Abstract
We analyzed 71 clinical and environmental Cryptococcus gattii strains that had been isolated before or after the advent of azole antifungals to determine their level of heteroresistance to fluconazole (LHF). All strains of C. gattii manifested heteroresistance, with LHFs that ranged between 4 μg/ml and 32 μg/ml. A considerably higher proportion of the C. gattii strains (86%) than Cryptococcus neoformans strains (46%) exhibited LHFs that were ≥16 μg/ml. No significant correlation was observed between the molecular type or serotypes of strains and their respective LHF. The strains which expressed a higher LHF were also more resistant to xenobiotics than the strains with a low LHF, and the level of resistance to xenobiotics was significantly higher than that reported for C. neoformans. The heteroresistant subpopulation, whose level of drug resistance had been raised in a stepwise manner to 64 μg/ml, reverted to the original LHF upon daily transfers in drug-free medium. Importantly, the strains with high LHFs were significantly more virulent than those with low LHFs. Since all the clinical isolates that had not been exposed to azole drugs as well as the environmental strains manifested heteroresistance to fluconazole, heteroresistance of C. gattii to azoles is an intrinsic mechanism as in C. neoformans and is associated with the strain's virulence.
Cryptococcosis is caused by two species, Cryptococcus neoformans and Cryptococcus gattii (15); fluconazole (FLC), a triazole, is widely used for the treatment of cryptococcosis regardless of the causative species (29). Fluconazole-resistant strains of C. neoformans have been increasingly reported from cases of therapy failure in AIDS patients undergoing long-term maintenance therapy (1-3, 27, 36). In 1999, we reported heteroresistance to fluconazole, an adaptive mode of drug resistance, in strains of C. neoformans recovered from two patients. One patient had AIDS and suffered from a series of recurrent cryptococcosis episodes during azole maintenance therapy, while the other was a non-AIDS patient who had never been treated with azole drugs (26). Our recent expanded characterization of heteroresistance using over 100 strains of C. neoformans that had been isolated at least a decade prior to the advent of azole antifungals revealed that C. neoformans is intrinsically heteroresistant to fluconazole. The innate level of heteroresistance to fluconazole (LHF) ranged from 4 μg/ml to 64 μg/ml, and 50% of serotype A strains and 21% of serotype D strains manifested high LHFs (≥16 μg/ml). Importantly, the LHF was observed to correlate with both the virulence of C. neoformans strains in mice and resistance to xenobiotics unrelated to fluconazole (31).
Strains of C. gattii infect immunocompetent patients more often than C. neoformans strains (25), and animal studies have revealed that some strains of C. gattii are more virulent than strain H99, a well-characterized highly virulent C. neoformans strain of serotype A (10). Although the clinical manifestations of the disease caused by C. neoformans and C. gattii are not significantly different, the two species can readily be differentiated by their biochemical, genetic, ecological, and epidemiological characteristics (14). In light of the fact that strains of C. gattii infect immunocompetent individuals more often than C. neoformans strains and the two species are biologically and genetically distinguishable, we investigated the LHFs of C. gattii strains to compare them with those of the C. neoformans strains.
In this study, we have analyzed the LHFs of 71 clinical and environmental strains of C. gattii, which include serotypes B and C and are comprised of four major molecular types, VGII, VGIII, VGI, and VGIV. The strains were collected from different geographical areas, and all serotype B and the majority of serotype C clinical strains had been isolated at least 10 years prior to the availability of triazole antifungals for therapy of mycosis. As with C. neoformans, all strains of C. gattii manifested heteroresistance regardless of their chronology or source of isolation. Interestingly, a considerably higher percentage of the C. gattii strains (86%) expressed LHFs of ≥16 μg/ml than that for C. neoformans (46%) (27). The strains with high LHFs were more virulent in mice and more resistant to xenobiotics that are unrelated to fluconazole, as reported for C. neoformans. However, the degree of resistance to xenobiotics in the C. gattii strains with high LHFs was significantly higher than that for the strains of C. neoformans with high LHFs.
MATERIALS AND METHODS
Cryptococcus gattii strains.
A total of 71 clinical and environmental C. gattii strains of serotypes B and C that had been previously isolated from different geographic areas were included in this study (Table 1). All strains were purified by isolating a single colony, and their identity as C. gattii was confirmed according to routine diagnostic tests that included serotyping (Iatron Crypto serotyping kit), melanin formation, urease activity, growth at 37°C, and growth on l-canavanine-glycine-bromothymol blue medium (14). A set of standard strains for the molecular typing of C. gattii species were used as a reference: WM 179 (serotype B, VGI), WM 178 (serotype B, VGII), WM 161 (serotype B, VGIII), and WM 779 (serotype C, VGIV) (24). The isolates were stored in glycerol (25%) at −80°C until use and were maintained on YPD (2% glucose, 1% yeast extract, 2% peptone) agar at 25°C during this study.
TABLE 1.
