Abstract
The M230L mutation in HIV-1 reverse transcriptase (RT) is associated with resistance to first-generation nonnucleoside reverse transcriptase inhibitors (NNRTIs). The present study was designed to determine the effects of M230L on enzyme function, viral replication capacity (RC), and the extent to which M230L might confer resistance to the second-generation NNRTI etravirine (ETR) as well as to the first-generation NNRTIs efavirenz (EFV) and nevirapine (NVP). Phenotyping assays with TZM-bl cells confirmed that M230L conferred various degrees of resistance to each of the NNRTIs tested. Recombinant viruses containing M230L displayed an 8-fold decrease in RC compared to that of the parental wild-type (WT) virus. Recombinant HIV-1 WT and M230L mutant RT enzymes were purified; and both biochemical and cell-based phenotypic assays confirmed that M230L conferred resistance to each of EFV, NVP, and ETR. RT that contained M230L was also deficient in regard to each of minus-strand DNA synthesis, both DNA- and RNA-dependent polymerase activities, processivity, and RNase H activity, suggesting that this mutation contributes to diminished viral replication kinetics.
Highly active antiretroviral therapy (HAART) has been the standard of treatment for HIV infection since 1996 and has substantially increased the rates of survival of HIV-infected patients (20). Nonnucleoside reverse transcriptase inhibitors (NNRTIs), which include first-generation drugs such as nevirapine (NVP) and efavirenz (EFV), are important components of HAART, as is a newer agent, etravirine (ETR), which retains activity against HIV type 1 variants containing common drug resistance mutations, such as K103N, associated with the diminished activity of NVP and EFV. ETR seems to be able to adapt its orientation and, by so doing, overcome common NNRTI resistance-associated mutations (26, 39).
HIV-1 reverse transcriptases (RTs) are versatile DNA polymerases endowed with several properties essential for viral replication, i.e., RNA- and DNA-dependent DNA polymerases (RDDPs and DDDPs, respectively), RNase H, strand transfer, and strand displacement activities (37). NNRTIs inhibit RT by binding to a hydrophobic pocket adjacent to the active site of the enzyme (35). NNRTI resistance is due to mutations within the NNRTI binding pocket, often at amino acid positions 100 to 110, 180 to 190, and 220 to 240, that substantially decrease susceptibility to all first-generation NNRTIs (21), yet conflicting results on the role that this mutation may play in regard to ETR have emerged (36, 38, 39). It is also important to determine whether M230L impairs viral replication and to delineate any underlying molecular mechanisms that might be involved.
Therefore, we expressed and purified a recombinant HIV-1 RT enzyme containing M230L and performed both RNA- and DNA-dependent DNA polymerase assays to determine the impact of M230L on viral enzymatic capacity. As an additional control, we also studied dapivirine (DAP), a compound that has been licensed for possible development as a vaginal microbicide by Tibotec Pharmaceuticals to the International Partnership for Microbicides (IPM). Drug susceptibility was also determined in cell culture phenotyping assays with both wild-type (WT) viruses and recombinant viruses containing M230L.
MATERIALS AND METHODS
Chemicals, cells, and nucleic acids.
ETR was a gift from Tibotec Inc. DAP was obtained from the International Partnership for Microbicides. EFV and NVP were obtained from Bristol-Myers Squibb Inc. and Boehringer Ingelheim Inc., respectively. The HEK293T cell line was obtained from the American Type Culture Collection. The following reagents and cells were obtained through the NIH AIDS Research and Reference Reagent Program: infectious molecular clone pNL4-3 from Malcolm Martin and TZM-bl (JC53-bl) cells from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc.
The following oligonucleotides, which were synthesized by Integrated DNA Technologies Inc. and purified by 6% polyacrylamide-7 M urea gel electrophoresis, were used in this study: PPT17D (5′-TTAAAAGAAAAGGGGGG-3′), PPT19D (5′-TTAAAAGAAAAGGGGGGAC-3′), PPT57D (5′-CGTTGGGAGTGAATTAGCCCTTCCAGTCCCCCCTTTTCTTTTAAAAAGTGGCTAAGA-3′), Kim40R (5′-AAGCTTGGCTGCAGAATATTGCTAGCGGGAATTCGGCGCG-3′), Kim17D (5′-CGCGCCGAATTCCCGCT-3′), Kim32D (5′-CGCGCCGAATTCCCGCTAGCAATATTCTGCAG-3′), and 75D (5′-ATTGTAATACGACTCACTATAGCCGAATTCCCGCTAGCAATATTCTGCAGCCAAGCTTCCACCTGCAGGCATGCA-3′).
Site-directed mutagenesis.
The M230L mutation was introduced into the pNL4-3 proviral clone (1) by use of a QuikChange II XL site-directed mutagenesis kit (Stratagene) and subtype B HIV-1 RT heterodimer expression plasmid pRT6H-PROT (27). DNA sequencing was performed in both directions across the entire RT-coding region to verify the absence of spurious mutations and the presence of the desired mutation.
Preparation of virus stocks.
WT HIV-1 (HIV-1WT) and HIV-1 with the M230L mutation (HIV-1M230L) were generated by transfection of plasmids pNL4-3 and pNL4-3M230L into HEK293T cells by the use of Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. The viral supernatants were harvested at 48 h posttransfection, centrifuged at 800 × g for 5 min to remove the cellular debris, filtered through a 0.45-μm-pore-size filter, aliquoted, and stored at −80°C. The levels of p24 in the viral supernatant were measured by an HIV-1 p24 antigen enzyme-linked immunosorbent assay (Perkin-Elmer). Virus stocks were quantified for RT activity prior to infections (14). Virion-associated RT activity was measured as described previously (17), but with the following modifications: 50 μl of the RT reaction mixture contained 10 μl of culture supernatants, 6.25 μg/ml of poly(rA)-poly (dT)12-18 template-primer (T-P) in 50 mM Tris-HCl, pH 7.8-75 mM KCl-5 mM dithiothreitol-5 mM MgCl2-0.05% Triton X-100-2% ethylene glycol-0.3 mM reduced glutathione-5 μCi of [3H]dTTP (70 to 80 Ci/mmol, 2.5 mCi/ml). Following a 180-min incubation at 37°C, the reaction mixture was quenched by adding 0.2 ml of 10% cold trichloroacetic acid (TCA)-20 mM sodium pyrophosphate, and the mixture was incubated for at least 30 min on ice; the precipitated products were filtered onto 96-well MultiScreen high-throughput screening (HTS) FC filter plates (MSFCN6B; Millipore) and sequentially washed with 200 μl of 10% TCA and 150 μl of 95% ethanol. The radioactivity of the incorporated products was analyzed by liquid scintillation spectrometry with a 1450 MicroBeta TriLux microplate scintillation and luminescence counter (Perkin-Elmer).
Determination of relative replication capacity in TZM-bl cells.
