Abstract
A clinical strain of Acinetobacter calcoaceticus resistant to carbapenems was isolated from a blood culture sample from an inpatient in a hospital in Madrid (Spain) during a large outbreak of infection (affecting more than 300 inpatients), caused by a multidrug-resistant Acinetobacter baumannii clone. The carbapenem resistance in both the A. calcoaceticus and A. baumannii clones was due to a blaOXA-24 gene harbored in different plasmids. The plasmids were fully sequenced, revealing the presence of site-specific recombination binding sites putatively involved in mobilization of the blaOXA-24 gene. Comparison of plasmids contained in the two strains revealed possible horizontal transmission of resistance genes between the Acinetobacter species.
Since 1986, members of the genus Acinetobacter have been identified by Southern hybridization. Genospecies 1 (Acinetobacter calcoaceticus), 2 (Acinetobacter baumannii), 3, and 13TU are genetically closely related and are commonly known as the A. calcoaceticus-A. baumannii complex. With the exception of genospecies 1, the other members of this complex have been reported to be involved in nosocomial infections and are known to have the ability to spread within hospitals (1, 12). Acinetobacter calcoaceticus has traditionally been considered an environmental species and has never been associated with serious clinical infections.
Among β-lactamases, the most prevalent carbapenemases in Acinetobacter spp. are the class D β-lactamases, which are divided into 4 different groups: OXA-23, OXA-24, and OXA-58 with all their variants (17) and the OXA-51 family, which has been described as being intrinsic to A. baumannii (6). Although these β-lactamases have mainly been isolated from A. baumannii, recent studies have demonstrated the presence of OXA-58 in Acinetobacter genomic species 3, as well as in Acinetobacter phenon 6/ct13TU (10, 11) and in Acinetobacter genomospecies 13TU (8). None of these class D β-lactamases have been described in A. calcoaceticus (7).
During an outbreak of infection caused by a multidrug-resistant (including carbapenems) A. baumannii clone (named AbH12O-A2), which occurred in the 12 de Octubre Hospital (Madrid, Spain) and affected more than 300 patients, an A. calcoaceticus strain (named Acal H12O-07) was isolated from blood cultures from a 58-year-old man. The patient was admitted to the hospital with a cranial encephalic trauma. After admission and during the course of his stay, the patient presented several nosocomial infections. The patient was empirically treated with different cycles of antibiotics, starting with ceftriaxone plus levofloxacin, then piperacillin-tazobactam, meropenem plus vancomycin, and finally linezolid plus imipenem. After 4 months of hospitalization and in light of persistent fever, two sets of blood cultures were drawn and yielded isolation of a Gram-negative bacillus. The microorganism was identified with the automated WIDER system as a member of the A. baumannii-A. calcoaceticus complex. The antibiotic susceptibility profile obtained by microdilution revealed the following MICs (μg/ml): piperacillin-tazobactam, >64/4; ceftazidime, >16; cefepime, >16; imipenem, >8; meropenem, >8; tobramycin, ≤2; amikacin, >16; gentamicin, ≤2; ciprofloxacin, 1; trimethoprim-sufamethoxazole, ≤2/38. After clinical evaluation, the case was considered a primary bacteremia and the patient was administered a course of antimicrobial treatment with ciprofloxacin and gentamicin for 2 weeks, with a favorable clinical response.
The 16S rRNA of the isolate was also sequenced and yielded identification of an A. calcoaceticus strain. A putative β-lactamase enzyme with carbapenemase activity was detected by the Hodge test (9). A PCR amplification with primers designed from class D carbapenemase genes and metallo-β-lactamase genes (blaIMP, blaVIM, and blaSIM) from Acinetobacter (18) yielded isolation of a blaOXA-24 gene. A plasmid, named pMMCU1, was isolated from the clinical A. calcoaceticus strain. This plasmid was used to transform an A. baylyi ADP1 isolate. Imipenem and meropenem MICs for the A. baylyi ADP1 strain increased from 0.094 and 0.25, respectively, to >32 μg/ml in both cases, when the strain was transformed with plasmid pMMCU1. Moreover, a band corresponding to the blaOXA-24 gene was also obtained by PCR, with the plasmid isolated from the transformed A. baylyi ADP1 strain as template. The gene product revealed 100% identity with the previously described blaOXA-24 gene (2). The full plasmid was sequenced and was found to be 8,771 bp in size (GenBank accession code GQ342610). Moreover, the A. baumannii clone that caused the large outbreak was found to carry the pMMA2 plasmid (GenBank accession code GQ377752), which was also isolated, analyzed, and found to be 10,679 bp in size.