Clinical and environmental Cryptococcus gattii strains of serotypes B and C used in this study
| Strain | Date of isolation | VG type | Geographic area | Origin | Reference or source | LHF (μg/ml) |
|---|---|---|---|---|---|---|
| Serotype B, clinical (n = 25) | ||||||
| NIH76 | 1965 | III | USA | Patient | This study | 32 |
| NIH179 | <1970a | III | USA | Patient | This study | 16 |
| NIH184 | 1965 | III | USA | Patient | This study | 16 |
| NIH189 | 1970 | III | USA | Patient | This study | 16 |
| NIH190 | 1970 | III | USA | Patient | This study | 16 |
| NIH198 | 1970 | III | USA | Patient | This study | 16 |
| NIH200 | 1970 | II | USA | Patient | This study | 4 |
| NIH254 | 1970 | I | USA | Patient | This study | 32 |
| NIH281 | 1970 | II | USA | Patient | This study | 4 |
| NIH286 | 1968 | I | USA | Patient | This study | 32 |
| NIH367 | 1970 | II | USA | Patient | This study | 16 |
| NIH408 | 1970 | I | USA | Patient | This study | 16 |
| NIH409 | 1970 | II | USA | Patient | This study | 32 |
| NIH412 | 1970 | II | USA | Patient | This study | 4 |
| NIH435 | 1971 | I | USA | Patient | This study | 16 |
| NIH444 | 1972 | II | USA | Patient | 14a | 32 |
| NIH487 | 1970 | I | USA | Patient | This study | 32 |
| NIH535 | 1972 | IV | USA | Patient | This study | 4 |
| NIH653 | <1975 | I | USA | Patient | This study | 32 |
| NIH744 | 1975 | III | USA | Patient | This study | 32 |
| NIH754 | 1976 | II | USA | Patient | This study | 4 |
| NIH767 | 1975 | II | USA | Patient | This study | 32 |
| NIH771 | 1976 | III | USA | Patient | This study | 32 |
| B3939 | <1970 | I | Africa | Patient | 15 | 16 |
| NIH1123 | <1970 | I | USA | Patient | This study | 32 |
| Serotype B, environmental (n = 25) | ||||||
| RB-1 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 32 |
| RB-3 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 8 |
| RB-4 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 32 |
| RB-5 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 32 |
| RB-9 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 32 |
| RB-11 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 32 |
| RB-13 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 32 |
| RB-14 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 32 |
| RB-15 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 32 |
| RB-17 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 32 |
| RB-18 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 32 |
| RB-22 | 1999-2002 | II | Vancouver, Canada | Alder | 13 | 16 |
| RB-26 | 1999-2002 | II | Vancouver, Canada | Alder | 13 | 32 |
| RB-33 | 1999-2002 | II | Vancouver, Canada | Douglas Fir | 13 | 32 |
| RB-34 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 32 |
| RB-35 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 32 |
| RB-8 | 1999-2002 | II | Vancouver, Canada | Douglas fir | 13 | 32 |
| RAM-002 | II | Australia | 13 | 32 | ||
| RAM-005 | II | Australia | 13 | 32 | ||
| RAM-15 | II | Australia | W. Meyer | 32 | ||
| VPB571-058 | II | Australia | W. Meyer | 32 | ||
| B4506 | 1989 | III | Borosa Valley, Australia | Eucalyptus | 17 | 8 |
| B5763 | I | India | Eucalyptus | 5 | 8 | |
| B5765 | I | India | Eucalyptus | 5 | 16 | |
| B5788 | I | India | Eucalyptus | 5 | 8 | |
| Serotype C, clinical (n = 17) | ||||||
| NIH113 | <1966 | I | USA | Patient | This study | 32 |
| NIH139 | <1966 | I | USA | Patient | This study | 32 |
| NIH178 | <1966 | I | USA | Patient | This study | 32 |
| NIH187 | <1966 | I | USA | Patient | This study | 32 |
| NIH191 | <1970 | I | USA | Patient | This study | 32 |
| NIH257 | <1970 | III | USA | Patient | This study | 4 |
| NIH298 | <1970 | III | USA | Patient | This study | 32 |
| NIH312 | <1970 | III | USA | Patient | This study | 32 |
| NIH401 | <1970 | III | USA | Patient | This study | 32 |
| NIH403 | <1970 | I | USA | Patient | This study | 16 |
| NIH642 | 1973 | III | USA | Patient | This study | 32 |
| NIH18 | 1971 | I | USA | Patient | This study | 32 |
| NIH34 | 1962 | I | USA | Patient | This study | 16 |
| NIH917 | 1977 | I | USA | Patient | This study | 32 |
| H0058-1-78 | 1989 | III | Arauca, Colombia | Patient | E. Castaneda | 32 |
| H0058-1-2023 | 2003 | III | N. Santander, Colombia | Patient | E. Castaneda | 32 |
| H0058-1-2086 | 2004 | III | Caldas, Colombia | Patient | E. Castaneda | 32 |
| Serotype C, environmental (n = 4) | ||||||
| H0058-1-818 | 1998 | III | Cacuta, Colombia | Almond | E. Castaneda | 32 |
| H0058-1-1686 | 2003 | IV | El Colegio, Colombia | Almond | E. Castaneda | 32 |
| H0058-1-1875 | 2003 | II | Bogota, Colombia | Eucalyptus | E. Castaneda | 32 |
| H0058-1-1941 | 2003 | II | Santa Martha, Colombia | Almond | E. Castaneda | 32 |
<, strain was isolated before the year indicated.
Genotyping of strains.
Yeast cultures were grown overnight in YPD broth at 30°C, and the genomic DNA was extracted as previously described (35). The molecular genotype of the strains was determined by PCR analysis of the microsatellite DNA fingerprint patterns generated by the M13 primer and the restriction fragment length polymorphisms (RFLPs) of the URA5 gene sequence. PCR fingerprints using the microsatellite specific primer M13 were generated as described previously (24). Amplification products of the reaction were analyzed by electrophoresis on 1.4% agarose gels stained with ethidium bromide (10-mg/ml stock) and visualized under UV light. Based on the major bands in the patterns of the C. gattii standard reference strains, the strains were assigned to molecular types VGI to VGIV. To confirm the molecular type determined by M13 fingerprint patterns, the URA5 gene sequence was amplified by PCR and digested with enzymes HhaI and Sau96I prior to analysis of RFLPs (6, 24).
Analysis of the level of heteroresistance to fluconazole (LHF).
FLC powder was provided by Pfizer Global Research & Development (Groton, CT). Stock solutions were prepared in the solvent dimethyl sulfoxide (Sigma) at a concentration of 50 mg/ml. Etests (AB BIODISK, Solna, Sweden) were performed to determine the level of fluconazole susceptibility using YPD agar (31). The LHF was determined for all strains by spot testing on YPD agar supplemented with different concentrations of FLC (4 to 128 μg/ml). The lowest concentration of the drug at which minor resistant subpopulations emerged was identified as each strain's LHF. To determine the proportion of each strain's population that could tolerate different concentrations of FLC, representative strains from each LHF category were tested (Table 2). Briefly, cell suspensions (1 × 103 to 4 × 103 CFU/ml) of each selected strain were suspended in sterile saline and plated on either plain YPD agar or YPD agar supplemented with various concentrations of FLC. The growth patterns were recorded after incubation at 30°C for 72 h. The percentages of the resistant subpopulations that emerged at the lowest concentration of FLC were recorded (31).
