Abstract
Human beta-defensins (hBDs) are antimicrobial peptides of human innate immunity. The antibacterial activities of hBDs 1, 2, and 4 but not the activity of hBD3 are impaired by high salt levels. We have designed and synthesized seven novel hBD analogs, constituted by different domains of hBD1 (which is constitutively expressed in humans) and of hBD3 (which is induced by microorganisms and inflammatory factors in humans), that would maintain and potentially increase the wild-type antimicrobial activities and be salt resistant. We have compared the antibacterial, antiviral, and chemotactic activities of the analogs with those of hBD1 and hBD3. We show that the hBD1 internal region and the hBD3 C-terminal region are critical for antibacterial activity also at high salt concentrations, whereas deletion of the N-terminal region of hBD3 results in an increase in antibacterial activity. All analogs inhibited herpes simplex virus; antiviral activity was enhanced by the hBD1 internal region and the hBD3 C-terminal region. Wild-type and analog peptides were chemotactic for granulocytes and monocytes, irrespective of the salt concentrations. These new peptides may have therapeutic potential.
Beta-defensins (BDs) are highly conserved small peptides produced by plants, invertebrates, and vertebrates that developed as part of the primordial immune protective mechanism (19). Four of these peptides, called human BD1 (hBD1; DEFB1), hBD2 (DEFB4), hBD3 (DEFB103A), and hBD4 (DEFB104), are mainly expressed by respiratory, gastrointestinal, and urogenital epithelial cells either constitutively (hBD1) or after induction by microorganisms or inflammatory factors (hBD2 to hBD4) (19). All four hBDs are cationic and 36 to 45 amino acids long and show similar folding and an invariable six-cysteine motif that gives rise to three disulfide bonds (2, 11, 12, 25, 26).
Human beta-defensins 1 to 4 exert different bactericidal and antiviral activities against various pathogens (8, 15, 27). The antibacterial effects of hBD1 (9), hBD2 (33), and hBD4 (5) are attenuated by high NaCl concentrations, such as those in the airway surface fluid of patients with cystic fibrosis (CF) (21, 29). Human beta-defensin 3 can withstand NaCl concentrations as high as 150 mM, thanks to its peculiar structural characteristics and charge (10). In the field of viral diseases, hBD2 and -3 inhibit human immunodeficiency virus (HIV) type 1 (HIV-1) replication and virion infectivity (20, 31) and modulate HIV-1 coreceptor expression (20). Human herpes simplex virus (HSV) type 1 (HSV-1), HSV-2, and other viruses preincubated with alpha human neutrophil peptide 1 (hNP1) to hNP3 (6, 28) or theta (37) defensins lose their ability to infect target cells (28). As yet, there are no data on the effect of hBDs on HSV-1 and -2. In addition to direct antimicrobial activity, hBDs also exert chemotactic activity: hBD1, -2, and -3 are chemotactic for monocytes and dendritic and T cells. Human beta-defensin 3 is the only beta-defensin chemotactic for macrophages (4, 18, 19), whereas the chemotactic effect of hBDs on granulocytes has yet to be elucidated (4, 18).
The two natural defensins hBD1 and hBD3 were chosen for use in the experiments described in this paper for the following reasons: hBD1 is constitutively expressed but its antibacterial activity is greatly impaired by NaCl, while hBD3 is insensitive to salt. Thus, we designed and synthesized hBD analogs that, in principle, would maintain the antibacterial and antiviral activities of hBD1 and possess a resistance capability in the presence of high NaCl concentrations, like hBD3 does. We then compared the antibacterial, chemotactic, and antiviral activities of the novel synthetic analogs with those of wild-type hBD1 and hBD3. Our data show that some of the synthetic analogs have higher antimicrobial activity than the wild type, also at high NaCl concentrations.
MATERIALS AND METHODS
Peptide design.
The beta-defensin sequences used in this study were taken from the SWISS-Prot database. Graphics analyses were run on a Silicon Graphics Indigo2 workstation. The InsightII/Discover program (Biosym Technologies, San Diego, CA) was used to compare the structures of the beta-defensin molecules obtained from the Protein Data Bank (Brookhaven National Laboratory, Upton, NY). Structural parameters, such as net positive charge, hydrophobicity, and the hydrophobic moment, and the analysis of the hydrophobic moment/hydrophilic moment ratio were also evaluated in order to design new analogs that could have enhanced activity.
Peptide synthesis.
Peptides were synthesized by the standard solid-phase 9-fluorenylmethoxycarbonyl (Fmoc) method. NovaSyn TGA (Merck, Darmstadt, Germany) resin (substitution, 0.25 mmol/g) was used as solid-phase support, and syntheses were performed on a scale of 100 μmol. Ten equivalents of the first amino acid were coupled on the resin according to the N,N′-diisopropylcarbodiimide (DIC)/4-dimethylaminopyridine (DMAP) method (with 10 equivalents of Fmoc-amino acid, 5 equivalents of DIC, 0.1 equivalent of DMAP). All successive amino acids, 4 equivalents relative to the resin loading, were coupled by using 1 equivalent of Fmoc-amino acid, 1 equivalent of 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium tetrafluoroborate, 1 equivalent of N-hydroxybenzotriazole (HOBT; 0.5 mM HOBT in N-methylpyrrolidone [NMP]) and 2 equivalents of N,N-diisopropylethylamine (DIPEA; 1 mM DIPEA in NMP). The Fmoc protecting group was removed with 30% (vol/vol) piperidine in NMP. The peptides were fully deprotected and cleaved from the resin with trifluoroacetic acid (TFA) with 5% thioanisole, 3% ethandithiol, and 2% anisole as scavengers. The crude peptides were precipitated with ice-cold ethyl ether, filtered, dissolved in water, lyophilized, and reduced with dithiothreitol (DTT). The peptides were purified to homogeneity by preparative reverse-phase high-pressure liquid chromatography (RP-HPLC). The samples were injected on a Phenomenex (Torrance, CA) C18 column (22 mm by 25 cm, 5 mm) eluted with an H2O-0.1% TFA (solvent A) and CH3CN-0.1% TFA (solvent B) solvent mixture. A linear gradient from 5 to 50% solvent B over 17 min at a flow rate of 20 ml/min was used. The collected fractions were lyophilized to dryness and analyzed by analytical RP-HPLC on a Shimadzu class LC10 column equipped with a diode array detector (SPD-M10AV) and by use of a Phenomenex C18 analytical column (4.6 by 250 mm, 5 mm). The identity and the oxidation state of the purified peptides were confirmed by electron spray ionization liquid chromatography-mass spectrometry (ESI LC-MS) with a Thermo electron MSQ surveyor. The peptides were dissolved in phosphate-buffered saline (PBS) for all experiments.
