Abstract
AmpG is an inner membrane permease which transports products of murein sacculus degradation from the periplasm into the cytosol in Gram-negative bacteria. This process is linked to induction of the chromosomal ampC beta-lactamase gene in some members of the Enterobacteriaceae and in Pseudomonas aeruginosa. In this study, the ampG homologue of Stenotrophomonas maltophilia KJ was analyzed. The ampG homologue and its upstream ampN gene form an operon and are cotranscribed under the control of the promoter PampN. Expression from PampN was found to be independent of β-lactam exposure and ampN and ampG products. A ΔampN allele exerted a polar effect on the expression of ampG and resulted in a phenotype of null β-lactamase inducibility. Complementation assays elucidated that an intact ampN-ampG operon is essential for β-lactamase induction. Consistent with ampG of Escherichia coli, the ampN-ampG operon of S. maltophilia did not exhibit a gene dosage effect on β-lactamase expression. The AmpG permease of E. coli could complement the β-lactamase inducibility of ampN or ampG mutants of S. maltophilia, indicating that both species have the same precursor of activator ligand(s) for β-lactamase induction.
Many Gram-negative bacteria have a chromosomally encoded ampR-β-lactamase module and display an inducible β-lactamase activity, including Citrobacter freundii (20), Enterobacter cloacae (11), Pseudomonas aeruginosa (24), Burkholderia cepacia (33), and Stenotrophomonas maltophilia (28). The ampR-β-lactamase module contains a divergently transcribed control unit, in which the AmpR protein regulates the expression of the β-lactamase gene (19, 23). Two types of β-lactamase genes have been reported in such ampR-β-lactamase modules, i.e., ampC-like and class A β-lactamase genes (19).
The paradigm for regulated β-lactamase production is the ampR-ampC system of C. freundii (27, 29). The induction of the AmpC β-lactamase is controlled by the activities of AmpG, NagZ, AmpD, AmpR, and PBP4 (26). Upon challenge with β-lactam, the major murein sacculus degradation products, GlcNAc-anhMurNAc-peptide (tri-, tetra-, and pentapeptide) (also referred to as anhydromuropeptides), are transported into the cytoplasm by the cytoplasmic membrane-bound permease AmpG (17, 21). In the cytoplasm, the anhydromuropeptides can be hydrolyzed either by AmpD (N-acetylanhydromuramyl-l-alanine amidase) (10, 14) to be further recycled into UDP-MurNAc-pentapeptide or by NagZ (β-N-acetylglucosaminidase) (1, 35) to release the 1,6-anhydro-MurNAc-peptide. AmpR, a LysR family transcriptional regulator, regulates the expression of the ampC gene by the type of ligand interacting with the AmpR protein (7, 15). 1,6-anhydro-MurNAc-peptide (5, 6) and UDP- MurNAc-pentapeptides (34) act as an activator and repressor ligand, respectively.
The role of AmpG in the AmpC induction system of some ampC-bearing microorganisms has been elucidated. AmpG works as an inner membrane permease which can transport the precursors of the actual inducer from the periplasm into cytosol (21). The substrate specificity of the AmpG permease of Escherichia coli was revealed to be for the structural unit of the disaccharide, N-acetylglucosaminyl-β-1,6-anhydro-N-acetylmuramic acid (GlcNAc-anhMurNAc) (2).
S. maltophilia has an ampR-linked class A β-lactamase (L2) gene and an ampR-unlinked class B β-lactamase (L1) gene. Due to production of these two chromosomally encoded β-lactamases, S. maltophilia is inherently resistant to a wide range of β-lactams (25). To date, the genetic apparatus responsible for chromosomal class A β-lactamase induction has not been fully characterized. In silico analysis of the recently released genome sequence of S. maltophilia K279a (4) revealed five homologs of the ampC induction system, including Smlt0413, Smlt3538, Smlt1562, Smlt0154, and Smlt3723 for ampG, nagZ, ampDI, ampDII, and ampR, respectively. The functions of AmpR, AmpDI, and AmpDII of S. maltophilia were elucidated in our previous study (19, 36).