TABLE 2.
Percent heteroresistant subpopulation of clinical and environmental C. gattii strains at different fluconazole concentrationsa
| Strain | Serotype | Source | Fluconazole concn (μg/ml) |
VG type | ||||
|---|---|---|---|---|---|---|---|---|
| 2 | 4 | 8 | 16 | 32 | ||||
| NIH200 | B | Clinical | 100 | 0.1 | II | |||
| NIH754 | B | Clinical | 100 | 0.04 | II | |||
| NIH412 | B | Clinical | 100 | 0.07 | II | |||
| NIH535 | B | Clinical | 100 | 0.1 | III | |||
| NIH198 | B | Clinical | 100 | 100 | 100 | 1.9 | I | |
| B3939 | B | Clinical | 100 | 100 | 100 | 0.36 | III | |
| NIH184 | B | Clinical | 100 | 100 | 100 | 35 | II | |
| NIH444 | B | Clinical | 100 | 100 | 100 | 100 | 0.5 | III |
| NIH189 | B | Clinical | 100 | 100 | 100 | 0.7 | III | |
| NIH190 | B | Clinical | 100 | 100 | 100 | 0.8 | III | |
| NIH367 | B | Clinical | 100 | 100 | 100 | 9 | III | |
| NIH409 | B | Clinical | 100 | 100 | 100 | 100 | 0.4 | III |
| NIH286 | B | Clinical | 100 | 100 | 100 | 100 | 0.06 | I |
| NIH771 | B | Clinical | 100 | 100 | 100 | 100 | 9 | III |
| B4506 | B | Env | 100 | 100 | 12 | III | ||
| B5788 | B | Env | 100 | 100 | 5.7 | I | ||
| B5763 | B | Env | 100 | 100 | 0.03 | I | ||
| B5765 | B | Env | 100 | 100 | 100 | 0.05 | I | |
| RB3 | B | Env | 100 | 100 | 55 | II | ||
| RB14 | B | Env | 100 | 100 | 100 | 100 | 0.8 | II |
| RB22 | B | Env | 100 | 100 | 100 | 0.15 | II | |
| RAM15 | B | Env | 100 | 100 | 100 | 100 | 10 | II |
| RAM002 | B | Env | 100 | 100 | 100 | 100 | 0.53 | II |
| VPB572-058 | B | Env | 100 | 100 | 100 | 100 | 1.6 | II |
| NIH18 | C | Clinical | 100 | 100 | 100 | 100 | 0.44 | I |
| NIH34 | C | Clinical | 100 | 100 | 100 | 0.22 | I | |
| NIH257 | C | Clinical | 100 | 2 | III | |||
| NIH113 | C | Clinical | 100 | 100 | 100 | 100 | 0.99 | I |
| H0058-1-78 | C | Clinical | 100 | 100 | 100 | 100 | 2.3 | III |
| H0058-1-2023 | C | Clinical | 100 | 100 | 100 | 100 | 7.3 | III |
| NIH191 | C | Clinical | 100 | 100 | 100 | 100 | 20 | I |
| NIH298 | C | Clinical | 100 | 100 | 100 | 100 | 9 | III |
| H0058-1-1941 | C | Env | 100 | 100 | 100 | 100 | 1.4 | II |
| H0058-1-818 | C | Env | 100 | 100 | 100 | 100 | 16 | III |
| H0058-1-1686 | C | Env | 100 | 100 | 100 | 100 | 0.75 | IV |
Strains in bold indicate those selected for resistance assays to different xenobiotics as shown in Fig. 5.
Stability of FLC resistance in vitro.
Subclones from the representative strains within each category of LHF that had adapted to tolerate 64 μg/ml FLC were grown and suspended in YPD broth. Fifty microliters of the suspension was then repeatedly transferred into 5 ml of fresh YPD broth daily and incubated at 30°C for 24 h. The proportions of the subpopulations expressing resistance to FLC (16 to 64 μg/ml) were determined by periodically plating a 50 μl suspension of each subculture on either YPD agar alone or YPD agar plus FLC and incubated at 30°C for 72 h. The number of colonies resistant at each level of FLC concentration was recorded.
Resistance to other xenobiotics.
Four to five strains from different LHF categories were randomly chosen for analysis of their resistance to xenobiotics unrelated to azole drugs (shown in bold in Table 2). Each strain was first inoculated in YPD broth and incubated overnight at 30°C, washed, serially diluted in sterile saline, and spotted (3 μl) onto YPD agar alone and YPD agar containing different xenobiotics. The compounds tested were trichostatin A, rhizoxin, and gliotoxin at various concentrations. All compounds were obtained from Sigma (Sigma-Aldrich, St. Louis, MO), unless specified.
Virulence in mice.
Virulence among strains with different LHFs was compared using a murine model of pulmonary cryptococcosis. Cultures of the strains to be tested were grown overnight in standard broth medium (YPD) at 30°C. The cultures were then diluted to the desired concentration in a 0.9% NaCl solution. Six-week-old anesthetized female BALB/c mice (weight, 20 g) were infected by intranasal inoculation of a 20-μl droplet containing 5 × 107 yeast cells. Representative strains from Canada, Australia, and India that had exhibited different LHFs were used for comparisons of virulence. The strains used had been determined to have comparable levels of production of capsule, melanin, urease, and growth at 37°C. Ten animals were used for each strain. The survival of mice was recorded daily for 35 days, and the survival data were statistically analyzed using the log rank method.
RESULTS AND DISCUSSION
Relationship between serotype, molecular type, and LHF of C. gattii strains.