Folding and disulfide formation.
Oxidation in 20% dimethyl sulfoxide (DMSO) was performed by dissolving samples in 0.1 M Tris HCl (pH 8.2)-1 mM EDTA at a peptide concentration of 1 mM. The folding reaction was monitored by RP-HPLC at 8, 16, and 48 h. The folded product was analyzed and purified, and its molecular mass was determined by LC-MS.
CD measurements.
Circular dichroism (CD) spectra were recorded with a Jasco J-715 spectropolarimeter in a 0.1-cm quartz cell at room temperature. The spectra of reduced and oxidized hBD1 (0.1 mM) and hBD3 and of reduced analogs 1C and 3I were obtained in 5 mM HEPES buffer at pH 7.5 in the presence or the absence of 40% trifluoroethanol (TFE). The spectra were the averages of three consecutive scans from 250 to 195 nm, recorded with a bandwidth of 3 nm, a time constant of 16 s, and a scan rate of 10 nm/min. The spectra were recorded and corrected for the blank.
Antibacterial activity assay.
A CFU assay of the antibacterial activities of the hBDs against Pseudomonas aeruginosa ATCC 27853 (American Type Culture Collection, Manassas, VA), Enterococcus faecalis ATCC 29212, and Escherichia coli ATCC 25922 was performed. The strains were grown under aerobic conditions in tryptic soy broth (Difco Laboratories, Detroit, MI) at 37°C and incubated in the presence of the hBDs for 2 h at 37°C. We used two concentrations of peptides, i.e., 2.5 μM, at which 100% of the P. aeruginosa strains were killed by wild-type hBD3 (minimal bactericidal concentration [MBC]), and 12.5 μM. For the salt dependence assay, NaCl was included in the incubation buffer over a range of concentrations potentially present in the airway surface fluid of healthy subjects and patients with CF (20), i.e., 0, 50, 100, and 200 mM NaCl. Each assay was performed in triplicate. Bactericidal activity (the mean and standard deviation [SD] of three assays) was expressed as the ratio between the number of colonies counted and the number of colonies on a control plate. The MICs of the wild-type peptides and their analogs were determined by use of a modified version (3a) of the broth microdilution assay of the National Committee for Clinical Laboratory Standards with a final inoculum of 105 CFU/ml. The following peptide concentrations were used: 100, 50, 25, 12.5, 6.25, 3.12, and 1.56 μM.
Antiviral activity.
Vero cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum. HSV-1 carrying a lacZ gene driven by the cytomegalovirus (CMV) immediate-early 1 promoter to express beta-galactosidase was propagated as described previously (7). All experiments were conducted in parallel with no-peptide controls. To assess the effects of the peptides on the inhibition of HSV infectivity, we treated cell monolayers as follows. (i) For the coexposure experiments, the cells were incubated with increasing concentrations of the peptides (1, 5, 10, 20, 50, 100, and 250 μM) and with the viral inoculum for 45 min at 37°C. Nonpenetrated viruses were inactivated by citrate buffer at pH 3.0. Monolayers were fixed and stained with 5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside (X-gal), and the plaque numbers were scored. The experiments were performed in triplicate, and the percent inhibition was calculated with respect to the number of plaques in the no-peptide control experiments. (ii) For the virus preexposure experiments, approximately 2 × 104 PFU of HSV-1 was incubated in the presence of 20 μM peptides for 45 min at 37°C and then titrated on Vero cell monolayers. (iii) For the cell preexposure experiments, Vero cells were incubated with 20 μM peptides for 30 min at 4°C and infected with serial dilutions of HSV-1 for 45 min at 37°C.
Preparation of leukocytes.
Monocytes and neutrophils were isolated as described in the StemSep Cell Technical Manual (30a). Neutrophils were purified from lysed buffy coats by use of a StemSep negative selection system. Cell viability was assessed by analysis of the light-scatter properties, and the cells were counted by use of a logical gate strategy based on Trucount tubes (Becton Dickinson) before and after the negative selection.
Measurement of leukocyte migration.
The chemotaxis assay of monocytes and neutrophils (90 to 95% purity) was performed in 96-well disposable chemotaxis chambers (Neuroprobe, Gaithersburg, MD) with polyvinylpyrrolidone-free polycarbonate membranes (pore sizes, 8 mm for monocytes and 5 mm for neutrophils). Chemotaxis was measured by use of the calcein fluorescence signal, and the results were read in a microplate fluorescence reader (Victor 3 1420 multilabel counter; Perkin-Elmer). We calculated the migration index as the ratio between the sample mean fluorescence (after subtraction of the negative-control mean fluorescence)/positive-control mean fluorescence. Each assay was performed in triplicate.
Toxicity.
We assessed the cytotoxicities of wild-type hBDs and analogs on neutrophils and monocytes at different concentrations (i.e., 2.5 and 12.5 μM) and on Vero cells, also at different concentrations (i.e., 10 to 500 μM), by a lactate dehydrogenase assay, which was carried out according to the manufacturer's instructions by use of a cytotoxicity detection kit (Roche Diagnostic, Milan, Italy).
Statistical methods.
All data are summarized by using the mean and the standard deviation computed for three independent replicates. Differences in means were tested by the two-tailed unpaired or paired t test, as appropriate.
RESULTS
Structural features of wild-type peptides hBD1 and hBD3.
