% to 18% (9, 10, 19). The presence of the V362I mutation in patients could adversely affect responses to BVM treatment.
Three patient isolates with >100-fold reduced susceptibility to BVM (isolates 2, 4, and 29) did not have the V362I mutation or any change at position 369, 370, or 371. These viruses had a few polymorphisms in SP1 that could potentially be responsible for the resistance observed: isolate 2 had the S373A, A374G, and T375A polymorphisms; isolate 4 had the N372A, A374T, and T375N polymorphisms; and isolate 29 had the T375A polymorphism. The presence of the T375A polymorphism by itself in isolate 29 is unlikely to play a role in resistance to BVM, as it is also found in an isolate with nearly wild-type susceptibility to BVM (isolate 36). As a consequence, it is likely that the genotypic determinants of resistance in this isolate reside outside of SP1 and the polymorphism T303V in the capsid protein, which was uniquely found in that isolate, may be involved in that role. While it is conceivable that the combination of polymorphisms in SP1 may be responsible for the loss of susceptibility to BVM in isolates 2 and 4, most of these SP1 polymorphisms—S373A, A374T, T375A, and T375N—were also found in isolates or mutants constructed by site-directed mutagenesis with nearly wild-type BVM susceptibility and are therefore unlikely to play a role in resistance to BVM, leaving only A374G in isolate 2 and N372A in isolate 4 with potential roles in resistance to BVM. However, as with isolate 29, it may be that the genetic basis of resistance lies in the capsid gene. The capsid polymorphisms H219P and R264K, unique to isolate 2, and the capsid polymorphisms K162R, Q182S, T186I, N252G, and E260D, unique to isolate 4, may play a role in resistance to BVM. In addition, protease inhibitor resistance-conferring mutations could also play a role in BVM resistance in these isolates, as they carry mutations such as L38W, P39Q, I50V, T74S, T80A, and N83H that are not found in isolates that are sensitive to BVM. Further studies would be required to address the potential role of these polymorphisms in resistance to BVM.
In conclusion, we have found that nearly 60% of the patient isolates in our study had innate resistance to BVM of >10-fold. Interestingly, none of the in vitro-selected resistance-conferring mutations to BVM were found to exist in the 31 patient isolates included in our study. Instead, we showed that a previously undescribed polymorphism, V362I (CA protein-V230I), is a major mutation conferring resistance to BVM, and we confirmed that the V370A polymorphism (SP1-V7A) also plays a major role in resistance to BVM. These genotypic and phenotypic data extend the existing data set on BVM resistance and should help with the development of diagnostics for predicting the response to BVM treatment in HIV-1-infected patients.
Acknowledgments
We thank Kirsten White, Damian McColl, and Tomas Cihlar for their careful review of the manuscript, as well as Katyna Borroto-Esoda and Jenny Svarovskaia for their helpful discussions.
Footnotes
Published ahead of print on 22 March 2010.
REFERENCES
- 1.Adamson, C. S., S. D. Ablan, I. Boeras, R. Goila-Gaur, F. Soheilian, K. Nagashima, F. Li, K. Salzwedel, M. Sakalian, C. T. Wild, and E. O. Freed. 2006. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (bevirimat). J. Virol. 80:10957-10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson, C. S., K. Waki, S. D. Ablan, K. Salzwedel, and E. O. Freed. 2009. Impact of human immunodeficiency virus type 1 resistance to protease inhibitors on evolution of resistance to the maturation inhibitor bevirimat (PA-457). J. Virol. 83:4884-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloch, M., N. Bodsworth, G. Mather, C. Workman, A. Balach, H. Byakagwa, and A. Beelen. 2009. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother., abstr. H-1230. American Society for Microbiology, Washington, DC.