It is well known that C. neoformans (serotypes A, D, and AD) and C. gattii (serotypes B and C) strains differ from each other in several characteristics that include epidemiology, ecology, biochemistry, and genetics. Differences in the two species are also recognized by the risk factors of the host. While HIV infection is the major risk factor for cryptococcosis due to C. neoformans strains, this is not the case for infection by strains of C. gattii (16, 28). In the present study, we report differences between the two species in the LHFs. A total of 71 clinical and environmental C. gattii isolates of serotypes B and C from different geographic sources were used in this study (Table 1). As was the case with C. neoformans, all 71 strains of C. gattii exhibited innate levels of heteroresistance to fluconazole regardless of serotype, source, or time of isolation relative to the advent of azoles for the treatment of fungal diseases. All the clinical strains of serotype B (25 strains) and C (17 strains), excluding the three serotype C strains obtained from Colombia, had been isolated in or before 1977, at least a decade prior to the availability of triazole drugs. These strains had been maintained in our laboratory as lyophilized stocks. Of the 25 environmental isolates of serotype B, 17 were from Canada (13), 5 from Australia (17), and 3 from India (5). All 4 environmental strains of serotype C were isolated in Colombia.
Heteroresistance to fluconazole is the phenomenon described for C. neoformans as the emergence of a minor subpopulation of resistant cells within a single colony of the susceptible strain that can tolerate FLC concentrations higher than the strain's MIC levels (31). The levels of resistance to fluconazole were determined by Etest analysis (Fig. 1) and spot test analysis of serial dilutions (Fig. 2) for each strain on growth media supplemented with different concentrations of fluconazole to determine the LHF. Emergence of resistant subpopulations within the patch was indicative of heteroresistance at that concentration of fluconazole (Fig. 2, lanes 1A to C). In C. neoformans, 79% of the serotype D strains (22 of 28) and 50% of the serotype A strains (51 of 102) which had previously not been exposed to azole drugs showed LHFs of ≤8 μg/ml fluconazole (31). As shown in Table 1, 86% of the C. gattii strains (61 of 71) which had not been exposed to azole drugs had LHFs of ≥16 μg/ml. All clinical strains isolated before 1990 were considered to be fluconazole-naive strains. Of the serotype B clinical strains isolated prior to the advent of triazole antifungals, 80% (20 of 25) exhibited LHFs of ≥16 μg/ml, while 92.9% of the serotype C clinical strains isolated during the same era (16 of 17) exhibited similar LHFs (Tables 3 and 4). The LHFs of a majority of the serotype C strains (18 of 21), regardless of their origin, was 32 μg/ml (Table 4). These results indicated that overall, more strains of C. gattii had high LHFs than did strains of C. neoformans (31).
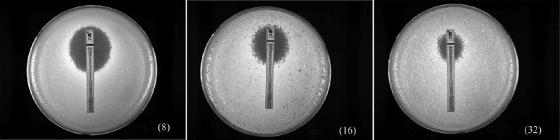
FIG. 1.
Determination of fluconazole resistance by Etest analysis. Three representative cultures, exhibiting resistance to 8, 16, and 32 μg/ml fluconazole, are shown.
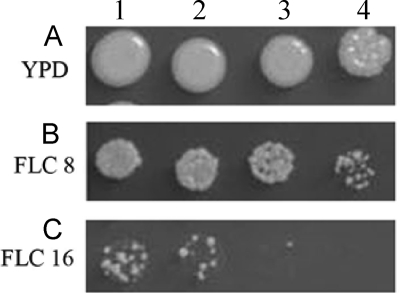
FIG. 2.
Determination of LHF by spot test analysis. Spot test analysis representing growth of C. gattii strains on YPD agar medium alone (A) and YPD agar medium supplemented with 8 μg/ml (B) or 16 μg/ml (C) fluconazole. Lanes 1 to 4 represent 10-fold serial dilutions of culture. This example shows the spot test of a strain determined to have the LHF of 16 μg/ml FLC.
TABLE 3.
Population analysis profile of C. gattii serotypes B isolates screened on FLC-containing medium
| Isolate source | Level of heteroresistance to fluconazole (μg/ml) | No. of isolates tested (%) |
|---|---|---|
| Clinical | 4 | 5 (20) |
| 16 | 9 (36) | |
| 32 | 11 (44) | |
| Environmental | 8 | 4 (16) |
| 16 | 2 (8) | |
| 32 | 19 (76) |
aA total of 25 clinical isolates and 25 environmental isolates were tested.
TABLE 4.
Population analysis profile of C. gattii serotype C isolates screened on FLC-containing medium
| Isolate source | Level of heteroresistance to Fluconazole (μg/ml) | No. of isolates tested (%) |
|---|---|---|
| Clinical | 4 | 1 (5.8) |
| 16 | 2 (11.8) | |
| 32 | 14 (82.4) | |
| Environmental | 32 | 4 (100) |
aA total of 17 clinical isolates and 4 environmental isolates were tested.
In C. neoformans, strains of serotype A tend to have a higher LHF than that of the strains of serotype D (31). In C. gattii, though, there were more serotype C than serotype B strains that had high LHFs. However, the difference was not as great as that between serotype A and D strains of C. neoformans (31). The genetic diversity between strains of serotype A and serotype D is well documented, and these strains are clearly distinguishable by the commonly used molecular typing tools, such as M13-based PCR fingerprint patterns or multilocus sequence typing (4, 18, 22). The molecular types of 71 C. gattii strains were determined by their M13 PCR fingerprint patterns and their URA5 RFLPs. The results showed that all four major molecular types of C. gattii, VGII, VGIII, VGI, and VGIV, were present in a decreasing order of frequency (Table 1). Figure 3 shows the different molecular types among the representative isolates from five different geographical areas as discerned from their M13 PCR fingerprint patterns (Fig. 3A) as well as their corresponding URA5 RFLPs (Fig. 3B). The C. gattii isolates from Vancouver used in this study were all of the VGIIa molecular type. No apparent correlation was observed between molecular type and LHF, although the three environmental strains of the VGI molecular type isolated from India consistently showed lower LHFs than those of the VGI strains from other geographical regions. A correlation between serotype and the LHF was not significant, although a slightly higher proportion of serotype C had ≥16 μg/ml LHFs than did the strains of serotype B. Strains of serotype A are comprised of molecular types VNI and VNII, while strains of serotype D are exclusively of the VNIV molecular type (24). However, the molecular types of C. gattii strains, VGI to VGIV, are not serotype associated and can occur in strains of serotype B or C (23). This may explain the correlation between the serotype and the LHF of C. neoformans strains and an absence of such a correlation between molecular type and either serotype or LHF in strains of C. gattii.