In this study, the homologue of AmpG of S. maltophilia was characterized. The ampG gene of S. maltophilia and an upstream open reading frame (ORF), named ampN, formed an operon that was shown to be critical for L1 and L2 β-lactamases induction in S. maltophilia.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
Table 1 contains a list of strains and plasmids used in this study. Tetracycline (50 μg/ml) was added to maintain the pRK415 derivatives (16). PCR primers were designed based on the S. maltophilia K279a genome sequence (4) and are listed in Table 2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or properties | Reference |
|---|---|---|
| Stenotrophomonas maltophilia | ||
| KJ | Wild type, a clinical isolate from Taiwan | 13 |
| KJN1xylEΩ | S. maltophilia KJ ampN isogenic mutant, ampN::xylE-GmΩ, inserting a xylE-GmΩ cassette into the SphI site upstream of the ampN gene | This study |
| KJN2xylEΩ | S. maltophilia KJ ampN isogenic mutant, ampN::xylE-GmΩ, inserting an xylE-GmΩ cassette into the EcoRI site of the ampN gene | This study |
| KJGxylEΩ | S. maltophilia KJ ampG isogenic mutant, ampG::xylE-GmΩ, inserting an xylE-GmΩ cassette into the SalI site of the ampG gene | This study |
| KJΔN2GxylEΩ | S. maltophilia KJ ampN deletion and ampG insertion mutant, ΔampN, ampG::xylE-GmΩ | This study |
| KJΔN2 | S. maltophilia KJ ampN deletion mutant, ΔampN | This study |
| Escherichia coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rk− mk+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
| S17-1 | recA pro thi hsdR with integrated RP4-2-tc::Mu-Kan::Tn7; Tra+ Trr Smr | 32 |
| W3110 | F−trpA23 xyl glySL | 8 |
| Plasmids | ||
| pEX18Tc | sacB oriT, Tcr | 9 |
| pRK415 | Mobilizable broad-host-range plasmid cloning vector, RK2 origin; Tcr | 16 |
| pX1918GT | Plasmid containing the xylE-gentamicin resistance cassette; Ampr Gmr | 31 |
| pTxylE | T vector with a complete xylE gene; Apr | 13 |
| pNG | pEX18Tc with a 2,692-bp ampN-ampG operon; Apr | This study |
| pN1xylEΩ | pEX18Tc with an ampN-ampG operon in which a xylE-GmΩ cassette was inserted into the SphI site upstream of ampN; Tcr | This study |
| pN2xylEΩ | pEX18Tc with a ampN-ampG operon in which a xylE-GmΩ cassette was inserted into the EcoRI site of ampN; Tcr | This study |
| pGxylEO | pEX18Tc with an ampN-ampG operon in which a xylE-GmΩ cassette was inserted into the SalI site of ampG; Tcr | This study |
| pΔN2 | Derived from pNG, deleted an internal NotI-SmaI DNA fragment of the ampN gene; Tcr | This study |
| pRK-NG | pRK415 with a 2,721-bp ampN-ampG operon; Tcr | This study |
| pRK-G | pRK415 with a complete ampG gene; Tcr | This study |
| pRK-N | pRK415 with a complete ampN gene; Tcr | This study |
| pRK-EcG | pRK415 with a complete ampG gene of E. coli; Tcr | This study |
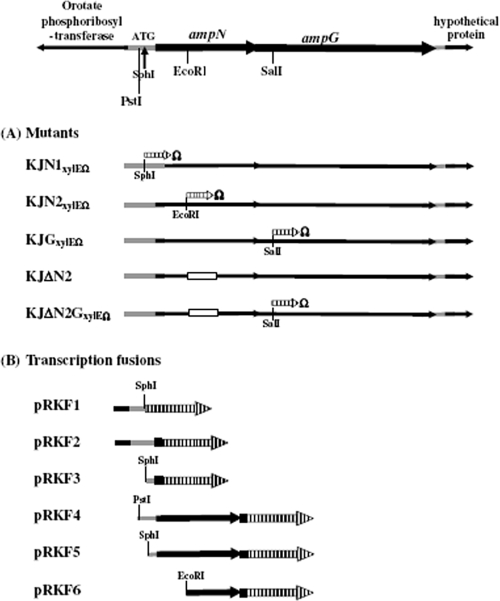