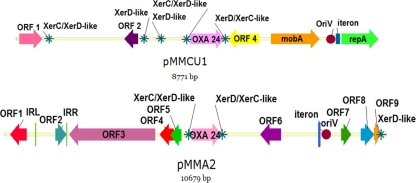
Comparative analysis among the sequences of the two plasmids revealed different scaffolds and coding regions, as shown in Fig. 1 and Tables 1 and 2. The pMMCU1 plasmid showed the highest homology with plasmid pABO2 (GenBank accession code AY228470) and carried the mobilization region derived from the previously described pMAC plasmid (GenBank accession code AY541809). The pMMA2 plasmid displayed the highest homology with the previously described p2ABAYE plasmid (GenBank accession code CU459138.1).
FIG. 1.
(Top) Diagram of the 8,771-bp pMMCU1 plasmid obtained from the A. calcoaceticus clinical strain (Acal H12O-07). (Bottom) Diagram of the 10,679-bp pMMA2 plasmid obtained from the A. baumannii strain (AbH12O-A2) that caused the large outbreak of infection. The asterisks indicate the Xer-like recombination sites.
TABLE 1.
Description of the pMMCU1 plasmid isolated from A. calcoaceticus (AcalH2O-O7)
| Featureb | Positions | Properties and/or putative function | GenBank/EMBL accession no. of match | Plasmid or chromosomal homology (reference) |
|---|---|---|---|---|
| ORF 1 | 74-631 | Hypothetical protein | AY228470 | pAB02 |
| XerC/XerD-like | 782-809 | Recombination sites | FM210331.1 | pABVA01 |
| ORF 2 | 2621-2941 | Hypothetical protein | CU468230 | ABSDF |
| XerD-like | 3090-3100 | Recombination sites | FM210331.1 | pABVA01 |
| XerD-like | 3503-3513 | Recombination sites | FM210331.1 | pABVA01 |
| XerC/XerD-like | 4116-4143 | Recombination sites | AJ239129 | RYC52763/97 (2)a |
| blaOXA-24 | 4217-5044 | Carbapenem-hydrolyzing oxacillinase | AJ239129 | RYC52763/97 (2)a |
| XerD/XerC-like | 5055-5082 | Recombination sites | AJ239129 | RYC52763/97 (2)a |
| ORF 4 | 5113-5874 | Hypothetical protein | AY541809 | pMAC |
| MobA | 6174-7343 | Plasmid mobilization protein | AY541809 | pMAC |
| OriV | 7524-7724 | Origin of DNA replication | AY228470 | pAB02 |
| Iteron | 7741-7836 | Imperfect 4-repeat iterons; control of DNA replication | AY228470 | pAB02 |
| RepA | 7881-8768 | repA_AB; DNA replication protein | AY228470 | pAB02 |
TABLE 2.