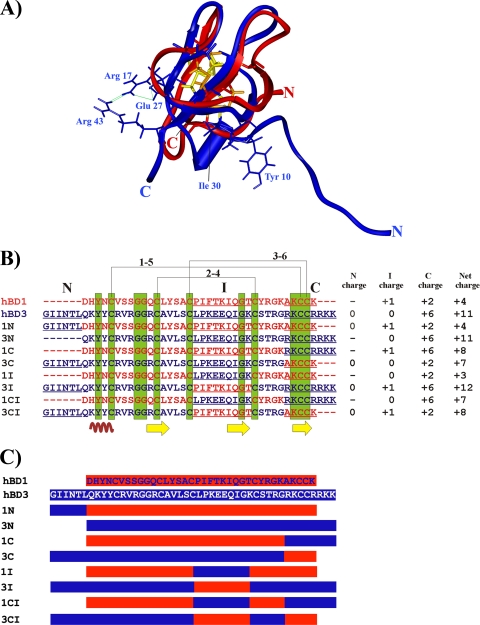
To identify the domains responsible for the antibacterial, antiviral, and chemotactic effects of hBD1 and hBD3, we looked for differences in their structures. Superimposition of the three-dimensional structures of hBD1 and hBD3 showed differences between the two molecules in the N-terminal region, the C-terminal region, and the internal domain between the third and the fourth Cys residues (Fig. 1A to C). In particular, the N-terminal helix in hBD3 starts from Lys8, whereas residues 1 to 7 are disordered. Furthermore, the N-terminal helix in hBD3 is strongly connected to the β sheet with several van der Waals contacts between Ile30 and Tyr10, whereas there are no strong, well-defined interactions in hBD1. The N-terminal region of hBD3 has four positive charges (Lys8, Arg12, Arg14, and Arg17) that are lacking in hBD1. The C-terminal region in hBD3 is packed against the internal domain, which contains two glutamic acid residues (Glu27 and Glu28), and in fact, Arg43 interacts with Arg17 of β-sheet 1, which also interacts with Glu27 of β-sheet 2. This interaction does not occur in hBD1. Furthermore, the C-terminal domain (8 amino acids) of hBD3 has more positive charges (net charge, +6) than the C-terminal domain (5 amino acids) of hBD1 (net charge, +2). The charged residues in the internal domain between the third and the fourth Cys residues also differ between hBD3 and hBD1. In particular, hBD3 has two Glu and two Lys residues, whereas hBD1 has only one Lys residue. Finally, hBD3 has more net total positive charges (+11) than hBD1 (+4).
FIG. 1.
(A) Superimposition of hBD1 (red) and hBD3 (blue) polypeptide chains. The side chains of some charged residues (blue) and disulfide bonds (orange and yellow) are also shown. (B) Sequence alignments and net charges of hBD1, hBD3, and synthetic analogs. The disulfide connectivities among the consecutively numbered cysteines are shown at the top. The hBD1 and hBD3 regions that have been substituted in the analogs and that are underlined are N (amino-terminal peptide segment), I (internal domain between Cys3 and Cys4), and C (C-terminal peptide segment). The net charges for these segments are reported on the right. Conserved residues are marked by the vertical green bands; secondary structure elements are shown at the bottom in brown (alpha helix) and yellow (beta sheet). The colored segments of sequences derive from hBD1 (red) or hBD3 (blue). (C) The colored drawings in the boxed bars indicate the regions exchanged between hBD1 and hBD3.
Design and synthesis of novel peptides.
To produce variants that contain the regions described above in different combinations, we synthesized eight analogs (the hBD1 and hBD3 regions that have been changed in the analogs are underlined or evidenced in color in Fig. 1B and C, respectively). The analogs synthesized (Fig. 1B) have net charges ranging from +3 to +12. All peptides were obtained from solid-phase peptide synthesis, and the yields of the purified linear products were good (approximately 25 to 30%). Mass spectrometry revealed the formation of three disulfide bonds in all the analogs, as in the wild-type defensins.
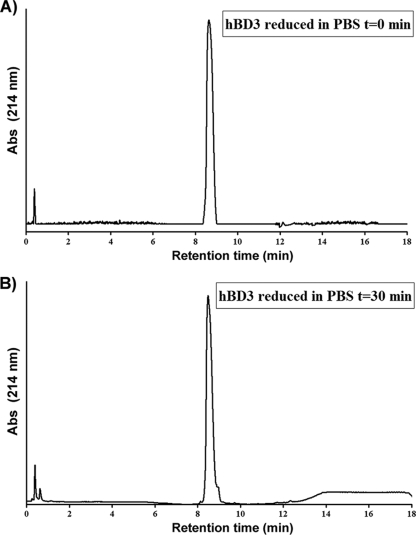
All peptides were dissolved in PBS (at pH 7.4) immediately before the start of the experiments. To verify that the dissolved peptides did not undergo any oxidation process before the assays, we analyzed the reduced hBD1 dissolved in PBS at times 0 and 30 min by RP-HPLC (Fig. 2). No oxidation occurred during the observation period.
FIG. 2.
Reduced hBD3 was dissolved in PBS (pH 7.4) and analyzed by RP-HPLC at times (t) of 0 and 30 min with a linear gradient of 10 to 50% acetonitrile over 15 min. Abs, absorbance.
Circular dichroism measurements.
We probed the structures of hBD1 and hBD3 by CD spectroscopy in an aqueous buffer and in the presence of the organic modifier TFE (Fig. 3A to D). The spectra of the reduced peptides differed greatly under the two conditions. The spectra in buffer were typical of a random coil conformation, whereas in the presence of TFE, there was a broad negative band at about 205 to 210 nm and a small negative shoulder at about 220 nm. The negative band at 205 to 210 nm indicates a β-sheet conformation. On the contrary, the spectra of the oxidized peptides in buffer and in TFE (Fig. 3B and D) are very similar, which confirms that the folding of oxidized hBD1 and hBD3 is mainly stabilized by the three disulfide bonds. The TFE spectra of the reduced and the oxidized forms are very similar. Our results confirm that, because of the absence of disulfide bridges, the reduced peptides have a less constrained structure than the oxidized peptides; however, the reduced peptides appear to fold correctly in the presence of an organic modifier. We also probed the structures of the reduced peptides 1C and 3I (Fig. 3E and F), which were the most active analogs in the antibacterial experiments; the change in the folding of the two peptides occurred in the presence of an organic modifier.
FIG. 3.
Circular dichroism spectra in HEPES buffer in the presence or absence of 40% TFE of reduced (A) and oxidized (B) hBD1, reduced (C) and oxidized (D) hBD3, reduced 1C (E), and reduced 3I (F).