| pRKF1 | pRK415 with a 330-bp DNA fragment upstream from the correspondent start codon of S. maltophilia K279a Smlt0412 and a transcriptional fusion xylE gene | This study |
| pRKF2 | pRK415 with a 468-bp DNA fragment upstream from the amended ampN start codon and an ampN::xylE transcriptional fusion | This study |
| pRKF3 | pRK415 with a 140-bp DNA fragment upstream from the amended ampN start codon and an ampN::xylE transcriptional fusion | This study |
| pRKF4 | pRK415 with a 187-bp DNA fragment upstream from the amended ampN start codon, a complete ampN gene, and an ampG::xylE transcriptional fusion | This study |
| pRKF5 | pRK415 with a 140-bp DNA fragment upstream from the amended ampN start codon, a complete ampN gene, and an ampG::xylE transcriptional fusion | This study |
| pRKF6 | pRK415 with a 297-bp DNA fragment upstream from the ampG start codon and an ampG::xylE transcriptional fusion | This study |
TABLE 2.
Primers used in this study
| Primer name | Target gene(s) (region) | Sequence (5′ → 3′)a | Amplicon length (bp) | Purpose |
|---|---|---|---|---|
| AmpNG-F | ampN, ampG | CTGGCAAGCTTGGCATCGGCATAGCAC | 2,692 | Cloning |
| AmpNG-R | ampN, ampG | CCCTGCTCTAGAAAGGCGGCACAG | ||
| AmpG-Ec-F | ampG (E. coli) | GCCTGCAGGAATTCATCAAGCAGAAC | 1,661 | Cloning |
| AmpG-Ec-R | ampG (E. coli) | CAAACTCTAGACCGCACAAAAGAAC | ||
| F12-F | Upstream ampN and partial ampN | CTGGCGAGCTCGGCATCGGCATAGCAC | 362 | Transcriptional fusion assay |
| F1-R | Upstream ampN and partial ampN | TTGGCGTcTAGACTGATGATGCGCATG | ||
| F12-F | Upstream ampN and partial ampN | CTGGCGAGCTCGGCATCGGCATAGCAC | 509 | Transcriptional fusion assay |
| F23-R | Upstream ampN and partial ampN | AAGCTCTAGAGGCCGTCCGGGCGG | ||
| F35-F | Upstream ampN and partial ampN | CATGGAGCTCATCAGTTTCAACGCCAA | 168 | Transcriptional fusion assay |
| F23-R | Upstream ampN and partial ampN | AAGCTCTAGAGGCCGTCCGGGCGG | ||
| F4-F | Upstream ampN, ampN, and partial ampG | AGCCCTGCAGTCAACCGGTATGCTTGC | 885 | Transcriptional fusion assay |
| F456-R | Upstream ampN, ampN, and partial ampG | TGCCAAGCTTGGCGCGGCTTGGCAG | ||
| F35-F | Upstream ampN, ampN, and partial ampG | CATGGAGCTCATCAGTTTCAACGCCAA | 833 | Transcriptional fusion assay |
| F456-R | Upstream ampN, ampN, and partial ampG | TGCCAAGCTTGGCGCGGCTTGGCAG | ||
| F6-F | Partial ampN, and partial ampG | GAACTGCAGCTGCCTGCCGGAGGAACG | 335 | Transcriptional fusion assay |
| F456-R | Partial ampN, and partial ampG | TGCCAAGCTTGGCGCGGCTTGGCAG | ||
| 16rDNAQ-F | 16rDNA | GACCTTGCGCGATTGAATG | 75 | qRT-PCR |
| 16rDNAQ-R | 16rDNA | CGGATCGTCGCCTTGGT | ||
| NQ-F | ampN | TGCGGCGACTGGAACATC | 181 | qRT-PCR |
| NQ-R | ampN | GGTTGAGCAGGCGGTAGG | ||
| AmpGQ-F | ampG | CGCAAGGAAGGCATCGAG | 151 | qRT-PCR |
| AmpGQ-R | ampG | ACAGCAGCATCCAGCCAC | ||
| 0414Q-F | Smlt0414-homolog | GGACCCGCTGCTGCTGGAAC | 160 | qRT-PCR |
| 0414Q-R | Smlt0414-homolog | GGCATGGGACAGGGCATCCAC | ||
| L1Q-F | L1 β-lactamase gene | ACCCCTGGCAGATCGGCAC | 257 | qRT-PCR |