The replicative capacities of competent clonal wild-type and M230L HIV-1 were evaluated in a noncompetitive infectivity assay with TZM-bl cells in a manner similar to that described previously (40). In parallel experiments, recombinant viruses were serially diluted 2-fold from stock suspensions, prior to quantification of a part of each dilution for RT activity, and added to 20,000 TZM-bl cells per well in a 96-well cell culture plate. The virus and cells were cocultured for 48 h, after which the cells were washed with phosphate-buffered saline and lysed with 1× cell culture lysis reagent diluted from 5× cell culture lysis reagent (Promega, Madison, WI). This time point had been identified in previous experiments to be optimal for the performance of comparisons (data not shown), since earlier times did not indicate exponential growth and later times resulted in reduced activity due to cell culture saturation. Luciferase activity was measured with a luciferase assay system (Promega), as recommended by the manufacturer. Luminescence was measured with a 1450 MicroBeta TriLux microplate scintillation and luminescence counter (Perkin-Elmer). The viral replication level was expressed as percentage of relative light units (RLU).
Phenotypic assay in TZM-bl cells.
The antiviral activities of the NNRTIs against HIV-1WT and HIV-1M230L derived from pNL4-3 molecular clones were evaluated in TZM-bl cells, as described previously (43). Serial dilutions (1:2) of the test compounds were prepared in dimethyl sulfoxide (DMSO) and then in culture medium (Dulbecco modified Eagle medium) to yield a final DMSO concentration of 0.2% in the cultures. Serially diluted compounds were added to 10,000 cells/well (96-well plates), and the cells were then infected with either the M230L or the WT viral variant. Standardization of the virus infections was accomplished by always adding 200 50% tissue culture infective doses, as calculated in TZM-bl cells (i.e., ∼180,000 ± 30,000 RLU) and as detected by luminescence. All cultures were maintained at 37°C under 5% CO2 for 48 h. Drug efficacy was determined by quantifying the luciferase activity as a measure of viral replication by using the luciferase reagent (Promega). RLU were detected with a 1450 MicroBeta TriLux microplate scintillation and luminescence counter (Perkin-Elmer). The 50% effective concentrations (EC50s) were determined by nonlinear regression with GraphPad Prism (version 5.01) software.
Purification of recombinant HIV-1 RTs and determination of enzyme activity.
The polymerase activity of each recombinant RT preparation was evaluated in duplicate by using various amounts of RTs and a synthetic homopolymeric poly(rA)-poly(dT)12-18 template-primer (Midland Certified Reagent Company). Each 50-μl reaction mixture contained 0.1 U/ml (5 μg/ml) poly(rA)-poly(dT)12-18, 5 mM dithiothreitol (DTT), 50 mM Tris-HCl, pH 7.8, 60 mM KCl, and 6 mM MgCl2. The reactions were initiated by adding 5 μM dTTP with 5 μCi [3H]dTTP (70 to 80 Ci/mmol; Perkin-Elmer). Aliquots of 15 μl were removed at 3, 7, and 15 min to ensure the linearity of the reaction and were quenched by the addition of ice-cold 10% trichloroacetic acid containing 20 mM sodium pyrophosphate. After a 30-min incubation on ice, aliquots were filtered through 1.2-μm-pore-size glass fiber type C filter multiwell plates (Millipore) and sequentially washed with 10% trichloroacetic acid and ethanol. The extent of radionucleotide incorporation was then determined by liquid scintillation spectrometry. The amount of [3H]dTTP incorporated was plotted as the numbers of cpm versus time, and the specific activities were determined from the slopes of the linear regression analyses. An active unit of RT was defined as the amount of enzyme that incorporated 1 pmol of dTTP in 10 min at 37°C.
NNRTI inhibition of DDDP activity.
The T-P substrates used to study the inhibition of DNA synthesis by NNRTIs (primer ppt17D, template ppt57D) were derived from the polypurine tract (PPT) of the HIV-1 genome. Primer ppt17D was radiolabeled at its 5′ end with [γ-32P]ATP and annealed to the ppt57D template, as described previously (11). The levels of catalysis by recombinant WT and mutant RT enzymes were determined by measuring the percentage of extension of the labeled ppt17D primer on the ppt57D template. Each of the NNRTI compounds to be evaluated, including ETR, DAP, EFV, and NVP, was serially diluted in 50% DMSO. The reaction mixtures contained 150 nM labeled primer-template (calculated according to the primer concentration), 42 to 105 nM recombinant RTs at similar activities of primer extension, 50 mM Tris-HCl (pH 7.8), 5 mM MgC12, 60 mM KCl, 5 mM DTT, and 5% DMSO in a total volume of 20 μl. Initiation of the reaction was performed by adding 100 μM each dATP, dCTP, dTTP, and ddGTP; ddGTP was used so that primer extension would be restricted to 4 nucleotides (nt) for better resolution and convenient quantification. After 15 min at 37°C, an equal volume of formamide sample buffer (96% formamide, 0.05% each bromophenol blue and xylene cyanol, 20 mM EDTA) was added, and the heat-denatured samples were resolved in a 6% polyacrylamide-7 M urea gel, followed by phosphorimaging. The percentage of extension was analyzed with ImageQuant software (GE Healthcare). The 50% inhibitory concentration (IC50) of each NNRTI was determined with GraphPad Prism (version 5.01) software from plots of the percentages of the level of primer extension relative to the logarithm of the inhibitor concentration.
NNRTI inhibition of RDDP activity.
NNRTI inhibition of RDDP activity was performed essentially as reported previously (29, 33, 48). The reaction mixture (50 μl) contained 50 mM Tris-HCl (pH 7.8), 5 mM MgC12, 60 mM KCl, 5 mM DTT, 10 μM dTTP with 2.5 μCi of [3H]dTTP (70 to 80 Ci/mmol; Perkin-Elmer), 5 μg/ml of template-primer poly(rA)-oligo (dT)12-18 (Midland Certified Reagent Company), 10 U of recombinant RTs, and various amounts of the RT inhibitors (ETR, DAP, EFV, and NVP). After incubation at 37°C for 15 min, the reactions were terminated by adding 0.2 ml of 10% cold TCA-20 mM sodium pyrophosphate, and the mixture was incubated for at least 30 min on ice. The precipitated products were filtered onto a 96-well MultiScreen HTS FC filter plate (Millipore). The filter plate was prewet with 150 μl assay buffer prior to use and sequentially washed with 200 μl of 10% TCA and 150 μl of 95% ethanol. The radioactivity of the incorporated products was analyzed by liquid scintillation spectrometry. The IC50s of each NNRTI were determined by nonlinear regression analysis with GraphPad Prism (version 5.01) software.
Efficiency of minus-strand ssDNA synthesis.