Description of the pMMA2 plasmid isolated from the A. baumannii strain that caused a large outbreak of infection (AbH12O-A2)a
| Feature | Positions | Properties and/or putative function | GenBank/EMBL accession no. of match(es) | Plasmid or chromosomal homology (reference) |
|---|---|---|---|---|
| ORF 1 | 141-602 | Septicolysin (endotoxin) | CU459138.1 | p2ABAYE |
| IRL | 855-872 | IS4 inverted repeat left | YP_001957893.1 | NA |
| ORF 2 | 1418-1719 | Putative IS4 transposase family | YP_001957893.1 | NA |
| IRR | 1722-1739 | IS4 inverted repeat right | YP_001957893.1 | NA |
| ORF 3 | 1793-4204 | Putative TonB-dependent receptor | CU459138.1 | p2ABAYE |
| ORF 4 | 4332-4715 | Putative cytoplasmic protein | CU459138.1 | p2ABAYE |
| ORF 5 | 4633-4929 | Putative inner membrane protein | CU459138.1 | p2ABAYE |
| XerC/XerD-like | 5128-5155 | Recombination sites | AJ239129, AY228470, NC_010481 | RYC52763/97 (2)b pABO2 pABIR |
| blaOXA-24 | 5229-6056 | Carbapenem-hydrolyzing oxacillinase | AJ239129 | RYC52763/97 (2)b |
| XerC/XerD-like | 6069-6096 | Recombination sites | AJ239129, AY228470, NC_010481 | RYC52763/97 (2)b pABO2 pABIR |
| ORF 6 | 7145-7714 | DNA replication protein | ZP_04663577 | AB900 |
| Iteron | 8791-8878 | Imperfect 4-repeat iterons; control of DNA replication | CU459138.1 | p2ABAYE |
| OriV | 8896-9100 | Replication origin | CU459138.1 | p2ABAYE |
| ORF 7 | 9410-9682 | Hypothetical protein | CU459138.1 | p2ABAYE |
| ORF 8 | 9953-10324 | Hypothetical protein | CU459138.1 | p2ABAYE |
| ORF 9 | 10324-10497 | Hypothetical protein | CU459138.1 | p2ABAYE |
| XerD-like | 10521-10531 | Recombination sites | CU459138.1 | p2ABAYE |
The blaOXA-24 gene was detected in both the pMMCU1 and pMMA2 plasmids and was found to be flanked by 11-bp conserved inverted repeats separated by a 6-bp variable region (GenBank accession codes GQ342610 and GQ377752). The 5′ sequence flanking the blaOXA-24 gene in the pMMCU1 plasmid was ATTTCGCATAACGCCCATTATGTTAAAT, and the 3′ sequence was AATTAACATAATACGCCTTATGCGAAAT. Similarly, in the case of the pMMA2 plasmid, the sequence located at the 5′ position was ACTTCGGATAACGCCCATTATGTTAAAT, and that located at the 3′ position was TTAACATAATACACCTTATACGAAATGC. The Xer-like binding site sequences described in the pMMCU1 and pMMA2 plasmids showed 79.5, 76, 76, 91, 74, 81.5, and 90.5% and 76.5, 73, 73, 88.5, 80, 76.5, and 88.5% identity, respectively, with their counterparts found in plasmids pABVA01, p2ABAYE, pAB0057, pAB02, pAB2, pAV1, and pABIR, respectively, and in both cases they showed the highest identity, 100% and 88.5%, respectively, with the XerC/XerD-like binding sites located in the chromosomal region of the blaOXA-24 gene previously described in the RYC52763/97 strain (2, 5).
The blaOXA-24 gene was already observed as part of one of the discrete DNA modules flanked by XerC/XerD-like sites within Acinetobacter plasmids. These Xer-like binding sites have been suggested to be involved in the mobilization of discrete DNA modules within Acinetobacter plasmids and chromosomes by site-specific recombination mechanisms (5).
Moreover, other plasmids were also isolated from minor A. baumannii clones that appeared during the outbreak (GenBank accession codes GQ904226, GQ476987, and GQ904227). These plasmids also harbored the blaOXA-24 gene integrated in different locations flanked by XerC/XerD-like binding sites. These results show that during the outbreak there was no exchange of a common plasmid carrying the blaOXA-24 gene among the strains isolated. On the contrary, the plasmid structures provide evidence supporting the hypothesis that different plasmids exchange the blaOXA-24 gene comprised within a very limited region between the two closest XerC/XerD-like binding sites.