| L1Q-R | L1 β-lactamase gene | CAGCAGCACCGCCGTTTC | ||
| L2Q-F | L2 β-lactamase gene | 5′-AACGCACCCACCGATGCC-3′ | 221 | qRT-PCR |
| L2Q-R | L2 β-lactamase gene | 5′-CGCCTGTCCAGCAATGCC-3′ | ||
| 415-F | Backbone of pRK415 | CGACGACACCCGAAAAAAG | 281 | Check |
| 415-R | Backbone of pRK415 | CATTAGCAACATTATCGCACAG |
Underlining indicates the restriction sites introduced for cloning.
Cloning of ampN-ampG operon of S. maltophilia KJ.
For cloning the whole ampN-ampG operon, the PCR primers AmpNG-F and AmpNG-R (Table 2) were designed to target amplification from 331 bp upstream of the putative endonuclease gene (Smlt0412, here named ampN) to 181 bp downstream of the putative ampG gene (Smlt0413). The primers were designed with HindIII and XbaI restriction sites at each end. The PCR conditions and reaction parameters can be found elsewhere (19). Upon restriction with HindIII and XbaI, the 2.7-kb PCR product was ligated into pEX18Tc (9), previously digested with the same enzymes. The resulting plasmid was denoted as pNG.
Mutant construction.
For mutant construction, a gene replacement approach was carried out in which mutants were obtained by double-crossover homologous recombination with mutagenic plasmids (Table 1). For mutagenic plasmid construction, a xylE-GmΩ cassette retrieved from pX1918GT (31) was inserted into SphI-treated, EcoRI-treated, and SalI-treated pNG to create the mutagenic plasmids pN1xylEΩ, pN2xylEΩ, and pGxylEΩ, respectively. Plasmid pΔN2 was derived from pNG by two-round restriction digestion and ligation, in which the NotI-SmaI fragment of the ampN gene was deleted. For all mutagenic plasmids, the orientation of the xylE gene was checked by DNA sequencing to confirm that xylE and the inserted gene had the same orientation. The procedures for conjugation and mutant selection can be found elsewhere (19). The authenticity of the mutants was checked by colony PCR amplification (18) and sequencing. The double mutant was constructed from the single mutant sequentially through the same procedure.
Complementation assays.
The intact ampG gene of E. coli W3110 was amplified from the chromosome of E. coli W3110 with the primers AmpG-Ec-F and AmpG-Ec-R, which carry the embedded PstI and XbaI sites, respectively (Table 2). The 1.6-kb ampG-containing PCR amplicon was cloned into PstI-XbaI-restricted pRK415 to yield pRK-EcG. The plasmids pRK-NG, pRK-N, and pRK-G were subcloned from pNG with the compatible cloning restriction enzyme sites. The orientation of these complementary genes is the same as that of the resident lac promoter of pRK415. Each complementation plasmid was introduced into S. maltophilia via conjugation as described previously (19). As a control, the same strain was also transformed with an empty plasmid, pRK415. Acquisition of the appropriate plasmid was confirmed by a colony PCR method (18) using the paired primers 415-F and 415-R (Table 2).