Using a cell-free system, the efficiencies of synthesis of minus-strand strong-stop DNA (ssDNA) by WT and M230L mutant enzymes were monitored with human natural tRNA3Lys (Bio S&T, Lachine, Quebec, Canada) in an HIV primer binding site (PBS) RNA primer-template system (4). The PBS RNA was transcribed in vitro from BssHII-linearized pHIV-PBS DNA by using a T7-Megashortscript kit (Ambion, Austin, TX), as described previously (11). The tRNA primer was heat annealed to the RNA template prior to the initiation of DNA synthesis, to ensure complete hybridization. The procedure was performed in a reaction mixture containing 50 mM Tris-HCl (pH 7.8), 50 mM NaCl, 40 nM tRNA3Lys, and 120 nM template PBS RNA. This mixture was incubated for 2 min at 95°C, followed by incubation for 20 min at 70°C and slow cooling to room temperature. The synthesis of minus-strand ssDNA was initiated by the addition of an equal amount of RT in the presence of 6 mM MgCl2 and 10 μM deoxynucleoside triphosphates (dNTPs). Aliquots (4 μl) were removed at different times, and the reactions were stopped in 4 μl of formamide sample buffer (96% formamide, 0.05% each bromophenol blue and xylene cyanol FF, 20 mM EDTA). The reactions were monitored by including 10 μCi of [α-32P]dCTP (3,000 Ci/mmol; Perkin-Elmer) in the reaction mixture. The products were separated on 6% polyacrylamide-7 M urea gels and were exposed to phosphorimager screens after the gels were dried.
RNA- and DNA-dependent polymerase activity.
The processive polymerization by HIV-1 RT on both DNA and RNA templates was carried out by using a heparin enzyme trap to ensure a single round of binding, primer extension, and dissociation. The T-Ps were prepared by annealing 32P-end-labeled 19-mer primer ppt19D or 17-mer primer kim17D to a 3-fold molar excess of DNA (ppt57D) or RNA (kim40R) template prior to the reactions. The RT (10 U) and T-Ps were preincubated for 5 min at 37°C. The reactions were initiated by the addition of 50 μM dNTPs and the heparin trap (final concentration, 2 mg/ml), and the reaction mixture was incubated at 37°C; 2 μl of the reaction mixture was removed and mixed with 8 μl of stop solution (90% formamide, 10 mM EDTA, 0.1% each xylene cyanol and bromophenol blue) at different time points for a total of 15 min. The ability of the heparin trap to limit polymerization on RNA and DNA templates was verified in control reactions in which the enzymes were preincubated with the heparin trap before the addition of the template-primer and dNTPs. The reaction products were analyzed by 6% denaturing polyacrylamide gel electrophoresis and phosphorimaging.
Processivity assays.
The processivities of various recombinant RT proteins were analyzed by using a heparin enzyme trap to ensure a single round of binding, primer extension, and dissociation. The T-Ps were prepared by annealing 32P-end-labeled oligo(dT) (GE Healthcare) to an equimolar concentration of poly(rA) homopolymeric RNA template (GE Healthcare) prior to the reactions. The RT (10 U) and T-Ps were preincubated for 5 min at 37°C. The reactions were initiated by the addition of dTTP and the heparin trap (final concentration, 2 mg/ml), and the reaction mixture was incubated at 37°C for 10 min; 2 μl of the reaction mixture was removed and mixed with 8 μl of stop solution (90% formamide, 10 mM EDTA, 0.1% each xylene cyanol and bromophenol blue). The ability of the heparin trap to limit polymerization on the RNA template was verified in control reactions in which the heparin trap was preincubated with substrate before the addition of RT and dTTP. The reaction products were analyzed by 6% denaturing polyacrylamide gel electrophoresis and phosphorimaging.
RT-catalyzed RNase H activity.
Assays for RNase H activity were performed with 41-mer 5′-labeled 32P-heteropolymeric RNA template kim40R annealed to complementary 32-mer DNA oligomer kim32D at a 1:4 molar ratio, as described previously (18). The reactions were conducted at 37°C in mixtures containing 200 nM RNA-DNA duplex substrate with equal amounts of RT enzymes in assay buffer (50 mM Tris-HCl, pH 7.8, 60 mM KCl, 5 mM MgCl2) in the absence or the presence of the heparin trap (final concentration, 2 mg/ml). Aliquots were removed at different time points after the initiation of the reactions, and the reactions were quenched by use of an equal volume of formamide loading dye. The samples were heated to 90°C for 3 min, cooled on ice, and electrophoresed through 6% polyacrylamide-7 M urea gels. The gels were analyzed by phosphorimaging. The efficacy of the heparin trap was verified by preincubation experiments done by use of a 10-min preincubation of the enzymes with the kim17D-kim40R substrate and various concentrations of the heparin trap, followed by initiation of the reaction with magnesium and dNTPs.
RESULTS
Purification of recombinant HIV-1 RT enzymes.
Recombinant WT heterodimeric RT (p66 and p51 subunits) and the RT enzyme containing M230L were purified by single-step nickel chelate chromatography to >95% homogeneity; the RT p66 and p51 subunits were processed to similar molar ratios, on the basis of SDS-PAGE analysis (data not shown). The M230L mutation introduced into the recombinant HIV-1 RT did not interfere with either heterodimer formation or enzyme purification.
Inhibitory effects of NNRTIs determined by RDDP and DDDP assays.
The inhibitory effects of various NNRTIs on RDDP activity were measured by a filter-based filtration RT assay. The sensitivities of the WT and mutant RTs to each of NVP, EFV, ETR, and DAP were determined. The results show that the mutated RT displayed high-level resistance to all these NNRTIs. The IC50s for each drug tested with each of the RTs are shown in Table 1. The fold change (FC) of the IC50s ranged from approximately 10 for ETR and DAP to 16 for EFV and 45 for NVP.
TABLE 1.
Sensitivities of WT and M230L RTs to NNRTIs
| Polymerase and RT | IC50 (nM)a |
|||
|---|---|---|---|---|
| ETR | DAP | EFV | NVP | |
| DDDP | ||||
| WT | 126 ± 28 | 131 ± 23 | 57 ± 7 | 3.2 × 103 ± 4 × 102 |
| M230L | 1,320 ± 31 | 1,290 ± 32 | 910 ± 26 | 156 × 103 ± 8 × 102 |
| Fold change | 10.5 | 9.9 | 16 | 48.8 |
| RDDP | ||||
| WT | 68 ± 9 | 19 ± 3 | 30 ± 6 | 4.1 × 103 ± 3 × 102 |
| M230L | 696 ± 27 | 172 ± 31 | 740 ± 35 | 185 × 103 ± 13 × 103 |
| Fold change | 10.2 | 9.1 | 24.7 | 45.1 |
Data represent the means ± standard deviations of three separate determinations.