Antibacterial activities of hBD1 and hBD3 and their analogs.
We tested the antibacterial activities of the hBD1 and hBD3 peptides in both their reduced and their oxidized forms. The antibacterial activities of reduced and oxidized hBD1 against P. aeruginosa were superimposable (Fig. 4A). The same result was obtained with both reduced and oxidized hBD3 (data not shown). Again, the results were similar when hBD1 and hBD3 were tested against E. coli and E. faecalis (data not shown). Therefore, in all subsequent experiments, we used only the reduced forms of hBD1 and hBD3 and their analogs.
FIG. 4.
Antibacterial activity at increasing NaCl concentrations of reduced and oxidized hBD1 (2.5 μM) against P. aeruginosa (A); of analogs 3N, 1C, and 3I (2.5 μM) against P. aeruginosa (B); and of hBD3 (2.5 μM) against P. aeruginosa, E. faecalis, and E. coli (C). Error bars show the SDs of experiments performed in triplicate.
The MICs of hBD1, hBD3, and their analogs were determined by conventional broth microdilution assays. The MIC values ranged from 12.5 to 25.0 μM for all the microorganisms tested. Peptides hBD1, hBD3, 3N, 1C, 3I, and 1CI (12.5 μM) exerted strong antibacterial effects against P. aeruginosa, E. coli, and E. faecalis.
The antibacterial activities of hBD1, hBD3, and their analogs against P. aeruginosa, E. coli, and E. faecalis, expressed as the percentage of CFU killed, are reported in Fig. 5A, B, and C, respectively. Two concentrations of each peptide (2.5 and 12.5 μM) and four concentrations of NaCl (0, 50, 100, and 200 mM) were used.
FIG. 5.
Activities (means and SDs of three replicates for each assay) of wild-type hBD1 and hBD3 and their novel analogs against P. aeruginosa (A), E. coli (B), and E. faecalis (C) at different NaCl concentrations.
As reported in Fig. 5A, at a concentration of 2.5 μM, the antibacterial activity of hBD1 against P. aeruginosa was strongly inhibited at an NaCl concentration as low as 50 mM (paired t test versus the result for medium without NaCl, P < 0.001).
On the contrary, the activity of hBD3 against P. aeruginosa was not inhibited by 50 mM NaCl; was slightly, albeit significantly, inhibited by 100 mM NaCl (paired t test versus the result for hBD3 activity at 0 mM NaCl, P < 0.01), and was greatly inhibited by 200 mM NaCl (paired t test versus the result for hBD3 activity at 0 mM NaCl, P < 0.001). At a concentration of 12.5 μM, the activity of hBD1 was again inhibited by 50 mM NaCl (paired t test versus the result for medium without NaCl, P < 0.001), while the antibacterial activity of hBD3 against P. aeruginosa was significantly inhibited only by 200 mM NaCl (paired t test versus the result for hBD3 activity at 0 mM NaCl, P < 0.001). Similarly, in the presence of 200 mM NaCl, the antibacterial activity of hBD3 at 2.5 and 12.5 μM against E. coli (Fig. 5B) and E. faecalis (Fig. 5C) was significantly inhibited compared to its activity in the absence of NaCl.
When hBD3 is considered to be the reference wild-type defensin because it had greater antibacterial activity than hBD1, the most active analogs against P. aeruginosa were 3N, 1C, and 3I (Fig. 5A). These peptides maintained good antibacterial activity at increasing NaCl concentrations and were the most active peptides at 200 mM NaCl. This can be seen more clearly in Fig. 4B, in which we compare the percentages of CFU killed by 3N, 1C, and 3I with the percentage killed by hBD3. As shown in Fig. 5A, analog 1CI also displayed good antibacterial activity at 200 mM NaCl, but it was less active than hBD3 at 50 and 100 mM NaCl. The other analogs, namely, 1N, 1I, and 3CI, had low levels of activity at increasing NaCl concentrations, particularly at 200 mM NaCl. At a peptide concentration of 12.5 μM, the pattern of antibacterial activity against P. aeruginosa was similar to that at a peptide concentration of 2.5 μM, which shows that the three analogs (3N, IC, and 3I) also had the highest levels of activity against P. aeruginosa at 200 mM NaCl. It is interesting to note that at 200 mM NaCl, the antibacterial activities of the three peptides were much higher (90% or more) than the antibacterial activity of hBD3 (10.4%).
Figures 5B and C show the activities of the analogs against the other bacterial strains, i.e., E. coli and E. faecalis, respectively. Again, peptides 3N, 1C, and 3I exerted the highest levels of antibacterial activity also at high NaCl concentrations; this was true at peptide concentrations of both 2.5 and 12.5 μM. Figure 5 also shows that the trend of the activities of all wild-type peptides and analogs at increasing NaCl concentrations against the three bacteria were similar; however, there was some difference in the level of activity. For example, as shown in Fig. 4C, at 200 mM NaCl, hBD3 at a concentration of 2.5 μM displayed a significantly higher level of activity against E. coli and E. faecalis than against P. aeruginosa (paired t test, P < 0.01 in both cases).
Antiviral activities of hBD1 and hBD3 and their analogs against herpes simplex virus type 1.
We first evaluated, using the lactate dehydrogenase assay, the viabilities of cells exposed to the wild-type defensins and to their analogs and found that they did not differ significantly from those of cells not exposed to defensins (data not shown). We then compared the antiviral effects of reduced and oxidized hBD1 and found no difference between the two forms (Fig. 6A). Therefore, we used the reduced forms of hBD1 and hBD3 and of analogs 1N, 3N, 1C, 3I, and 3C in all subsequent experiments.
FIG. 6.
Antiviral activities, tested with herpes simplex virus type 1, of reduced and oxidized wild-type hBD1 with different peptide concentrations (A); antiviral activities (cell, virus, and peptide at zero time) with increasing concentrations of hBD1 and hBD3 and analogs 1C, 1N, 3C, 3N, and 3I (B); and antiviral activities of peptides hBD1 and hBD3 and analogs 1C, 1N, 3C, 3N, and 3I (20 μM) after different treatments (see also Materials and Methods): coexposure, cell, virus, and peptide at time zero; virus pretreatment, virus and peptide before cell infection; and cell preexposure, cell and peptide before virus infection (C). Error bars show the SDs of experiments performed in triplicate.