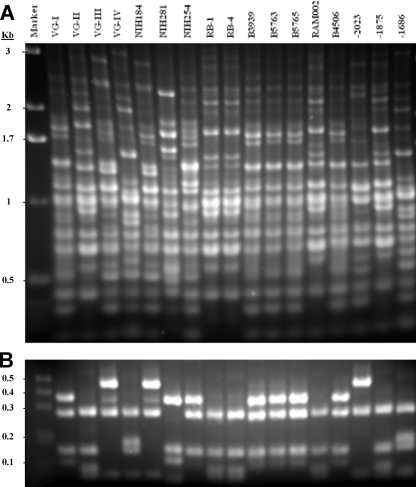
FIG. 3.
(A) Examples of PCR fingerprint patterns obtained by using the M13 primer for clinical and environmental strains of C. gattii that represent the four different molecular types. Lane 1, size marker; lanes 2 to 5, reference strains representing the four molecular types VGI to VGIV. (B) Examples of URA5 gene restriction fragment length polymorphism profiles from clinical and environmental isolates of C. gattii representing the four different molecular types. Lanes are as in Fig. 1A.
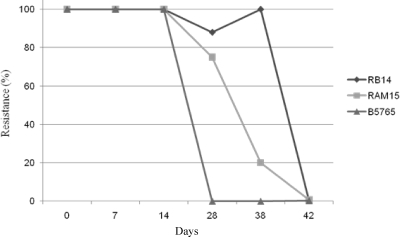
Heteroresistance by definition is the ability of a subpopulation to adapt to higher concentrations of the drug in a stepwise manner, resulting in homogeneous resistant populations. Furthermore, resistance acquired in such a manner is also lost in a stepwise manner upon repeated transfer into drug-free medium, resulting in reversion to the original LHF (31). To determine whether such elevations in drug resistance were transitional in nature, cultures of representative strains from Canada (RB-14, 32 μg/ml LHF), Australia (RAM-15, 32 μg/ml LHF), and India (B-5765, 8 μg/ml LHF) were grown at the fluconazole concentration of each one's LHF and then sequentially transferred to media with increasing concentrations of fluconazole. The subpopulations of Canadian and Australian isolates with an LHF of 32 μg/ml were able to adapt to levels of fluconazole as high as 256 μg/ml, while the Indian isolates with an LHF of 8 μg/ml could only achieve a maximum resistance level of 64 μg/ml (data not shown). The strains that had adapted to 64 μg/ml FLC were cultured in YPD medium supplemented with 64 μg/ml fluconazole and then transferred daily into drug-free YPD broth and incubated at 30°C. Aliquots of these cultures were removed daily and plated on YPD media with different concentrations of the drug to monitor the loss of resistance. All the strains lost the elevated level of resistance to fluconazole and returned to their original, innate levels of resistance. The numbers of transfers in drug-free medium that were required for the loss of resistance correlated with the original LHFs: strains with LHF of 32 μg/ml that had adapted to 64 μg/ml FLC required 42 transfers to return to the original level of resistance, while only 28 transfers were required for the strain from India with 8 μg/ml LHF (Fig. 4). As represented in Table 2, all strains exhibited heteroresistance to fluconazole but at various concentrations, 4 μg to 32 μg, indicating that C. gattii strains are innately heteroresistant to fluconazole. While for C. neoformans, only 50% and 21% of serotype A and serotype D strains, respectively, exhibited LHFs of ≥16 μg/ml, for C. gattii, 82% and 95% of serotype B and serotype C strains, respectively, exhibited LHFs of ≥16 μg/ml. The overall LHFs of C. gattii strains, therefore, were significantly higher than those observed for strains of C. neoformans.
FIG. 4.
Loss of the acquired tolerance to fluconazole by heteroresistant subpopulations of C. gattii strains upon daily transfer of the cultures grown at 30°C in YPD medium without fluconazole.
Relationship between LHF and resistance to toxins and antibiotics produced by environmental microorganisms.
In C. neoformans, a correlation between the LHF of a strain and its resistance to several xenobiotics has been reported (31). We tested several of the xenobiotics produced by microorganisms that C. gattii strains are likely to confront in the environment. Strains of C. gattii most commonly inhabit tree hollows (8) where numerous other microorganisms may cohabitate. The trees with which C. gattii strains are found to be associated include alder, cedar, Douglas fir, almond, and particularly, the hollows of the Eucalyptus species (2, 9, 10, 15). Since all environmental isolates were recovered from trees such as alder and Douglas fir, prior exposure to agricultural fungicides, such as clotrimazole and ketoconazole, in areas of their recovery is not likely.