The inhibitory effects of the NNRTIs on DDDP activity were measured by a gel-based primer extension assay (Fig. 1). Figure 1 presents representative gels showing the dose-dependent inhibition of DNA polymerase activity by NNRTIs. The IC50s were determined and are summarized in Table 1. For ETR and DAP, the difference in the IC50s between the WT and the mutant RT was approximately 10-fold. The IC50 of EFV for the mutant RT was increased by 16-fold relative to that for the WT RT. The highest level of resistance was observed with NVP, the IC50 of which was approximately 50-fold greater for the mutant RT than for the WT RT (Table 1). These results suggest that the M230L mutation confers high-level resistance to both first- and second-generation NNRTIs.
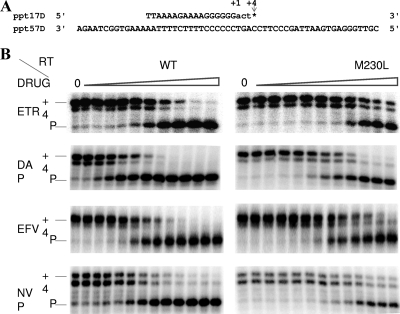
FIG. 1.
Inhibition of DNA-dependent DNA polymerase RT activity by NNRTIs determined by a fixed-time gel-based RT assay. (A) Graphic representation of the primer-template system (ppt17D-ppt57D) used to monitor the inhibition of HIV-1 RT DNA polymerase activity by NNRTIs. The 17-mer DNA primer ppt17D was labeled with 32P at the 5′ terminus and annealed to the 57-mer DNA template ppt57D. +1 and +4, positions of the first and the last nucleotides incorporated, respectively; *, position of the incorporated ddGTP. (B) Dose-dependent inhibition of DNA polymerase activity by NNRTIs. All reactions were resolved by denaturing 6% polyacrylamide gel electrophoresis, visualized by phosphorimaging, and quantified with ImageQuant software (GE Healthcare). The positions of the labeled primer (P) and the full-length extension product (+4) are indicated on the left. The concentrations of the NNRTIs used are as follows: for ETR, 0, 0.6, 1.7, 5, 15.2, 45.6, 137, and 411 nM and 1.23, 3.7, and 11.1 μM; for DAP, 0, 5, 15, 45.7, 137, and 410 nM; 1.23, 3.7, 11.1, 100, and 500 μM; and 1 mM; for EFV, 0, 0.56, 1.69, 5, 15, 45.7, 137, and 410 nM and 1.23, 3.7, 11.1, 33.3, and 100 μM; and for NVP, 0, 10, 31, 95, 285, 857 nM; 2.5, 7.7, 23, 69, 208, and 625 μM; and 1.87 mM.
Inhibitory activities of NNRTIs determined by phenotypic assay in TZM-bl cells.
TZM-bl cells were infected with HIV-1WT and HIV-1M230L in the presence of various concentrations of NNRTIs (ETR, DAP, EFV, and NVP). Relative luciferase activity was used as a measure of inhibition of viral growth, and the EC50s for each inhibitor were calculated (Table 2). The EC50s of ETR, DAP, EFV, and NVP were 1.25 nM, 0.91 nM, 3.57 nM, and 163 nM, respectively, for the WT virus. The HIV-1 variant with the M230L mutation showed reduced sensitivities to each of these NNRTIs, i.e., approximately 6-fold higher EC50s for ETR and DAP, 18-fold higher EC50s for EFV, and 16-fold higher EC50s for NVP. Thus, NVP was the least potent of the NNRTIs tested against both WT virus and viruses containing M230L.
TABLE 2.
EC50 values for WT viruses and viruses containing M230L
| Virus | EC50 (nM)a |
|||
|---|---|---|---|---|
| ETR | DAP | EFV | NVP | |
| WT | 1.25 ± 0.007 | 0.91 ± 0.366 | 3.57 ± 3.25 | 1.63 × 102 ± 70 |
| M230L | 7.27 ± 0.014 | 5.8 ± 2.27 | 64.3 ± 5.29 | 26.9 × 102 ± 4.1 × 102 |
| Fold change | 5.8 | 6.4 | 18 | 16.5 |
Data represent the means ± standard deviations of two independent experiments run in duplicate.
Evaluation of relative replication capacity.
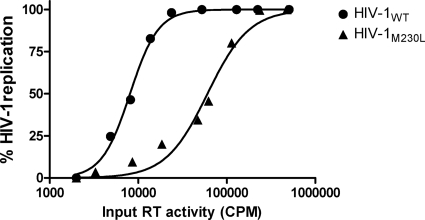
To determine the effect of the M230L mutation on replication capacity, we employed an infectivity assay using TZM-bl cells and serially diluted clonal virus stocks. The relative infectivities of the WT and M230L viruses were determined by measuring the luciferase activity at 48 h postinfection. The results presented in Fig. 2 show that HIV-1M230L had an 8-fold replication disadvantage compared with the replication capacity for HIV-1WT.
FIG. 2.
Effect of the M230L mutation on viral replicative capacity. Virus stocks were prepared through transfection of HEK 293T cells with proviral clones pNL4-3WT and pNL4-3M230L, normalized for RT activity, and used to infect TZM-bl cells to monitor viral replication. Luciferase activity was measured at 48 h postinfection. The relative infectivities of the WT and M230L viruses are shown on the y axis, while the x axis denotes the input virus stocks expressed as RT activity. These values translate to an 8-fold virus replication disadvantage for HIV-1M230L compared with the replication capacity of HIV-1WT.
Effect of the M230L mutation on efficiency of minus-strand ssDNA synthesis from the natural tRNA3Lys primer.
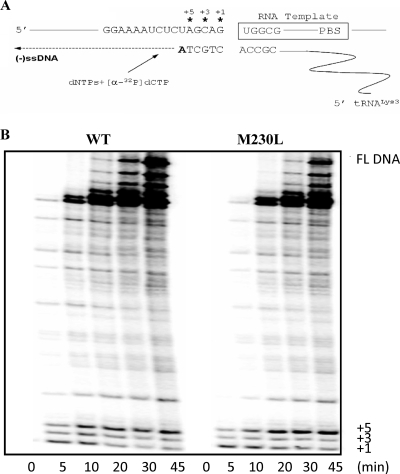
The first step in the reverse transcription of HIV-1 requires tRNA3Lys as the primer, which is annealed to a region near the 5′ end of the viral RNA termed the PBS. This annealing primes the synthesis of minus-strand ssDNA catalyzed by reverse transcriptase, and this step can sometimes be impeded by the presence of drug resistance mutations (11, 13, 42, 44). We therefore investigated whether the M230L mutation affected the efficiency of minus-strand ssDNA synthesis by using HIV-1 PBS RNA template and a tRNA3Lys-specific primer. Reactions involving M230L RT yielded much less product than those performed with WT RT, and the decrease in the level of formation of full-length DNA products was pronounced at all points in these time course experiments (Fig. 3). Furthermore, a pause at the early stages of DNA synthesis, most obviously, a pause at position +5, was significantly increased with the use of the mutated RT. These results suggest that the M230L substitution in HIV-1 RT can lead to diminished levels of tRNA-primed synthesis of minus-strand ssDNA.