Construction of promoter-xylE transcriptional fusions.
To clarify the possible promoter regions of the ampN and ampG genes, six transcriptional fusions were constructed using the pRK415 vector. Six DNA fragments, of 168 bp, 335 bp, 362 bp, 509 bp, 833 bp, and 885 bp in size, were obtained by PCR using pNG as the template. The used primers (Table 2) included restriction sites for subsequent cloning. The PCR amplicons were treated with the restriction enzymes and cloned into pRK415 at the compatible restriction sites. A xylE gene was then inserted following the amplicon fragments to produce the plasmids pRKF1 to pRKF6 (Fig. 1 and Table 1). The orientation of the xylE gene in all constructs was opposite to that of PlacZ of the pRK415 vector.
FIG. 1.
Genomic organization surrounding the ampN-ampG operon of S. maltophilia K279a. The vertical arrow indicates the annotated start codon of the ampN gene in the K279a genome. The orientation of the gene is indicated by the arrow. The crosshatched arrows represent the xylE cassette. (A) The genetic organization of the constructed mutants. The white box indicates the deleted region. The symbol Ω indicates the inserted transcriptional terminator. (B) The structures of the transcription fusions. All plasmids are derivatives of pRK415.
Determination of β-lactamase activity.
A published procedure was used for studying β-lactamase induction (19), except that cefuroxime was added as the inducer at 20 μg/ml. Enzyme activity was calculated by using a molar absorption coefficient of 20,500 M−1 cm−1 at 486 nm for nitrocefin. The specific activity was expressed as nanomoles of nitrocefin hydrolyzed per minute per milligram of protein. The protein concentration was determined using the Bio-Rad protein assay reagent, with bovine serum albumin as a standard. The differential L1 and L2 β-lactamase activity was determined by the modified nitrocefin-EDTA method (12).
Determination of catechol 2,3-dioxygenase (C23O) activity.
Catechol-2,3-dioxygenase is encoded by the xylE gene, and its activity was measured in intact cells as described previously (19). The rate of hydrolysis was calculated by using 44,000 M−1 cm−1 as the extinction coefficient. One unit of enzyme activity (Uc) was defined as the amount of enzyme that converts 1 nmol substrate per min. The specific activity was expressed as Uc/optical density at 450 nm (OD450).
Antimicrobial susceptibility test.
MICs were determined in triplicate by a standard 2-fold serial agar dilution method according to the guidelines of the Clinical Laboratory Standards Institute (CLSI) (3). All antibiotics were purchased from Sigma. The MICs of imipenem and meropenem were determined using Etest strips (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions.
qRT-PCR.
The preparation of total RNA and cDNA and quantitative real-time PCR (qRT-PCR) were carried out as described previously (36). The mRNA of 16S rRNA genes was chosen as the endogenous reference RNA for relative quantification. Relative quantities of mRNA from each gene of interest were determined by the comparative ΔΔCT method (22). Individual targets were amplified with the primers listed in Table 2. Each experiment was performed three times, and the coefficient of variation of the results was <10%.
Nucleotide sequence accession number.
The nucleotide sequence of the S. maltophilia KJ ampN-ampG operon has been deposited in GenBank under accession no. GQ469998.
RESULTS AND DISCUSSION
ampG and its upstream gene (ampN) form an operon.
In silico analysis of the ampG locus of S. maltophilia K279a (4) revealed the presence of three genes in the same orientation, including Smlt0412 (annotated as an endonuclease/exonuclease/phosphatase family protein in the genome database; designated the ampN gene hereinafter), Smlt0413 (annotated as ampG), and Smlt0414 (annotated as a hypothetical protein; abbreviated as the hp gene hereinafter).