All peptides inhibited HSV infectivity in a dose-dependent manner (Fig. 6B). The most active peptides were hBD3 (no residual infectivity at 50 μM), analog 3N (10% residual infectivity at 50 μM), analog 1C (34% residual infectivity at 50 μM), and analog 3I (27% residual infectivity at 50 μM). The three other peptides tested (hBD1, 1N, and 3C) displayed lower levels of antiviral activity at 50 μM. HSV-1 infectivity was not inhibited in experiments carried out without peptide.
We next evaluated the antiviral activities of wild-type peptides and their analogs in coexposure experiments (with cell, virus, and peptide at zero time), virus preexposure experiments (virus plus peptide before cell infection), and cell preexposure experiments (cell plus peptide before virus infection). This allowed us to evaluate if and in which phase the peptides could inhibit virus infectivity. We choose a peptide concentration of 20 μM; at this concentration, each of the peptides tested inhibited HSV infectivity (Fig. 6B), albeit with different degrees of efficacy. The most efficient peptides were hBD3 and analog 3N under all three experimental conditions (Fig. 6C); analogs 1C and 3I were particularly active when they were tested in the cell preexposure experiment. The other peptides (hBD1, 1N, and 3C) displayed lower levels of antiviral activity than hBD3 (unpaired two-tailed t test with significance set at 0.05). Viral infectivity was not inhibited in experiments carried out without peptide.
Chemotactic activities of hBD1 and hBD3 and their analogs.
To better define the antimicrobial activities of hBD1 and hBD3 and their analogs, we evaluated their chemotactic effects against neutrophils and monocytes at various NaCl concentrations. We first carried out the lactate dehydrogenase cytotoxicity assay to exclude the possibility that the two hBDs and their analogs cause monocyte and neutrophil cytolysis. We then evaluated the chemotactic effects of hBD1 and hBD3 on neutrophils and monocytes in the presence of 100, 150, and 200 mM NaCl, with 100 mM NaCl being the minimal concentration required for the assay medium. As shown in Table 1, NaCl concentrations of 150 and 200 mM did not influence the chemotactic effect of hBD1 on neutrophils compared with the effect at 100 mM. NaCl concentrations of 150 and 200 mM significantly reduced the chemotactic effect of hBD3 on neutrophils but significantly increased the chemotactic effects of both hBD1 and hBD3 on monocytes (Table 1). However, at 200 mM NaCl, the chemotactic effects of both wild-type defensins on monocytes were comparable to the effect obtained at 100 mM NaCl, which indicates that the optimal NaCl concentration for monocyte chemotaxis is 150 mM NaCl.
TABLE 1.
Chemotactic activities of wild-type hBD1 and hBD3 against neutrophils and monocytes at different NaCl concentrationsa
| hBD | NaCl concn (mM) | % migrating neutrophils | % migrating monocytes |
|---|---|---|---|
| hBD1 | 100 | 13.5 ± 3.4 | 12.5 ± 2.8 |
| 150 | 12.5 ± 1.8 | 24.3 ± 2.8b | |
| 200 | 11.2 ± 2.8 | 10.1 ± 1.4 | |
| hBD3 | 100 | 8.7 ± 2.6 | 7.3 ± 2.1 |
| 150 | 5.4 ± 2.1b | 12.8 ± 3.2b | |
| 200 | 5.0 ± 1.0b | 7.2 ± 1.6 |
The peptide concentration was 1.25 μM. The percentage of migrating cells is in reference to the total number of cells in each assay. The data represent the means ± SDs of three replicates.
Significantly different (two-tailed unpaired t test, P < 0.05) from the activity of the same molecule against the same cells at 100 mM NaCl.
Table 2 shows the chemotactic effects of all peptides on neutrophils and monocytes, irrespective of the NaCl concentration. All peptides were chemotactic for neutrophils, and the effect was greater at a peptide concentration of 1.25 μM than at higher concentrations. Therefore, we compared the chemotactic activity of each peptide at 1.25 μM to that of 1.25 μM hBD1 using the two-tailed unpaired t test with significance set at 0.05. Wild-type hBD1 and hBD3 and their analogs were chemotactic for neutrophils, albeit with some slight, not significant, differences in the magnitudes of their effects. At concentrations above 1.25 μM, the chemotactic effects of hBD1 and hBD3 on neutrophils were dramatically reduced (Table 2). Hence, we did not make any comparison between the analog peptides at a concentration of 2.5 μM.
TABLE 2.
Chemotactic activities of wild-type hBD1 and hBD3 and analogs against neutrophils and monocytesa
| BD | % migrating neutrophils at peptide concn of: |
% migrating monocytes at peptide concn of 1.25 μM | ||
|---|---|---|---|---|
| 1.25 μM | 2.5 μM | 12.5 μM | ||
| hBD1 | 13.5 ± 3.4 | 5.6 ± 2.8 | 4.3 ± 2.2 | 12.5 ± 2.8 |
| hBD3 | 8.7 ± 2.6 | 6.2 ± 2.4 | 4.9 ± 2.1 | 24.3 ± 3.1b |
| 1N | 10.4 ± 3.3 | 10.8 ± 3.3 | 3.2 ± 2.1 | 12.6 ± 2.3 |
| 3N | 11.2 ± 3.8 | 6.7 ± 3.9 | 1.5 ± 0.9 | Not tested |
| 1C | 12.8 ± 3.1 | 8.9 ± 3.5 | 3.9 ± 1.5 | 1.27 ± 0.48b |
| 1I | 9.8 ± 4.3 | 9.4 ± 4.2 | 3.5 ± 2.2 | 10.3 ± 2.3 |
| 3I | Not tested | Not tested | Not tested | 13.6 ± 2.9 |
| 1CI | 6.3 ± 2.9 | 0.0b | 0.0b | 8.6 ± 2.7 |
| 3CI | 15.5 ± 3.6 | 11.4 ± 3.8 | 3.1 ± 1.0 | 8.4 ± 2.0 |
The percentage of migrating cells is in reference to the total number of cells in each assay. Data are means ± SDs for three replicates.
Significantly different (two-tailed unpaired t test, P < 0.05) from the activity of wild-type hBD1.