FIG. 3.
Efficiency of tRNA3Lys-primed minus-strand ssDNA synthesis in cell-free assays. The efficiencies of synthesis of full-length DNA with WT and M230L mutant RTs were compared in time course experiments. (A) Graphic representation of the cell-free system used for the synthesis of full-length minus-strand ssDNA. The PBS RNA template used in this system consists of 258 nt at the 5′ end of the HIV-1 genome, which contains the R, U5, and PBS regions. (B) Synthesis of full-length (FL) DNA by recombinant WT and mutant enzymes monitored in time course experiments. The reactions were initiated by the addition of 10 μM dNTPs and were monitored by measurement of the incorporation of [α-32P]dCMP into the extending DNA strand. The reactions were stopped at different time points during a period of 45 min. Time point zero indicates samples taken at 20 s after initiation. The full-length DNA product and the +1, +3, and +5 pausing sites are shown on the right.
Decreased rate of processive polymerization by RT containing M230L in regard to both RNA and DNA templates.
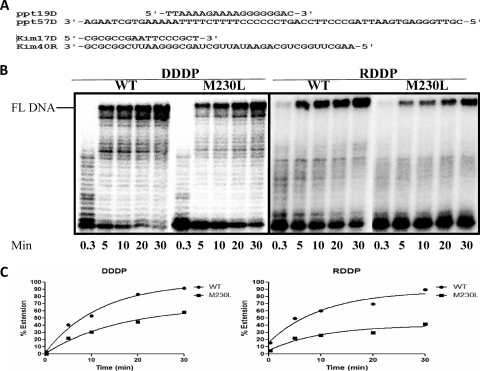
To investigate the effect of M230L on the rates of processive polymerization, we tested both WT and M230L RTs by using heteropolymeric DNA (57-mer) and an RNA template (40-mer) annealed to 5′ P32-labeled oligodeoxynucleotide primers (19-mer and 17-mer, respectively) in a time course experiment in the presence of a heparin trap to ensure that each DNA molecule synthesized resulted from a single processive cycle. Figure 4 shows that the M230L mutation caused decreased polymerization in both the DDDP and the RDDP assays; thus, RT containing M230L is less active than the WT enzyme.
FIG. 4.
Processive polymerization by WT and M230L mutant RTs. (A) Graphic representation of the T-P systems used to monitor the DDDP and RDDP activities of recombinant RTs. DNA primers ppt19D and kim17D were labeled with 32P and annealed to DNA template ppt57D and RNA template kim40R, respectively. (B) Polymerization activities were analyzed by monitoring the percentage of full-length (FL) DNA synthesis in time course experiments in the presence of the heparin trap. The position of full-length DNA is indicated on the left side of the panel. All reactions were resolved by denaturing 6% polyacrylamide gel electrophoresis, visualized by phosphorimaging, and quantified with ImageQuant software (GE Healthcare). (C) Graphic representation of the calculated percentage of full-length DNA synthesis in the time course experiments from the gel results. Band intensities were calculated by ImageQuant software, and the data were curve fit by GraphPad Prism (version 5.01) software.
Processivity defect of RT containing the M230L mutation.
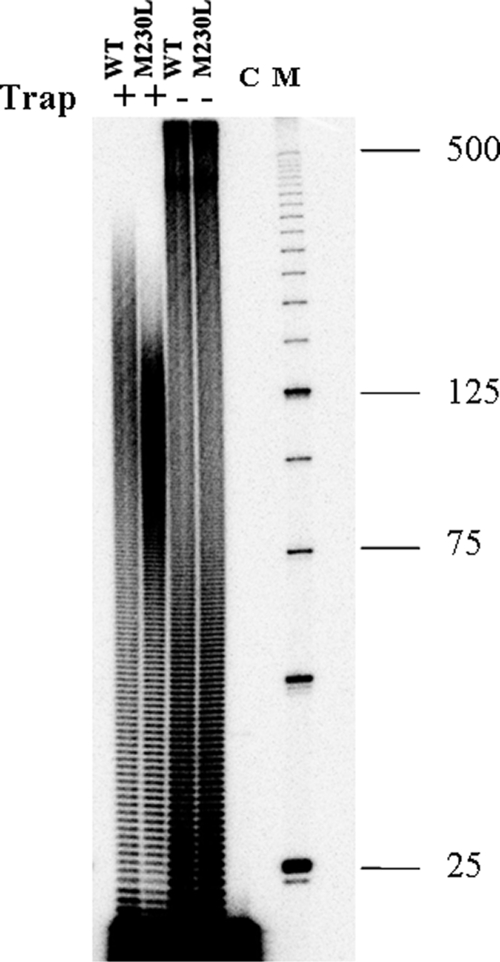
The processivity of a polymerase is defined as the number of nucleotides incorporated in a round of binding, elongation, and dissociation. Earlier studies have shown that HIV replication efficiency is related to RT processivity (3, 8), and some drug resistance mutations, such as M184V, has been shown to decrease RT processivity in in vitro processivity assays (6, 7, 41). To investigate the effect of M230L on enzyme processivity, we tested WT and M230L RTs by using a homopolymeric poly(rA) RNA template (average length, 500 nt) annealed to 5′ 32P-labeled oligo(dT) primers in a fixed-time experiment in the presence of a heparin trap to ensure that each synthesized DNA molecule resulted from a single processive cycle. Figure 5 shows that the M230L mutation caused decreased processivity within a size range of the longest products at 75 nt to 150 nt, whereas the corresponding product lengths for the WT enzyme were 100 nt to 225 nt. Thus, the RT containing M230L is less processive than the wild-type enzyme, consistent with the diminished replication capacity of viruses containing this mutation.
FIG. 5.
Reduced polymerase processivity of RT containing the M230L mutation. The processivities of the recombinant WT and M230L RT proteins were assessed with a homopolymeric RNA template poly(rA)-oligo (dT)12-18 DNA primer. The DNA primer was labeled with 32P at the 5′ terminus and was annealed to the poly(rA) RNA template at an equimolar ratio. Processivities were analyzed by monitoring the size distribution of the DNA products in fixed-time experiments in the presence of a heparin trap. Parallel reactions were run in the absence of the heparin trap to ensure that similar amounts of enzyme activities were present in the reactions. Lane C, control reaction to verify the efficiency of the heparin trap by preincubation with substrate prior to addition of RT; lane M, a 25-bp DNA ladder used as a size standard. The sizes (in kilobases) of some fragments of the standard are indicated on the right. All reaction products were resolved by denaturing 6% polyacrylamide gel electrophoresis and were visualized by phosphorimaging.