PCR of S. maltophilia KJ chromosomal DNA with the primers AmpNG-F/AmpNG-R (Table 2) yielded a 2,692-bp amplicon, which contained two ORFs, i.e., the 663-bp ORF encoding the AmpN protein and the 1,374-bp ORF encoding the AmpG protein (Fig. 1). Since insignificant signal peptide was predicted (http://www.cbs.dtu.dk/services/SignalP/), the AmpN protein seems to be located in the cytoplasm. The AmpN and AmpG proteins of S. maltophilia KJ displayed, respectively, 99% and 99% identities to those of S. maltophilia K279a and 98% and 96% identities to those of S. maltophilia R551-3, suggesting that ampN and ampG are highly conserved alleles in S. maltophilia. Moreover, the AmpG protein of strain KJ shared an identity of less than 58% to the corresponding homologues of the different species, such as a 33% identity to AmpG of E. coli. The AmpG protein of S. maltophilia KJ was predicted to have 12 transmembrane segments (TM) with the N and C termini in the cytoplasm (http://www.cbs.dtu.dk/services/TMHMM-2.0/).
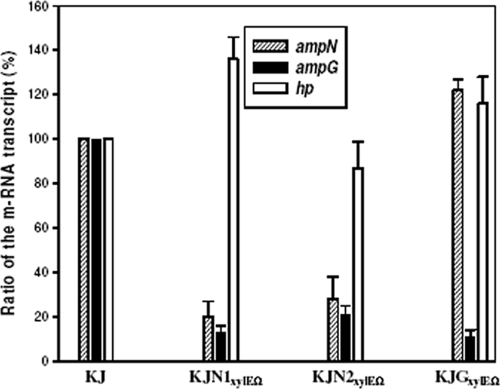
The ampN and ampG genes were found to overlap with four nucleotides, while a 77-bp intergenic region (IG) was found between ampG and the hp gene, suggesting that these three genes may form an operon. Therefore, a qRT-PCR experiment was used to measure the relative transcript ratio of the ampN, ampG, and hp genes in the parent KJ strain and in two polar mutants, KJN1xylEΩ and KJGxylEΩ. Insertion of a transcription terminator (Ω) in the ampG gene (polar mutant KJGxylEΩ) did not block the expression of the downstream hp gene (Fig. 2). Furthermore, the ampG transcript but not the hp gene-specific transcript markedly decreased in the polar mutant KJN1xylEΩ (Fig. 2), suggesting that the ampN and ampG genes can form an operon. In terms of genomic architecture, the ampN-ampG operon of S. maltophilia resembles the yajG-ampG operon of E. coli (21).
FIG. 2.
Relative ratios of ampN, ampG, and hp gene transcripts in strains KJ, KJN1xylEΩ, KJN2xylEΩ, and KJGxylEΩ. The bar indicates the ratio of a specific gene transcript of the different mutant strains to that of strain KJ, as measured by qRT-PCR. The transcript of each gene of wild-type strain KJ was assigned as 100%. The mean value is based on results of three independent experiments. The line indicates the SD.
Characterization of the ampN-ampG operon.
To further assess the relative amounts of ampN and ampG transcripts, the C23O activities of KJN1xylEΩ and KJGxylEΩ were determined. C23O activity in KJN1xylEΩ (8 Uc/OD450) was insignificant compared to the activity of 42 Uc/OD450 in KJGxylEΩ (Table 3). After careful inspection of the sequence of the ampN gene, another possible initiation codon (ATG) was noticed, 138 bp downstream from the original ATG designated by the K279a genome sequence project. The xylE-Ω cassette was inserted onto the original ATG, as indicated by the SphI site in Fig. 1, in the mutant KJN1xylEΩ, which might be the reason why no significant C23O activity was detected in KJN1xylEΩ. To verify this possibility, the mutant KJN2xylEΩ, with a xylE-Ω cassette inserted 500 bp downstream from the original ATG, was constructed, and the ampN, ampG, and hp gene transcripts and C23O activity of KJN2xylEΩ were determined. The qRT-PCR result for KJN2xylEΩ was consistent with that for KJN1xylEΩ (Fig. 2). Table 3 shows that strains KJN2xylEΩ and KJGxylEΩ exhibited equivalent C23O activities, regardless of inducers. These results support the aforementioned assumption of the initial codon and that AmpN is a 220-amino-acid (aa) polypeptide. In addition, the ampN and ampG transcripts are nearly equivalently expressed in the ampN-ampG operon. This behavior is different from that of the yajG-ampG operon of E. coli, in which the expression of ampG seems to be lower than that of the upstream yajG gene (21).