DNA recombination through the Xer system in plasmids requires XerC and XerD (recombinases); XerC/XerD-like binding sites; accessory proteins, such as PepA, ArgR, and ArcA; and an accessory sequence of about 180 bp located near the core site (3, 4, 13, 15). Recombination occurs via formation of a heterotetrameric complex in which each recombinase catalyzes the exchange of one pair of DNA strands in a reaction that proceeds through the Holliday junction intermediate. The accessory proteins bind the accessory sequence and induce formation of a synaptic complex that is required for recombination (3). These recombination events are involved in site-specific integration and excision of lysogenic genomes, transposition of conjugative transposons, termination of chromosome replication, and plasmid stability (3, 14, 16).
Sequence analysis revealed that all the plasmids isolated from the outbreak carried the same blaOXA-24 mobilization cassette, which contained the following regions, from 5′ to 3′: a region of 142 bp followed by XerC/XerD-like binding sites, a region of 72 bp, and the blaOXA-24 gene finally followed by 10 bp and the downstream XerD/XerC-like binding sites (GenBank accession codes GQ342610, GQ377752, GQ904226, GQ476987, and GQ904227, respectively). We suggest that both of these regions, of 142 and 72 bp, may act as targets for the accessory proteins required for Xer recombination. Moreover, we found that XerC and XerD recombinases are encoded in the A. baumannii chromosome of strains AB0057, SDF, AYE, and ACICU (GenBank accession codes for XerC: YP_002320364.1, YP_001706416.1, YP_001712817.1, and YP_001847531.1, respectively; for XerD: ZP_02977538.1, YP_001708368, YP_001715285.1, and YP_001844923.1, respectively). PepA has been also found in the genomes of A. baumannii AB0057, SDF, and AYE (GenBank accession codes YP_002317727.1, YP_001708379.1, and YP_001715297.1, respectively).
These findings suggest that Xer recombination may be responsible for mobilization of the blaOXA-24 gene, and experiments are in progress to confirm this hypothesis. As the Xer-like binding sites are located in opposite directions, recombination through the Xer system should occur by gene inversion. Therefore, the most likely explanation would seem to be that putative Xer-mediated recombination events led to the dissemination of the blaOXA-24 gene among different plasmids and that acquisition of the resistance gene by A. calcoaceticus was mediated by the transfer of one of these plasmids. Note that A. calcoaceticus is usually regarded as an environmental species and this is the first description of a plasmid-mediated carbapenem-hydrolyzing oxacillinase, OXA-24, isolated from an A. calcoaceticus strain causing bacteremia.
Dissemination of resistance genes via Xer recombination in plasmids has previously been suggested (5). The present study emphasizes the threat associated with this mechanism in relation to the dissemination of carbapenemase genes among different Acinetobacter species in hospital environments.
Nucleotide sequence accession numbers.
The sequences of plasmids pMMCU1 and pMMA2 were deposited in GenBank under nucleotide sequence accession numbers GQ342610 and GQ377752, respectively. Other plasmid sequences from outbreak clones were deposited under accession numbers GQ904226, GQ476987, and GQ904227.
Acknowledgments
This study was supported by the Spanish Network for Research on Infectious Diseases—REIPI (Instituto de Salud Carlos III, RD06/0008/0025 and RD006/0008/0011), Fondo de Investigaciones Sanitarias (PI061368, PI081368, and PS09/00687), and SERGAS (PS07/90) and by grants from Xunta de Galicia (07CSA050916PR). Margarita Poza and Alejandro Beceiro are in receipt of Isidro Parga Pondal and Angeles Alvariño grants, respectively (research contracts from Xunta de Galicia).
Footnotes
Published ahead of print on 12 April 2010.