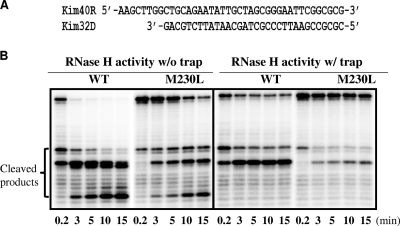
RNase H activity.
Previous studies have shown that substitutions at a number of residues in or adjacent to the primer grip region impair RNase H activity and contribute to reductions in HIV-1 replication fitness (15, 31). We performed an RNase H time course analysis in the absence or the presence of a heparin trap by using a recessed 32-mer DNA primer hybridized to a 5′-end-labeled 40-mer RNA to monitor 3′-DNA-directed RNase H activity (Fig. 6). The presence of the heparin trap permits the analysis of cleaved products from a single event of binding of RT to the substrate. Under both conditions, the RNase H activity of the M230L mutant RT was lower than that of the WT RT (Fig. 6). We also performed RNase H cleavage analysis in the absence and the presence of a heparin trap using a 75-mer DNA oligonucleotide (75D) annealed to 5′-end-labeled RNA substrate kim40R to monitor 5′ RNA-directed RNase H activity. The results demonstrated that M230L RT results in a decreased cleavage efficiency compared to that of WT RT, as observed in the 3′-DNA-directed RNase H activity assay (data not shown). Thus, the M230L mutation also impairs RNase H activity.
FIG. 6.
RNase H activities of WT and M230L recombinant RTs. (A) Graphic representation of the substrate RNA-DNA (kim40R-kim32D) duplex used to monitor the cleavage efficiency of the M230L and WT RTs. The 40-mer RNA kim40R was labeled at its 5′ terminus with 32P and annealed to the 32-mer DNA oligonucleotide kim32D. (B) RNase H activities were analyzed by monitoring substrate cleavage in time course experiments in the absence (left panel) or the presence (right panel) of a heparin trap. The positions of the cleaved products are indicated on the left side. All reactions were resolved by denaturing 6% polyacrylamide gel electrophoresis.
DISCUSSION
The present study was designed to determine the impact of the M230L mutation in the HIV RT on the antiviral activity of the second-generation NNRTIs ETR and DAP, as well as the first-generation NNRTIs EFV and NVP. M230L was first identified by in vitro passage of HIV in the presence of the NNRTI delavirdine (DLV) (24, 30, 34). This mutation has also been detected in clinical isolates from patients and has been shown to reduce the levels of susceptibility to each of NVP, DLV, and EFV by 23- to 58-fold (22).
M230 is located within the highly conserved primer grip region of RT, and this position is highly conserved among all HIV isolates. Therefore, the effects of the M230L mutation on virus replication and on the underlying properties of RT should not depend on the particular RT or virus strain that is used in the assays. In particular, defects in replication capacity associated with M230L have previously been documented by other groups (23) and confirmed here. Moreover, a previous paper by our group has revealed the absence of important differences in enzymatic function between the RT enzymes of HIV-1 subtype B and HIV-1 subtype C (47).
Recently, the DUET-1 and DUET-2 clinical trials have identified an array of 17 resistance-associated mutations (RAMs) that confer diminished sensitivity to the second-generation NNRTI ETR; these mutations include V90I, A98G, L100I, K101E/H/P, V106I, E138A, V179D/F/T, Y181C/I/V, G190A/S, and M230L (38). Our data confirm that M230L confers resistance to all currently available NNRTIs in both phenotypic and biochemical assays. In the former, recombinant viruses harboring M230L displayed similarly moderate levels (5- to 6-fold) of resistance to ETR and DAP and higher-level (>10-fold) resistance to EFV and NVP. The data that we obtained with ETR are consistent with those presented in previous reports, which showed that M230L-containing viruses were ≈3-fold more resistant to ETR than WT RT-containing viruses (38, 39). However, a different study reported that M230L was not associated with resistance to either ETR or EFV (36). Our RDDP and DDDP enzyme assays further confirm that M230L causes similar ≈10-fold higher levels of resistance to each of the second-generation NNRTIs ETR and DAP and over 16-fold higher levels of resistance to EFV and NVP. Interestingly, a recent study showed that a newer NNRTI termed rilpivirine (formerly TMC278) was not affected by the presence of most single NNRTI RAMs, including those at positions 100, 103, 106, 138, 179, 188, 190, 221, 236, and 230 (5).
The prevalence of the M230L mutation in clinical isolates obtained from patients failing NNRTI-based therapy is low (2, 28). The emergence of resistance mutations can also depend on the effect of such mutations on viral replicative fitness (10), with mutations that confer high-level resistance with minimal effects on enzyme activity theoretically developing first. The use of a TZM-bl cell infectivity assay has confirmed that M230L-containing virus replicates less efficiently than WT virus, in agreement with the findings presented previously (23). The impaired replication capacity of M230L-containing virus might explain the rarity of this mutation in clinical isolates. However, we cannot exclude the possibility that hypersusceptibility to nucleoside and/or nucleotide RT inhibitors might result from the presence of the M230L mutation and that resensitization to NNRTIs might also take place in the presence of other NRTI resistance mutations, including the thymidine analogue mutations (TAMs) K65R and M184V. As a consequence, hypersusceptibility/resensitization effects might contribute to the rarity of M230L in clinical samples. We are investigating the interaction of the M230L mutation with other NRTI resistance mutations. It remains to be determined whether the emergence of this mutation may require other compensatory mutations that might enhance NNRTI resistance and restore viral replicative fitness.
Our biochemical characterization has shown that M230L severely impaired each of minus-strand ssDNA synthesis, both DNA- and RNA-dependent polymerase activity, processivity, and RNase H activity. Previous studies by us and others have documented that the efficiency of each of these reactions can be correlated with viral replication efficiency (11-13, 42, 44, 47).
M230 is located at the tip of the β12-β13 hairpin comprising amino acids 227 to 235 (FLWMGYELH), which defines the so-called primer grip, a highly conserved motif of retroviral RT. The primer grip is involved in maintenance of the primer terminus in an orientation appropriate for nucleophilic attack on an incoming dNTP (25). Mutational analysis of primer grip residues has shown their influence on various RT functions, including dNTP binding (46), polypurine tract removal (32), RNase H activity (31), template-primer utilization (16, 25), and the fidelity of DNA synthesis (9, 19, 45). The biochemical data presented here show that the described impairment in enzyme function associated with M230L contributes to reduced viral replication.
Further research is needed to determine how the M230L mutation might affect the binding kinetics of NNRTIs. We are performing a biochemical investigation of the interplay of M230L with other NNRTI and/or NRTI resistance mutations. We are also studying whether other resistance-related mutations that may appear in association with M230L may be able to help restore viral replication capacity while also potentially increasing the overall levels of drug resistance.