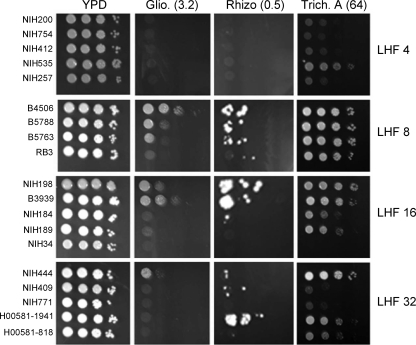
The tree hollows may harbor various ubiquitous saprophytic microorganisms, such as Penicillium, Aspergillus, and common soil bacteria. We tested trichostatin A, a histone deacetylase inhibitor produced by Streptomyces platensis, rhizoxin, a microtubule inhibitor produced by Burkholderia species, and gliotoxin, an antibiotic commonly produced by Aspergillus and Penicillium species (31). Figure 5 shows the susceptibilities of C. gatiii strains with various levels of heteroresistance against these three xenobiotics. All the strains of C. gattii were observed to be highly resistant to these xenobiotics at the minimum concentration which completely inhibited the growth of C. neoformans strains (31). We raised the concentration of gliotoxin (3.2 μg/ml) and trichostain A (64 μg/ml) 2-fold each and the concentration of rhizoxin (0.5 μg/ml) 5-fold in comparison to the minimum concentration that had been reported to inhibit growth of C. neoformans strains in order to achieve the minimum concentration at which C. gattii strains are inhibited. As was the case with strains of C. neoformans, the C. gattii strains with higher LHFs tended to be more resistant to the xenobiotics tested than the strains with low LHFs. C. gattii strains with a LHF of 4 μg/ml were generally more sensitive to all three compounds than strains with LHFs of 8 and 16 μg/ml FLC. The strains with a 32-μg/ml LHF, however, were not necessarily more resistant than the strains with a 16-μg/ml LHF and showed considerable variations in the degree of resistance (Fig. 5). These results show that the overall resistance of C. gattii to these xenobiotics is at least two times higher than that of the strains of C. neoformans. Although both species of Cryptococcus are recovered from soil samples and may share some common environmental niches, C. gattii strains are not known to be associated with pigeon droppings, the major environmental source of C. neoformans. With such ecological differences, C. gattii may have been predisposed to develop the ability to tolerate higher levels of certain xenobiotics than can C. neoformans strains.
FIG. 5.
Spot test analysis to determine resistance of the C. gattii strains to different xenobiotics: gliotoxin (Glio., 3.2 μg/ml), rhizoxin (Rhizo., 0.5 μg/ml), and trichostatin A (Trich. A, 64 μg/ml). The strains selected represent the four groups of strains that exhibited heteroresistance at 4, 8, 16, and 32 μg/ml fluconazole (boldface in Table 2).
Several other studies have also reported differences between the two species with regard to their susceptibilities to amphotericin B and various other azoles (7, 12, 25, 33, 34). Khan et al. (12) reported on antifungal susceptibilities based on the largest number of environmental isolates from both species and showed that C. gattii strains had significantly higher MICs for both amphotericin B and azoles. The contradictory reports regarding the in vitro susceptibility of C. neoformans and C. gattii strains to some antifungals may be due to the methods used by different laboratories. Although the Etest is recommended as a good alternative to the microbroth and broth macrodilution method of the Clinical and Laboratory Standards Institute (CLSI, formerly NCCLS) for antifungal susceptibilities of yeast, the results have sometimes been contradictory and do not always corroborate the MIC values (12). Larger inoculums with agitation have been proposed as an alternative method for MIC determinations (11). In Candida species, the standardized CLSI methodology has proven to be reliable, but not for Cryptococcus. It is noteworthy that drugs which show differences in resistance between the two species are those that inhibit or bind ergosterol in the cell membrane. Treatment with these drugs of cryptococcosis due to C. gattii has been more difficult than of disease caused by C. neoformans (25, 33). At present, the mechanism for the susceptibility difference between the two species against these antimycotics is unknown. In our experiments, a higher percentage of C. gattii isolates exhibited high heteroresistance levels to fluconazole than C. neoformans strains. The C. gattii strains with high LHFs were also found to express higher resistance to xenobiotics such as rhizoxin. This inherently higher LHF of C. gattii strains likely influences the MICs of the strains, resulting in the variability in resistance to different compounds between different studies. So, the LHF of the strains should be taken into account when investigating resistance of the strains for different compounds.
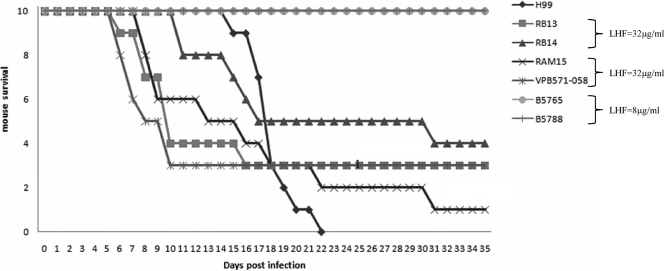
Virulence and LHF.
A total of six strains, including two strains each from Canada and Australia with an LHF of 32 μg/ml and two strains from India with an LHF of 8 μg/ml, were used for the virulence study. The highly virulent C. neoformans serotype A strain H99 was used as a control. Mice infected with C. gattii strains exhibiting a LHF of 32 μg/ml began to die significantly earlier than the mice infected with strains that had an LHF of 8 μg/ml (Fig. 6). Although the mice infected by C. gattii strains with a 32-μg/ml LHF started dying earlier than those infected by the H99 strain, their virulence was not found to be significantly different from that of H99, as determined by the log rank test (P ≥ 0.6). These results demonstrate a correlation between LHF and virulence in C. gattii strains which is similar to what has been reported for strains of C. neoformans (27). In the previous work, mice were infected with C. neoformans strains with high and low LHFs and then either left untreated or treated with fluconazole (31). The CFU of strains recovered from the tissues of infected mice were propagated on media with and without different concentrations of fluconazole and showed that the original LHF for the strains was maintained. The mice infected with strains with high LHFs were found to die earlier regardless of treatment, while those mice infected with strains with lower LHFs survived much longer if treated with fluconazole. Although mice were not treated with fluconazole in the present study, it is predicted that more mice infected with C. gattii strains will be unresponsive to fluconazole treatment than those infected with C. neoformans. Heteroresistance, an adaptive mechanism employed by the organism to counteract the stress of increasing drug concentration in the environment, may enhance the organism's fitness to survive under other stresses as well. This overall fitness may be translated into higher virulence.
FIG. 6.
Virulence in BALB/c mice following intranasal inoculation of 5 × 107 cells per mouse with the selected C. gattii strains from different geographic regions that exhibited different levels of heteroresistance to fluconazole.