Similarly, all peptides were chemotactic for monocytes when they were tested at a concentration of 1.25 μM (Table 2). In this case, we also compared the chemotactic activity of each analog at 1.25 μM to that of 1.25 μM hBD1. The chemotactic activity of peptide 1C was significantly lower than that of hBD1 (unpaired t test, P < 0.001). Wild-type hBD3 was significantly more chemotactic than hBD1 (unpaired t test, P < 0.001). The activities of the other molecules were comparable to the activity of hBD1. At a concentration above 1.25 μM, there was no further increase in chemotactic activity (data not shown).
DISCUSSION
The quest to exploit the antimicrobial activity of beta-defensins is hampered by their salt sensitivity. We have designed analogs of hBD1 and hBD3 that could have therapeutically meaningful properties.
There is some debate about whether oxidized or reduced molecules should be used to evaluate the antimicrobial activities of hBDs (32). In our hands, the reduced and oxidized forms of hBDs displayed the same antimicrobial activity. This finding is particularly interesting, considering that the three-disulfide-bond structure of hBDs is highly conserved (19). Our findings are in agreement with those of Hoover et al. (11), who showed that beta-defensins without disulfide bonds exert efficient antibacterial activity even at high salt concentrations. The latter observation is also in line with the findings described in a recent report that an hBD3 analog in which the six cysteines had been replaced with alanine residues maintained the strong antibacterial activity of the wild-type molecule (32). Defensins lack a distinct hydrophobic core, and thus, their folding is stabilized mainly by the presence of the three disulfide bonds. A previous report also indicated that linear analogs of hBD3 (1) have significantly higher levels of antimicrobial activity due to the increased structural flexibility of the linear forms. Although the mechanism whereby defensins exert their effect has yet to be established, it certainly involves contact with and permeabilization of the cell membrane through one of two models: the carpet model (several molecules sit on the surface of the cell) and the barrel stave model (the peptide oligomerizes and forms a multimeric pore in the cell membrane, thereby causing leakage of the cell contents). The CD spectra in buffer and in the presence of TFE of hBD1 and hBD3 in the reduced and oxidized forms indicate that the reduced forms of hBDs assume the correct folding and disulfide pattern, once they are placed in a solvent that mimics the membrane bilayer; thus, our results support the hypothesis that the defensins may assume the correct folding once they are in contact with the membrane bilayer. Therefore, our finding that the antimicrobial activity of hBD1 does not differ between the oxidized and the reduced forms suggests that either form may be used to evaluate their properties.
Wild-type hBD1 and hBD3 and their analogs exerted antibacterial activity over the same concentration ranges (2.5 to 12.5 μM, i.e., about 10 to 50 μg/ml) and at the same incubation time (2 h) found in previous hBD studies (16, 17, 23, 24, 30, 36). The antibacterial activity of hBD1 was inhibited by 50 mM NaCl, in agreement with previously published results (9), whereas the antibacterial activity of hBD3 was inhibited only by 200 mM NaCl. The latter observation contrasts with the notion that hBD3 is salt resistant, which emerged from studies that measured the activity of hBD3 in the presence of up to 150 mM NaCl (23, 30). Our study is the first to evaluate the antimicrobial activity of hBD3 at an NaCl concentration that is close to the mean concentration (182 mM) observed in the respiratory layer of patients with CF (29). Besides the fact that high NaCl levels in the airways of patients with CF inhibit the activity of hBDs (5, 9, 33), it is also not easy to eradicate respiratory infections in patients with CF because of the emergence of antibiotic resistance after long-term treatment (21). These considerations reinforce the need for more effective analogs.
Among the novel analogs that we synthesized in this study, the removal of the N-terminal domain of hBD3 (analog 3N) presumably changed the structure into a more compact conformation that would allow a more efficient interaction of the peptide with the bacterial surface even at the highest NaCl concentration tested. In fact, analog 1N (constituted by the hBD1 sequence plus the first 6 amino acids of the hBD3 terminus) had lower levels of antibacterial activity than hBD1 in the absence of salt. This result is in agreement with the low level of activity against E. coli and Staphylococcus aureus reported for a short peptide that contained the 17 N-terminal amino acids of hBD3 (13). More recently, a peptide of only 23 N-terminal amino acids of mouse beta-defensin 3 was found to exert strong activity against P. aeruginosa and S. aureus (22). However, it is not meaningful to compare these data with our results, given the homology of less than 70% with the human peptide.
Analog 1C (constituted by hBD1 and the C terminus of hBD3) had higher levels of antibacterial activity than the wild-type peptides at high NaCl levels, which suggests that the hBD3 C terminus is fundamental for antibacterial activity at high ionic strength. Therefore, it appears that the C-terminal region of hBD3 (which elongates the beta-3 structure of the defensin, thus making the molecule more compact) is critical for antibacterial activity at high NaCl concentrations. Interestingly, analogs spanning the C-terminal region of hBD1 to hBD3 were recently found to exert antifungal activity at various NaCl concentrations (14).
A third region that modulates antibacterial activity is the internal region of hBD1, as indicated by the high level of antibacterial activity of analog 3I (which is constituted by hBD3 with the internal region of hBD1) at high ionic strength. On the contrary, the two negatively charged Glu residues in the internal region of hBD3 impair efficient contact with the target surface, as indicated by the scarce antibacterial activity of analog 1I, in which the internal sequence of hBD1 was replaced with the hBD3 internal sequence.
The results obtained with the hBD analogs bearing two substitutions each confirm the results described above. In fact, analog 1CI, which contains the internal and the C-terminal regions of hBD3, maintains some antibacterial activity at high NaCl levels. Thus, the absence of the N terminus of hBD3, together with the presence of the C-terminal regions of hBD3, presumably makes the molecule more compact and also allows H bonding between Arg43 and Glu27, which occurs in wild-type hBD3. Analog 3CI contains the internal region and the C terminus of hBD1 but not the C terminus of hBD3 and therefore lacks the high charge and the more compact conformation conferred by those regions. Consequently, analog 3CI has a low level of antibacterial activity and low salt resistance.