REFERENCES
- 1.Bergogne-Berezin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bou, G., A. Oliver, and J. Martínez-Beltrán. 2000. OXA-24, a novel class D beta-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bui, D., J. Ramiscal, S. Trigueros, J. S. Newmark, A. Do, D. J. Sherratt, and M. E. Tolmasky. 2006. Differences in resolution of mwr-containing plasmid dimers mediated by the Klebsiella pneumoniae and Escherichia coli XerC recombinases: potential implications in dissemination of antibiotic resistance genes. J. Bacteriol. 188:2812-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornet, F., I. Mortier, J. Patte, and J. M. Louarn. 1994. Plasmid pSC101 harbors a recombination site, psi, which is able to resolve plasmid multimers and to substitute for the analogous chromosomal Escherichia coli site dif. J. Bacteriol. 176:3188-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Andrea, M. M., T. Giani, S. D'Arezzo, A. Capone, N. Petrosillo, P. Visca, F. Luzzaro, and G. M. Rossolini. 2009. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:3528-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heritier, C., L. Poirel, P. E. Fournier, J. M. Claverie, D. Raoult, and P. Nordmann. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwa, W. E., G. Subramaniam, P. Navaratnam, and S. D. Sekaran. 2009. Detection and characterization of class 1 integrons among carbapenem-resistant isolates of Acinetobacter spp. in Malaysia. J. Microbiol. Immunol. Infect. 42:54-62. [PubMed] [Google Scholar]
- 8.Koh, T. H., L.-H. Sng, G. C. Y. Wang, L.-Y. Hsu, and Y. Zhao. 2007. Carbapenemase and efflux pump genes in Acinetobacter calcoaceticus-Acinetobacter baumannii complex strains from Singapore. J. Antimicrob. Chemother. 60:1173-1174. [DOI] [PubMed] [Google Scholar]
- 9.Lee, K., Y. Chong, H. B. Shin, Y. A. Kim, D. Yong, and J. H. Yum. 2001. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 7:88-91. [DOI] [PubMed] [Google Scholar]
- 10.Marti, S., J. Sánchez-Céspedes, M. D. Blasco, M. Ruiz, P. Espinal, V. Alba, F. Fernández-Cuenca, A. Pascual, and J. Vila. 2008. Characterization of the carbapenem-hydrolyzing oxacillinase OXA-58 in an Acinetobacter genospecies 3 clinical isolate. Antimicrob. Agents Chemother. 52:2955-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marti, S., J. Sánchez-Céspedes, M. D. Blasco, P. Espinal, M. Ruiz, V. Alba, and J. Vila. 2008. Characterization of the carbapenem-hydrolyzing oxacillinase OXA-58 in an Acinetobacter phenon 6/ct13TU clinical isolate. Diagn. Microbiol. Infect. Dis. 61:468-470. [DOI] [PubMed] [Google Scholar]
- 12.Ribera, A., F. Fernández-Cuenca, A. Beceiro, G. Bou, L. Martínez-Martínez, A. Pascual, J. M. Cisneros, J. Rodríguez-Baño, J. Pachón, and J. Vila. 2004. Antimicrobial susceptibility and mechanisms of resistance to quinolones and beta-lactams in Acinetobacter genomic species 3. Antimicrob. Agents Chemother. 48:1430-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Summers, D. K., and D. J. Sherratt. 1984. Multimerization of high copy number plasmids causes instability: ColE1 encodes a determinant essential for plasmid monomerization and stability. Cell 36:1097-1103. [DOI] [PubMed] [Google Scholar]
- 14.Summers, D. K., and D. J. Sherratt. 1988. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 7:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tolmasky, M. E., S. Colloms, G. Blakely, and D. J. Sherratt. 2000. Stability by multimer resolution of pJHCMW1 is due to the Tn1331 resolvase and not to the Escherichia coli Xer system. Microbiology 146:581-589. [DOI] [PubMed] [Google Scholar]
- 16.Val, M.-E., M. Bouvier, J. Campos, D. Sherrat, F. Cornet, D. Mazel, and F.-X. Barre. 2005. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell 19:559-566. [DOI] [PubMed] [Google Scholar]
- 17.Walther-Rasmussen, J., and N. Hoiby. 2006. OXA-type carbapenemases. J. Antimicrob. Chemother. 57:373-383. [DOI] [PubMed] [Google Scholar]
- 18.Woodford, N., M. J. Ellington, J. M. Coelho, J. F. Turton, M. E. Ward, S. Brown, S. G. Amyes, and D. M. Livermore. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351-353. [DOI] [PubMed] [Google Scholar]