This study serves to highlight the stark differences between the two species with respect to the level of heteroresistance to fluconazole. It shows that a higher percentage of C. gattii strains manifest high LHFs than C. neoformans strains. Such an understanding would be beneficial for establishing therapeutic regimens and in predicting clinical outcomes with respect to developing resistance and relapses in cryptococcosis caused by C. gattii. Furthermore, strains of C. gattii were found to exhibit markedly higher levels of resistance to different xenobiotics than C. neoformans, which also mirrored their LHFs. This resistant property of the C. gattii strains most likely reflects an acquisition from its ecological niche.
The mechanism of heteroresistance in C. neoformans is yet to be clearly established (31). It is very different from the heteroresistance reported for Candida albicans, which is also most often treated with triazoles. Unlike in C. neoformans, heteroresistance in Candida albicans is reported to develop following short exposures to FLC, both in vitro and in vivo, and has been associated with disseminated infection in patients that had received bone marrow transplants (19, 21). However, as in C. albicans, heteroresistance in isolates of C. neoformans and C. gattii that became resistant to azole drugs was transient, since the strains became susceptible after serial transfers in the absence of the drug (19, 20). Recent studies have revealed an association between alterations in ploidy and azole resistance in C. albicans (30) and C. neoformans (32). In resistant strains, the presence of an extra whole chromosome or an isochromosome was detected, and such chromosomal gain or loss was strongly associated with increased and decreased azole resistance (30, 32). In the C. neoformans/C. gattii species complex, heteroresistance to azoles is innate and the level of heteroresistance to fluconazole varies between strains. The present study reveals that a significantly higher number of C. gattii strains than C. neoformans strains manifest a ≥16-μg/ml LHF that is associated with higher virulence. It remains to be elucidated whether a mechanism similar to that described for C. albicans is responsible for heteroresistance in the C. neoformans/C. gattii species complex.
Acknowledgments
We thank John E. Bennett for kindly providing C. gattii cultures in his collection that were deposited before 1980. We also thank Elizabeth Castaneda for her contribution of the Colombian strains of C. gattii.
This work was supported by funds from the intramural program of the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 12 April 2010.
REFERENCES
- 1.Armengou, A., C. Porcar, J. Mascaro, and F. Garcia-Bragado. 1996. Possible development of resistance to fluconazole during suppressive therapy for AIDS-associated cryptococcal meningitis. Clin. Infect. Dis. 23:1337-1338. [DOI] [PubMed] [Google Scholar]
- 2.Berg, J., C. J. Clancy, and M. H. Nguyen. 1998. The hidden danger of primary fluconazole prophylaxis for patients with AIDS. Clin. Infect. Dis. 26:186-187. [DOI] [PubMed] [Google Scholar]
- 3.Birley, H. D., E. M. Johnson, P. McDonald, C. Parry, P. B. Carey, and D. W. Warnock. 1995. Azole drug resistance as a cause of clinical relapse in AIDS patients with cryptococcal meningitis. Int. J. STD AIDS 6:353-355. [DOI] [PubMed] [Google Scholar]
- 4.Boekhout, T., B. Theelen, M. Diaz, J. W. Fell, W. C. Hop, E. C. Abeln, F. Dromer, and W. Meyer. 2001. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology 147:891-907. [DOI] [PubMed] [Google Scholar]
- 5.Chakrabarti, A., M. Jatana, P. Kumar, L. Chatha, A. Kaushal, and A. A. Padhye. 1997. Isolation of Cryptococcus neoformans var. gattii from Eucalyptus camaldulensis in India. J. Clin. Microbiol. 35:3340-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J., A. Varma, M. R. Diaz, A. P. Litvintseva, K. K. Wollenberg, and K. J. Kwon-Chung. 2008. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg. Infect. Dis. 14:755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S., T. Sorrell, G. Nimmo, B. Speed, B. Currie, D. Ellis, D. Marriott, T. Pfeiffer, D. Parr, K. Byth, et al. 2000. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Clin. Infect. Dis. 31:499-508. [DOI] [PubMed] [Google Scholar]
- 8.Ellis, D. H., and T. J. Pfeiffer. 1990. Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 28:1642-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escandon, P., A. Sanchez, M. Martinez, W. Meyer, and E. Castaneda. 2006. Molecular epidemiology of clinical and environmental isolates of the Cryptococcus neoformans species complex reveals a high genetic diversity and the presence of the molecular type VGII mating type a in Colombia. FEMS Yeast Res. 6:625-635. [DOI] [PubMed] [Google Scholar]
- 10.Fraser, J. A., S. S. Giles, E. C. Wenink, S. G. Geunes-Boyer, J. R. Wright, S. Diezmann, A. Allen, J. E. Stajich, F. S. Dietrich, J. R. Perfect, and J. Heitman. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437:1360-1364. [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Lopez, A., O. Zaragoza, M. Dos Anjos Martins, M. C. Melhem, J. L. Rodriguez-Tudela, and M. Cuenca-Estrella. 2008. In vitro susceptibility of Cryptococcus gattii clinical isolates. Clin. Microbiol. Infect. 14:727-730. [DOI] [PubMed] [Google Scholar]
- 12.Khan, Z. U., H. S. Randhawa, T. Kowshik, A. Chowdhary, and R. Chandy. 2007. Antifungal susceptibility of Cryptococcus neoformans and Cryptococcus gattii isolates from decayed wood of trunk hollows of Ficus religiosa and Syzygium cumini trees in north-western India. J. Antimicrob. Chemother. 60:312-316. [DOI] [PubMed] [Google Scholar]