TABLE 3.
Determination of C23O and β-lactamase activities of different S. maltophilia mutants and recombinant strains
| S. maltophilia strain | C23O activity (Uca/OD450) |
β-Lactamase activity (Unb/mg) |
||
|---|---|---|---|---|
| Basal | Inducedc | Basal | Inducedc | |
| KJ | 0 | 0 | 9 ± 0.7 | 1,522 ± 192 |
| KJN1xylEΩ | 8 ± 0.5 | 4 ± 0.3 | 4 ± 0.3 | 5 ± 0.3 |
| KJN2xylEΩ | 44 ± 5.8 | 39 ± 4.3 | 3 ± 0.2 | 5 ± 0.4 |
| KJGxylEΩ | 42 ± 7.9 | 41 ± 5.6 | 5 ± 0.5 | 6 ± 0.4 |
| KJΔN2GxylEΩ | 7 ± 1.5 | 4 ± 0.3 | 3 ± 0.3 | 3 ± 0.3 |
| KJΔN2GxylEΩ(pRK415) | 6 ± 0.6 | 5 ± 0.3 | ||
| KJΔN2GxylEΩ(pRK-N) | 4 ± 0.2 | 5 ± 0.4 | ||
| KJΔN2GxylEΩ(pRK-G) | 3 ± 0.2 | 4 ± 0.3 | ||
| KJΔN2GxylEΩ(pRK-NG) | 3 ± 0.4 | 5 ± 0.2 | ||
| KJΔN2 | 5 | 6 | ||
| KJ(pRK415) | 0 | 0 | ||
| KJ(pRKF1) | 5 ± 0.4 | 6 ± 0.5 | ||
| KJ(pRKF2) | 56 ± 7.0 | 50 ± 4.9 | ||
| KJ(pRKF3) | 7 ± 0.5 | 6 ± 0.5 | ||
| KJ(pRKF4) | 51 ± 5.6 | 45 ± 5.4 | ||
| KJ(pRKF5) | 9 ± 0.9 | 6 ± 0.5 | ||
| KJ(pRKF6) | 9 ± 0.7 | 8 ± 0.5 | ||
| KJ(pRK415) | 8 ± 4.6 | 1,735 ± 123.2 | ||
| KJ(pRK-NG) | 10 ± 4.8 | 1,615 ± 130.4 | ||
| KJN2xylEΩ(pRK415) | 7 ± 0.5 | 18 ± 3.0 | ||
| KJN2xylEΩ(pRK-NG) | 6 ± 0.5 | 1,453 ± 109.5 | ||
| KJN2xylEΩ(pRK-G) | 5 ± 0.3 | 12 ± 1.0 | ||
| KJN2xylEΩ(pRK-EcG) | 5 ± 0.4 | 882 ± 90.3 | ||
One unit of C23O activity is defined as 1 nmol of catechol hydrolyzed per minute. Results are expressed as the means ± SD of three independent determinations.
One unit of β-lactamase activity is defined as 1 nmol of nitrocefin hydrolyzed per minute. Results are expressed as the means ± SD of three independent determinations.
Twenty μg/ml cefuroxime as the inducer.