Several novel findings emerged from our study of the antiviral activities of beta-defensins against HSV-1. At a concentration of 50 μM, hBD3 and the 3N analog greatly inhibited virus infectivity, whereas 1C inhibited infectivity by about 60%. This finding is in agreement with data obtained in studies with HIV (3, 34). Antiviral activity is influenced by the same domains that modulate antibacterial activity. In fact, as emerges particularly from the virus preexposure experiments, the deletion of the N-terminal part in analog 3N results in an increase in antiviral activity, whereas analog 1N, which includes the N-terminal part, has poor antiviral activity. Similarly, the C-terminal domain of hBD3 enhances antiviral activity (note the lower level of activity of analog 3C compared with that of wild-type hBD3). On the basis of our findings, it appears that defensins, particularly thanks to some of their domains, exert potent antiviral activity. We also suggest that these two regions (the N- and C-terminal domains of hBD3) influence antiviral activity via the same mechanisms postulated for antibacterial activity (8). Indeed, the inhibitory activities of defensins differ among viruses, most likely depending on the features of the viral envelope structure, including its lipid composition (8).
Regarding the issue of the chemotactic effects of hBD1 (4, 18) and hBD3 (35), here we show that both hBD1 and hBD3 are chemotactic for neutrophils, irrespective of the NaCl concentration, whereas the chemotactic activity of hBD1 against monocytes was enhanced when the salt concentration was increased from 100 to 150 mM. Furthermore, on the basis of our overall data, it appears that the N-terminal, internal, and C-terminal domains do not affect the chemotactic activity against neutrophils and that hBD3 is particularly active against monocytes.
In conclusion, we designed and synthesized analogs of the innate immunity peptides hBD1 and hBD3. We then investigated their antibacterial, antiviral, and chemotactic activities and their salt resistance. Consequently, we were able to identify the peptides and/or their domains that exert the most potent antibacterial, antiviral, and chemotactic activities. Two of these peptides, i.e., analogs 1C and 3I, are the best suited, in terms of maximizing antimicrobial activity in the presence of high NaCl concentrations, and may be of benefit for the treatment of chronic infections like those observed in patients with CF.
Acknowledgments
This work was supported by grants L.502/94 2004 (to F.S.), 2005 (to G.C.), and 2006 (to F.S.) and grant L.5/95 (to F.S.) from the Ministero della Salute and Regione Campania; grant PS 35-126/Ind from the Ministry of University and Research (to F.S.); and grant DGRC 1901/09 (from the Regione Campania) (to F.S.).
We are indebted to Jean Ann Gilder for editing the text and to Danilo Ercolini for MIC determinations.
Footnotes
Published ahead of print on 22 March 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Bai, Y., S. Liu, P. Jiang, L. Zhou, J. Li, C. Tang, C. Verma, Y. Mu, R. W. Beuerman, and K. Pervushin. 2009. Structure-dependent charge density as a determinant of antimicrobial activity of peptide analogues of defensin. Biochemistry 48:7229-7239. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, F., K. Schweimer, E. Kluver, J.-R. Conejo-Garcia, W.-G. Frossmann, P. Rosch, K. Adermann, and H. Sticht. 2001. Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity. Protein Sci. 10:2470-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole, A. M., and R. I. Lehrer. 2003. Minidefensins: antimicrobial peptides with activity against HIV-1. Curr. Pharm. Des. 9:1463-1473. [DOI] [PubMed] [Google Scholar]
- 3a.Cole, A. M., and T. Ganz. 2000. Human antimicrobial peptides: analysis and application. Biotechniques 29:822-831. [DOI] [PubMed] [Google Scholar]
- 4.Conejo-Garcia, J. R., F. Jaumann, S. Schulz, A. Krause, J. Rodriguez-Jiménez, U. Forsmann, K. Andermann, E. Klüuver, C. Vogelmeier, D. Becker, R. Hedrich, W-G. Forsmann, and R. Bals. 2001. Identification of a novel, multifunctional ß-defensin (human ß-defensin 3) with specific antimicrobial activity. Cell Tissue Res. 306:257-264. [DOI] [PubMed] [Google Scholar]
- 5.Conejo-Garcia, J. R., A. Krause, S. Schulz, F.-J. Rodriguez-Jiménez, E. Kluver, K. Adermann, U. Forssmann, A. Frimpong-Boateng, R. Bals, and W.-G. Forssmann. 2001. Human β-defensin 4: a novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J. 15:1819-1821. [PubMed] [Google Scholar]
- 6.Daher, K. A., M. E. Selsted, and R. I. Lehrer. 1986. Direct inactivation of viruses by human granulocyte defensins. J. Virol. 60:1068-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 9.Goldman, M. J., G. M. Anderson, E. D. Stolzenberg, U. P. Kari, M. Zasloff, and J. M. Wilson. 1997. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 88:553-560. [DOI] [PubMed] [Google Scholar]
- 10.Harder, J., J. Bartels, E. Christophers, and J. M. Schroder. 2001. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707-5713. [DOI] [PubMed] [Google Scholar]
- 11.Hoover, D. M., O. Chertov, and J. Lubkowski. 2001. The structure of human beta-defensin-1: new insights into structural properties of beta-defensins. J. Biol. Chem. 276:39021-39026. [DOI] [PubMed] [Google Scholar]
- 12.Hoover, D. M., K. R. Rajashankar, R. Blumenthal, A. Puri, J. J. Oppenheim, O. Chertov, and J. Lubkovski. 2000. The structure of human beta-defensin-2 shows evidence of higher order oligomerization. J. Biol. Chem. 275:32911-32918. [DOI] [PubMed] [Google Scholar]