- 13.Kidd, S. E., F. Hagen, R. L. Tscharke, M. Huynh, K. H. Bartlett, M. Fyfe, L. Macdougall, T. Boekhout, K. J. Kwon-Chung, and W. Meyer. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. U. S. A. 101:17258-17263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon-Chung, K. J., I. Polacheck, and J. E. Bennett. 1982. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). J. Clin. Microbiol. 15:535-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Kwon-Chung, K. J. 1976. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68:942-946. [PubMed] [Google Scholar]
- 15.Kwon-Chung, K. J., and A. Varma. 2006. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 6:574-587. [DOI] [PubMed] [Google Scholar]
- 16.Kwon-Chung, K. J., A. Varma, J. C. Edman, and J. E. Bennett. 1992. Selection of ura5 and ura3 mutants from the two varieties of Cryptococcus neoformans on 5-fluoroorotic acid medium. J. Med. Vet. Mycol. 30:61-69. [PubMed] [Google Scholar]
- 17.Kwon-Chung, K. J., B. L. Wickes, L. Stockman, G. D. Roberts, D. Ellis, and D. H. Howard. 1992. Virulence, serotype, and molecular characteristics of environmental strains of Cryptococcus neoformans var. gattii. Infect. Immun. 60:1869-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Litvintseva, A. P., R. Thakur, R. Vilgalys, and T. G. Mitchell. 2006. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 172:2223-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marr, K. A., C. N. Lyons, K. Ha, T. R. Rustad, and T. C. White. 2001. Inducible azole resistance associated with a heterogeneous phenotype in Candida albicans. Antimicrob. Agents Chemother. 45:52-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marr, K. A., C. N. Lyons, T. R. Rustad, R. A. Bowden, and T. C. White. 1998. Rapid, transient fluconazole resistance in Candida albicans is associated with increased mRNA levels of CDR. Antimicrob. Agents Chemother. 42:2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marr, K. A., T. C. White, J. A. van Burik, and R. A. Bowden. 1997. Development of fluconazole resistance in Candida albicans causing disseminated infection in a patient undergoing marrow transplantation. Clin. Infect. Dis. 25:908-910. [DOI] [PubMed] [Google Scholar]
- 22.Meyer, W., D. M. Aanensen, T. Boekhout, M. Cogliati, M. R. Diaz, M. C. Esposto, M. Fisher, F. Gilgado, F. Hagen, S. Kaocharoen, A. P. Litvintseva, T. G. Mitchell, S. P. Simwami, L. Trilles, M. A. Viviani, and J. Kwon-Chung. 2009. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 47:561-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer, W., A. Castaneda, S. Jackson, M. Huynh, and E. Castaneda. 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 9:189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer, W., K. Marszewska, M. Amirmostofian, R. P. Igreja, C. Hardtke, K. Methling, M. A. Viviani, A. Chindamporn, S. Sukroongreung, M. A. John, D. H. Ellis, and T. C. Sorrell. 1999. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA: a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis 20:1790-1799. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell, D. H., T. C. Sorrell, A. M. Allworth, C. H. Heath, A. R. McGregor, K. Papanaoum, M. J. Richards, and T. Gottlieb. 1995. Cryptococcal disease of the CNS in immunocompetent hosts: influence of cryptococcal variety on clinical manifestations and outcome. Clin. Infect. Dis. 20:611-616. [DOI] [PubMed] [Google Scholar]
- 26.Mondon, P., Y. C. Chang, A. Varma, and K. J. Kwon-Chung. 2000. A novel episomal shuttle vector for transformation of Cryptococcus neoformans with the ccdB gene as a positive selection marker in bacteria. FEMS Microbiol. Lett. 187:41-45. [DOI] [PubMed] [Google Scholar]
- 27.Paugam, A., J. Dupouy-Camet, P. Blanche, J. P. Gangneux, C. Tourte-Schaefer, and D. Sicard. 1994. Increased fluconazole resistance of Cryptococcus neoformans isolated from a patient with AIDS and recurrent meningitis. Clin. Infect. Dis. 19:975-976. [DOI] [PubMed] [Google Scholar]
- 28.Perfect, J. R., and A. Casadevall. 2002. Cryptococcosis. Infect. Dis. Clin. North Am. 16:837-874, v-vi. [DOI] [PubMed] [Google Scholar]
- 29.Powderly, W. G., D. Finkelstein, J. Feinberg, P. Frame, W. He, C. van der Horst, S. L. Koletar, M. E. Eyster, J. Carey, H. Waskin, et al. 1995. A randomized trial comparing fluconazole with clotrimazole troches for the prevention of fungal infections in patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 332:700-705. [DOI] [PubMed] [Google Scholar]
- 30.Selmecki, A., A. Forche, and J. Berman. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sionov, E., Y. C. Chang, H. M. Garraffo, and K. J. Kwon-Chung. 2009. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob. Agents Chemother. 53:2804-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sionov, E., H. Lee, Y. C. Chang, and K. J. Kwon-Chung. 2009. Genome plasticity in Cryptococcus neoformans, a human pathogen, contributes to adaptive azole resistance. Abstr. 49th Annu. Intersci. Conf. Antimicrob. Agents Chemother., San Francisco, CA, abstr. M348. American Society for Microbiology, Washington, DC.
- 33.Speed, B., and D. Dunt. 1995. Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clin. Infect. Dis. 21:28-34; discussion, 35-36. [DOI] [PubMed] [Google Scholar]
- 34.Trilles, L., B. Fernandez-Torres, S. Lazera Mdos, B. Wanke, and J. Guarro. 2004. In vitro antifungal susceptibility of Cryptococcus gattii. J. Clin. Microbiol. 42:4815-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varma, A., S. Wu, N. Guo, W. Liao, G. Lu, A. Li, Y. Hu, G. Bulmer, and K. J. Kwon-Chung. 2006. Identification of a novel gene, URE2, that functionally complements a urease-negative clinical strain of Cryptococcus neoformans. Microbiology 152:3723-3731. [DOI] [PubMed] [Google Scholar]
- 36.Venkateswarlu, K., M. Taylor, N. J. Manning, M. G. Rinaldi, and S. L. Kelly. 1997. Fluconazole tolerance in clinical isolates of Cryptococcus neoformans. Antimicrob. Agents Chemother. 41:748-751. [DOI] [PMC free article] [PubMed] [Google Scholar]