Acknowledgments
We thank Stuart Le Grice for providing the pRT6H-PROT DNA construct and Daniela Moisi for technical assistance with the DNA sequencing reactions.
This research was supported by grants from the Canadian Institutes of Health Research (CIHR), the International Partnership for Microbicides, and Tibotec Inc.
The authors' contributions are as follows. M.A.W. provided resources for the research, supervised the project, and corrected the manuscript. H.-T.X. and Y.Q. performed the biochemical experiments and drafted the manuscript. S.M.S. and M.O. performed the phenotypic analyses. T.B.-M. performed some of the site-directed DNA mutagenesis work.
Footnotes
Published ahead of print on 22 March 2010.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antela, A., J. Casado, A. Moreno, F. Dronda, M. Perez-Elias, and S. Moreno. 2001. Clinical relevance of the m230l mutation in the reverse transcriptase gene in patients rescued with a regimen including NNRTIs, abstr. 571. 1st IAS Conf. HIV Pathog.
- 3.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 4.Arts, E. J., X. Li, Z. Gu, L. Kleiman, M. A. Parniak, and M. A. Wainberg. 1994. Comparison of deoxyoligonucleotide and tRNA(Lys-3) as primers in an endogenous human immunodeficiency virus-1 in vitro reverse transcription/template-switching reaction. J. Biol. Chem. 269:14672-14680. [PubMed] [Google Scholar]
- 5.Azijn, H., I. Tirry, J. Vingerhoets, M. P. de Bethune, G. Kraus, K. Boven, D. Jochmans, E. Van Craenenbroeck, G. Picchio, and L. T. Rimsky. 2010. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob. Agents Chemother. 54:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Back, N. K., M. Nijhuis, W. Keulen, C. A. Boucher, B. O. Oude Essink, A. B. van Kuilenburg, A. H. van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer, P. L., and S. H. Hughes. 1995. Analysis of mutations at position 184 in reverse transcriptase of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 39:1624-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caliendo, A. M., A. Savara, D. An, K. DeVore, J. C. Kaplan, and R. T. D'Aquila. 1996. Effects of zidovudine-selected human immunodeficiency virus type 1 reverse transcriptase amino acid substitutions on processive DNA synthesis and viral replication. J. Virol. 70:2146-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cases-Gonzalez, C. E., and L. Menendez-Arias. 2004. Increased G→A transition frequencies displayed by primer grip mutants of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 78:1012-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deeks, S. G. 2001. International perspectives on antiretroviral resistance. Nonnucleoside reverse transcriptase inhibitor resistance. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S25-S33. [DOI] [PubMed] [Google Scholar]
- 11.Diallo, K., B. Marchand, X. Wei, L. Cellai, M. Gotte, and M. A. Wainberg. 2003. Diminished RNA primer usage associated with the L74V and M184V mutations in the reverse transcriptase of human immunodeficiency virus type 1 provides a possible mechanism for diminished viral replication capacity. J. Virol. 77:8621-8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dykes, C., and L. M. Demeter. 2007. Clinical significance of human immunodeficiency virus type 1 replication fitness. Clin. Microbiol. Rev. 20:550-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frankel, F. A., C. F. Invernizzi, M. Oliveira, and M. A. Wainberg. 2007. Diminished efficiency of HIV-1 reverse transcriptase containing the K65R and M184V drug resistance mutations. AIDS 21:665-675. [DOI] [PubMed] [Google Scholar]
- 14.Freed, E. O., and M. A. Martin. 1994. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J. Virol. 68:2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerondelis, P., R. H. Archer, C. Palaniappan, R. C. Reichman, P. J. Fay, R. A. Bambara, and L. M. Demeter. 1999. The P236L delavirdine-resistant human immunodeficiency virus type 1 mutant is replication defective and demonstrates alterations in both RNA 5′-end- and DNA 3′-end-directed RNase H activities. J. Virol. 73:5803-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh, M., P. S. Jacques, D. W. Rodgers, M. Ottman, J. L. Darlix, and S. F. Le Grice. 1996. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry 35:8553-8562. [DOI] [PubMed] [Google Scholar]
- 17.Goff, S., P. Traktman, and D. Baltimore. 1981. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J. Virol. 38:239-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopalakrishnan, V., J. A. Peliska, and S. J. Benkovic. 1992. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc. Natl. Acad. Sci. U. S. A. 89:10763-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutierrez-Rivas, M., and L. Menendez-Arias. 2001. A mutation in the primer grip region of HIV-1 reverse transcriptase that confers reduced fidelity of DNA synthesis. Nucleic Acids Res. 29:4963-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirschel, B., and P. Francioli. 1998. Progress and problems in the fight against AIDS. N. Engl. J. Med. 338:906-908. [DOI] [PubMed] [Google Scholar]
- 21.Huang, W., A. Gamarnik, K. Limoli, C. J. Petropoulos, and J. M. Whitcomb. 2003. Amino acid substitutions at position 190 of human immunodeficiency virus type 1 reverse transcriptase increase susceptibility to delavirdine and impair virus replication. J. Virol. 77:1512-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, W., N. T. Parkin, Y. S. Lie, T. Wrin, R. Haubrich, S. Deeks, N. Hellman, C. J. Petropoulos, and J. M. Whitcomb. 2000. A novel HIV-1 RT mutation (M230L) confers NNRTI resistance and dose-dependent stimulation of replication. Antivir. Ther. 5(Suppl. 3):24-25. [Google Scholar]
- 23.Huang, W., T. Wrin, A. Gamarnik, J. Beauchaine, J. M. Whitcomb, and C. J. Petropoulos. 2002. Reverse transcriptase mutations that confer nonnucleoside reverse transcriptase inhibitor resistance may also impair replication capacity. Antivir. Ther. 7(Suppl. 1):S60. [Google Scholar]
- 24.Isaka, Y., S. Miki, S. Kawauchi, A. Suyama, H. Sugimoto, A. Adachi, T. Miura, M. Hayami, O. Yoshie, T. Fujiwara, and A. Sato. 2001. A single amino acid change at Leu-188 in the reverse transcriptase of HIV-2 and SIV renders them sensitive to non-nucleoside reverse transcriptase inhibitors. Arch. Virol. 146:743-755. [DOI] [PubMed] [Google Scholar]