- 4.Holz-Smith, S. L., I. C. Sun, L. Jin, T. J. Matthews, K. H. Lee, and C. H. Chen. 2001. Role of human immunodeficiency virus (HIV) type 1 envelope in the anti-HIV activity of the betulinic acid derivative IC9564. Antimicrob. Agents Chemother. 45:60-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanamoto, T., Y. Kashiwada, K. Kanbara, K. Gotoh, M. Yoshimori, T. Goto, K. Sano, and H. Nakashima. 2001. Anti-human immunodeficiency virus activity of YK-FH312 (a betulinic acid derivative), a novel compound blocking viral maturation. Antimicrob. Agents Chemother. 45:1225-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kashiwada, Y., F. Hashimoto, L. M. Cosentino, C. H. Chen, P. E. Garrett, and K. H. Lee. 1996. Betulinic acid and dihydrobetulinic acid derivatives as potent anti-HIV agents. J. Med. Chem. 39:1016-1017. [DOI] [PubMed] [Google Scholar]
- 7.Lalezari, J., S. McCallister, M. Gigliotti, C. Cohen, R. Elion, C. Brinson, G. Richmond, M. Thompson, and D. Martin. 2008. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC)-Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet., abstr. H-891. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 8.Li, F., R. Goila-Gaur, K. Salzwedel, N. R. Kilgore, M. Reddick, C. Matallana, A. Castillo, D. Zoumplis, D. E. Martin, J. M. Orenstein, G. P. Allaway, E. O. Freed, and C. T. Wild. 2003. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc. Natl. Acad. Sci. U. S. A. 100:13555-13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malet, I., B. Roquebert, C. Dalban, M. Wirden, B. Amellal, R. Agher, A. Simon, C. Katlama, D. Costagliola, V. Calvez, and A. G. Marcelin. 2007. Association of Gag cleavage sites to protease mutations and to virological response in HIV-1 treated patients. J. Infect. 54:367-374. [DOI] [PubMed] [Google Scholar]
- 10.Malet, I., M. Wirden, A. Derache, A. Simon, C. Katlama, V. Calvez, and A. G. Marcelin. 2007. Primary genotypic resistance of HIV-1 to the maturation inhibitor PA-457 in protease inhibitor-experienced patients. AIDS 21:871-877. [DOI] [PubMed] [Google Scholar]
- 11.Margot, N. A., J. M. Waters, and M. D. Miller. 2006. In vitro human immunodeficiency virus type 1 resistance selections with combinations of tenofovir and emtricitabine or abacavir and lamivudine. Antimicrob. Agents Chemother. 50:4087-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCallister, S., J. Lalezari, G. Richmond, M. Thompson, R. Harrigan, D. Martin, K. Salzwedel, and G. Allaway. 2008. Abstr. XVII Int. HIV Drug Resist., abstr. 8.
- 13.Sakalian, M., C. P. McMurtrey, F. J. Deeg, C. W. Maloy, F. Li, C. T. Wild, and K. Salzwedel. 2006. 3-O-(3′,3′-Dimethysuccinyl) betulinic acid inhibits maturation of the human immunodeficiency virus type 1 Gag precursor assembled in vitro. J. Virol. 80:5716-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi, C., and J. W. Mellors. 1997. A recombinant retroviral system for rapid in vivo analysis of human immunodeficiency virus type 1 susceptibility to reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 41:2781-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith, P. F., A. Ogundele, A. Forrest, J. Wilton, K. Salzwedel, J. Doto, G. P. Allaway, and D. E. Martin. 2007. Phase I and II study of the safety, virologic effect, and pharmacokinetics/pharmacodynamics of single-dose 3-O-(3′,3′-dimethylsuccinyl)betulinic acid (bevirimat) against human immunodeficiency virus infection. Antimicrob. Agents Chemother. 51:3574-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Baelen, K., K. Salzwedel, E. Rondelez, V. Van Eygen, S. De Vos, A. Verheyen, K. Steegen, Y. Verlinden, G. P. Allaway, and L. J. Stuyver. 2009. Susceptibility of human immunodeficiency virus type 1 to the maturation inhibitor bevirimat is modulated by baseline polymorphisms in Gag spacer peptide 1. Antimicrob. Agents Chemother. 53:2185-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verheyen, J., C. Verhofstede, E. Knops, L. Vandekerckhove, B. Diede, D. Kenny, A. Wensing, H. Pfister, R. Kaiser, and M. Nijuis. 2009. Abstr. 12th Eur. AIDS Conf., abstr. PE3.2/4.
- 18.Weislow, O. S., R. Kiser, D. L. Fine, J. Bader, R. H. Shoemaker, and M. R. Boyd. 1989. New soluble-formazan assay for HIV-1 cytopathic effects: application to high-flux screening of synthetic and natural products for AIDS-antiviral activity. J. Natl. Cancer Inst. 81:577-586. [DOI] [PubMed] [Google Scholar]
- 19.Yebra, G., and A. Holguin. 2008. The maturation inhibitor bevirimat (PA-457) can be active in patients carrying HIV type-1 non-B subtypes and recombinants. Antivir. Ther. 13:1083-1085. [PubMed] [Google Scholar]
- 20.Zhou, J., C. H. Chen, and C. Aiken. 2004. The sequence of the CA-SP1 junction accounts for the differential sensitivity of HIV-1 and SIV to the small molecule maturation inhibitor 3-O-{3′,3′-dimethylsuccinyl}-betulinic acid. Retrovirology 1:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou, J., L. Huang, D. L. Hachey, C. H. Chen, and C. Aiken. 2005. Inhibition of HIV-1 maturation via drug association with the viral Gag protein in immature HIV-1 particles. J. Biol. Chem. 280:42149-42155. [DOI] [PubMed] [Google Scholar]
- 22.Zhou, J., X. Yuan, D. Dismuke, B. M. Forshey, C. Lundquist, K. H. Lee, C. Aiken, and C. H. Chen. 2004. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J. Virol. 78:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]