- 13.Hoover, D. M., Z. Wu, K. Tucker, W. Lu, and J. Lubkowski. 2003. Antimicrobial characterization of human beta-defensin 3 derivatives. Antimicrob. Agents Chemother. 47:2804-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krishnakumari, V., N. Rangaraj, and R. Nagaraj. 2009. Antifungal activity of human beta defensins HBD-1 to HBD-3 and their C-terminal analogs Phd1-3. Antimicrob. Agents Chemother. 53:256-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehrer, R. I. 2004. Primate defensins. Nat. Rev. Microbiol. 2:727-738. [DOI] [PubMed] [Google Scholar]
- 16.Maisetta, G., G. Batoni, S. Esin, W. Florio, D. Bottai, F. Favilli, and M. Campa. 2006. In vitro bactericidal activity of human beta-defensin 3 against multidrug-resistant nosocomial strains. Antimicrob. Agents Chemother. 50:806-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maisetta, G., G. Batoni, S. Esin, G. Raco, D. Bottai, F. Favilli, W. Florio, and M. Campa. 2005. Susceptibility of Streptococcus mutants and Actinobacillus actinomycetemcomitans to bactericidal activity of human beta-defensin 3 in biological fluids. Antimicrob. Agents Chemother. 49:1245-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niyonsaba, F., H. Ogawa, and I. Nagaoka. 2004. Human beta-defensin-2 functions as a chemotactic agent for tumour necrosis factor-alpha-treated human neutrophils. Immunology 111:273-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pazgier, M., D. M. Hoover, D. Yang, W. Lu, and J. Lubkowski. 2006. Human beta-defensins. Cell. Mol. Life Sci. 63:1294-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinones-Mateau, M. E., M. M. Lederman, Z. Feng, B. Chakraborty, J. Weber, H. R. Rangel, M. L. Marotta, M. Mirza, B. Jiang, P. Kiser, K. Medvick, S. F. Sieg, and A. Weinberg. 2003. Human epithelial beta-defensins 2 and 3 inhibit HIV-1 replication. AIDS 17:39-48. [DOI] [PubMed] [Google Scholar]
- 21.Ratjen, F., and G. Doring. 2003. Cystic fibrosis. Lancet 361:681-689. [DOI] [PubMed] [Google Scholar]
- 22.Rohrl, J., D. Yang, J. J. Oppenheim, and T. Hehlgans. 2008. Identification and biological characterization of mouse beta-defensin 14, the orthologue of human beta-defensin 3. J. Biol. Chem. 283:5414-5419. [DOI] [PubMed] [Google Scholar]
- 23.Sahl, H. G., U. Pag, S. Bonness, S. Wagner, N. Antcheva, and A. Tossi. 2005. Mammalian defensins: structures and mechanism of antibiotic activity. J. Leukoc. Biol. 77:466-475. [DOI] [PubMed] [Google Scholar]
- 24.Sahly, H., S. Schubert, J. Harder, P. Rautenberg, U. Ullmann, J. Schroder, and R. Podschun. 2003. Burkholderia is highly resistant to human beta-defensin 3. Antimicrob. Agents Chemother. 47:1739-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawai, M. V., H. P. Jia, L. Liu, V. Aseyev, J. M. Wiencek, P. B. McCray, Jr., T. Ganz, W. R. Kearney, and B. F. Tack. 2001. The NMR structure of human beta-defensin-2 reveals a novel alpha-helical segment. Biochemistry 40:3810-3816. [DOI] [PubMed] [Google Scholar]
- 26.Schibli, D. J., H. N. Hunter, V. Aseyev, T. D. Starner, J. M. Wiencek, P. B. McCray, Jr., B. F. Tack, and H. J. Vogel. 2002. The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus. J. Biol. Chem. 277:8279-8289. [DOI] [PubMed] [Google Scholar]
- 27.Selsted, M. E., and A. J. Ouellette. 2005. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6:551-557. [DOI] [PubMed] [Google Scholar]
- 28.Sinha, S., N. Cheshenko, R. I. Lehrer, and B. C. Herold. 2003. NP-1, a rabbit alpha-defensin, prevents the entry and intercellular spread of herpes simplex virus type 2. Antimicrob. Agents Chemother. 47:494-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, J. J., S. M. Travis, E. P. Greenberg, and M. J. Welsh. 1996. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell 85:229-236. [DOI] [PubMed] [Google Scholar]
- 30.Starner, T. D., B. Agerberth, G. H. Gudmundsson, and P. B. McCray, Jr. 2005. Expression and activity of beta-defensins and LL-37 in the developing human lung. J. Immunol. 174:1608-1615. [DOI] [PubMed] [Google Scholar]
- 30a.Stem Cell Technologies. 2006. StemSep cell technical manual. Stem Cell Technologies, Vancouver, British Columbia, Canada.
- 31.Sun, L., C. M. Finnegan, T. Kish-Catalone, R. Blumenthal, P. Garzino-Demo, G. M. La Terra Maggiore, S. Berrone, C. Kleinman, Z. Wu, S. Abdelwahab, W. Lu, and A. Garzino-Demo. 2005. Human ß-defensins suppress human immunodeficiency virus infection: potential role in mucosal protection. J. Virol. 79:14318-14329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor, K., D. J. Clarke, B. McCullough, W. Chin, E. Seo, D. Yang, J. Oppenheim, D. Uhrin, J. R. W. Govan, D. J. Campopiano, D. MacMillan, P. Barran, and J. R. Dorin. 2008. Analysis and separation of residues important for the chemoattractant and antimicrobial activities of beta-defensin 3. J. Biol. Chem. 283:6631-6639. [DOI] [PubMed] [Google Scholar]
- 33.Tomita, T., S. Hitomi, T. Nagase, H. Matsui, T. Matsuse, S. Kimura, and Y. Ouchi. 2000. Effect of ions on antibacterial activity of human beta defensin 2. Microbiol. Immunol. 44:749-754. [DOI] [PubMed] [Google Scholar]
- 34.Weinberg, A., M. E. Quinones-Mateu, and M. M. Lederman. 2006. Role of human beta-defensins in HIV infection. Adv. Dent. Res. 19:42-48. [DOI] [PubMed] [Google Scholar]
- 35.Wu, Z., D. M. Hoover, D. Yang, C. Boulegue, F. Santamaria, J. J. Oppenheim, J. Lubkowski, and W. Lu. 2003. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human β-defensin 3. Proc. Natl. Acad. Sci. U. S. A. 100:8880-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yadava, P., C. Zhang, J. Sun, and J. A. Hughes. 2006. Antimicrobial activities of human beta-defensins against Bacillus species. Int. J. Antimicrob. Agents 28:132-137. [DOI] [PubMed] [Google Scholar]
- 37.Yasin, B., W. Wang, M. Pang, N. Cheshenko, T. Hong, A. J. Waring, B. C. Herold, E. A. Wagar, and R. I. Lehrer. 2004. Theta defensins protect cells from infection by herpes simplex virus by inhibiting viral adhesion and entry. J. Virol. 78:5147-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]