- 25.Jacques, P. S., B. M. Wohrl, M. Ottmann, J. L. Darlix, and S. F. Le Grice. 1994. Mutating the “primer grip” of p66 HIV-1 reverse transcriptase implicates tryptophan-229 in template-primer utilization. J. Biol. Chem. 269:26472-26478. [PubMed] [Google Scholar]
- 26.Johnson, L. B., and L. D. Saravolatz. 2009. Etravirine, a next-generation nonnucleoside reverse-transcriptase inhibitor. Clin. Infect. Dis. 48:1123-1128. [DOI] [PubMed] [Google Scholar]
- 27.Le Grice, S. F., and F. Gruninger-Leitch. 1990. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur. J. Biochem. 187:307-314. [DOI] [PubMed] [Google Scholar]
- 28.Llibre, J. M., J. R. Santos, T. Puig, J. Molto, L. Ruiz, R. Paredes, and B. Clotet. 2008. Prevalence of etravirine-associated mutations in clinical samples with resistance to nevirapine and efavirenz. J. Antimicrob. Chemother. 62:909-913. [DOI] [PubMed] [Google Scholar]
- 29.Munshi, V., M. Lu, P. Felock, R. J. Barnard, D. J. Hazuda, M. D. Miller, and M. T. Lai. 2008. Monitoring the development of non-nucleoside reverse transcriptase inhibitor-associated resistant HIV-1 using an electrochemiluminescence-based reverse transcriptase polymerase assay. Anal. Biochem. 374:121-132. [DOI] [PubMed] [Google Scholar]
- 30.Olmsted, R. A., D. E. Slade, L. A. Kopta, S. M. Poppe, T. J. Poel, S. W. Newport, K. B. Rank, C. Biles, R. A. Morge, T. J. Dueweke, Y. Yagi, D. L. Romero, R. C. Thomas, S. K. Sharma, and W. G. Tarpley. 1996. (Alkylamino)piperidine bis(heteroaryl)piperizine analogs are potent, broad-spectrum nonnucleoside reverse transcriptase inhibitors of drug-resistant isolates of human immunodeficiency virus type 1 (HIV-1) and select for drug-resistant variants of HIV-1IIIB with reduced replication phenotypes. J. Virol. 70:3698-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palaniappan, C., M. Wisniewski, P. S. Jacques, S. F. Le Grice, P. J. Fay, and R. A. Bambara. 1997. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J. Biol. Chem. 272:11157-11164. [DOI] [PubMed] [Google Scholar]
- 32.Powell, M. D., M. Ghosh, P. S. Jacques, K. J. Howard, S. F. Le Grice, and J. G. Levin. 1997. Alanine-scanning mutations in the “primer grip” of p66 HIV-1 reverse transcriptase result in selective loss of RNA priming activity. J. Biol. Chem. 272:13262-13269. [DOI] [PubMed] [Google Scholar]
- 33.Schiller, D. S., and M. Youssef-Bessler. 2009. Etravirine: a second-generation nonnucleoside reverse transcriptase inhibitor (NNRTI) active against NNRTI-resistant strains of HIV. Clin. Ther. 31:692-704. [DOI] [PubMed] [Google Scholar]
- 34.Slade, D. E., T. J. Dueweke, S. M. Poppe, S. M. Swaney, S. M. Wisniewski, V. Sharova, M. Stevenson, and W. G. Tarpley. 1995. HIV-1 acquires resistance to AZT and delavirdine in vitro by multiple RT substitutions, abstr. P-89. Abstr. 2nd Natl. Conf. Hum. Retrovir. Relat. Infect.
- 35.Smerdon, S. J., J. Jager, J. Wang, L. A. Kohlstaedt, A. J. Chirino, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1994. Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 91:3911-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su, G., J. Yan, Y. Li, A. Paul, J. Hang, S. Harris, H. Hogg, J. Dunn, C. N. K. Klumpp, and G. Heilek. 2007. In vitro selection and characterization of viruses resistant to R1206 a novel NNRTI. Antivir. Ther. 12(Suppl. 1):S35. [Google Scholar]
- 37.Telesnitsky, A., and S. P. Goff. 1998. Reverse transcriptase and the generation of retroviral DNA, p. 121-160. In J. Coffin, H. Varmus, and S. Hughes (ed.), Retroviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [PubMed]
- 38.Vingerhoets, J., L. Tambuyzer, H. Azijn, A. Hoogstoel, S. Nijs, M. Peeters, M. P. de Bethune, G. De Smedt, B. Woodfall, and G. Picchio. 2010. Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled phase III clinical studies. AIDS 24:503-514. [DOI] [PubMed] [Google Scholar]
- 39.Vingerhoets, J., H. Azijn, E. Fransen, I. De Baere, L. Smeulders, D. Jochmans, K. Andries, R. Pauwels, and M. P. de Bethune. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J. Virol. 79:12773-12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waheed, A. A., S. D. Ablan, M. K. Mankowski, J. E. Cummins, R. G. Ptak, C. P. Schaffner, and E. O. Freed. 2006. Inhibition of HIV-1 replication by amphotericin B methyl ester: selection for resistant variants. J. Biol. Chem. 281:28699-28711. [DOI] [PubMed] [Google Scholar]
- 41.Wainberg, M. A. 2004. The impact of the M184V substitution on drug resistance and viral fitness. Expert Rev. Anti Infect. Ther. 2:147-151. [DOI] [PubMed] [Google Scholar]
- 42.Wang, J., C. Dykes, R. A. Domaoal, C. E. Koval, R. A. Bambara, and L. M. Demeter. 2006. The HIV-1 reverse transcriptase mutants G190S and G190A, which confer resistance to non-nucleoside reverse transcriptase inhibitors, demonstrate reductions in RNase H activity and DNA synthesis from tRNA(Lys, 3) that correlate with reductions in replication efficiency. Virology 348:462-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei, X., C. Liang, M. Gotte, and M. A. Wainberg. 2002. The M184V mutation in HIV-1 reverse transcriptase reduces the restoration of wild-type replication by attenuated viruses. AIDS 16:2391-2398. [DOI] [PubMed] [Google Scholar]
- 45.Wisniewski, M., C. Palaniappan, Z. Fu, S. F. Le Grice, P. Fay, and R. A. Bambara. 1999. Mutations in the primer grip region of HIV reverse transcriptase can increase replication fidelity. J. Biol. Chem. 274:28175-28184. [DOI] [PubMed] [Google Scholar]
- 46.Wohrl, B. M., R. Krebs, S. H. Thrall, S. F. Le Grice, A. J. Scheidig, and R. S. Goody. 1997. Kinetic analysis of four HIV-1 reverse transcriptase enzymes mutated in the primer grip region of p66. Implications for DNA synthesis and dimerization. J. Biol. Chem. 272:17581-17587. [DOI] [PubMed] [Google Scholar]
- 47.Xu, H. T., J. L. Martinez-Cajas, M. L. Ntemgwa, D. Coutsinos, F. A. Frankel, B. G. Brenner, and M. A. Wainberg. 2009. Effects of the K65R and K65R/M184V reverse transcriptase mutations in subtype C HIV on enzyme function and drug resistance. Retrovirology 6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, Z., M. Walker, W. Xu, J. H. Shim, J. L. Girardet, R. K. Hamatake, and Z. Hong. 2006. Novel nonnucleoside inhibitors that select nucleoside inhibitor resistance mutations in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 50:2772-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]