To identify the promoter region of the ampN-ampG operon, the plasmid transcriptional fusion assay was performed. Table 3 shows the expressed C23O activity of transcriptional fusion constructs in the wild-type strain KJ. The background level of C23O activity was detected in KJ(pRKF1), reconfirming the results for the chromosomal transcriptional fusion construct KJN1xylEΩ. Strain KJ(pRKF4) displayed a significant C23O activity equivalent to that of KJ(pRKF2), indicating that the functional promoter PampN resides in the 187-bp DNA fragment upstream of the ampN gene, as shown by the transcriptional fusion construct pRKF4 (Fig. 1). In addition, KJ(pRKF6) displayed basal C23O activity, showing that ampG does not have its own promoter and ampG and ampN are codriven by PampN.
AmpG is involved in induction of the L1 and L2 β-lactamases.
To determine the role of AmpG in L1 and L2 β-lactamases expression, the phenotype of KJGxylEΩ was surveyed. KJGxylEΩ exhibited insignificant changes in growth rate or morphology relative to the parental strain (data not shown). The induced β-lactamase activity of the mutant KJGxylEΩ was low and was comparable to the basal activity(Table 3), suggesting that functional AmpG is required for beta-lactamase induction, likely by transporting the precursor of a possible induction ligand(s). Consistent with the fact that disruption of the ampN gene by a xylE-GmΩ cassette exerts a polar effect on the expression of the ampG gene, the KJN2xylEΩ mutant also displayed a phenotype of noninducibility (Table 3). The lack of induction of the L1 and L2 genes was also confirmed by measuring transcripts by qRT-PCR in the induced KJGxylEΩ strain (data not shown).
Characterization of ΔampN allele-mediated expression of ampG.
It has been shown that yajG of the yajG-ampG operon is not involved in either β-lactamase induction or the activity of AmpG in the Enterobacteriaceae (21). The mutant KJΔN2 resembled phenotypically the mutant KJGxylEΩ in β-lactamase activity (Table 3), indicating that the ampN gene can also be relevant to AmpG-involved β-lactamase expression.
To further elucidate whether the ΔampN allele exerts an influence on the transcriptional expression of the ampG gene, the strategy of an in vivo chromosomal transcriptional fusion study was employed. Strain KJΔN2GxylEΩ displayed a basal C23O activity of 7 Uc/OD450 (Table 3), suggesting that the transcription of ampG could be markedly impaired by the presence of the upstream ΔampN allele. To further confirm the negative effect of the ΔampN allele on ampG transcription, a qRT-PCR assay of strains KJ and KJΔN2, targeting the ampG transcript, was carried out. The qRT-PCR result showed the ampG transcript ratio of strain KJ to strain KJΔN2 was 5.27, which is in good agreement with results of the chromosomal transcriptional fusion assay.
The next task is to understand whether the basal C23O level in KJΔN2GxylEΩ was just a polar effect in the ampN-ampG operon or resulted from an AmpN- or AmpG-dependent regulation mechanism. The plasmids pRK-N, pRK-G, and pRK-NG were inserted into the mutant KJΔN2GxylEΩ for complementation, and the C23O activities of strains KJΔN2GxylEΩ(pRK415), KJΔN2GxylEΩ(pRK-N), KJΔN2GxylEΩ(pRK-G), and KJΔN2GxylEΩ(pRK-NG) were compared. The expression of the ampN and ampG genes in the complemented strains was confirmed by qRT-PCR (data not shown). Results (Table 3) showed that the complementation of ampN and/or ampG did not restore the C23O activity of KJΔN2GxylEΩ to the same level as that of KJGxylEΩ. Furthermore, the detectable C23O activity in strain KJN2xylEΩ (Table 3) confirmed that the expression of PampN is ampN and ampG independent.
AmpN and beta-lactamase induction.
Since a polar effect on the ampG gene occurs when the ampN gene is mutated, a complementation assay was performed to investigate whether ampN is involved in AmpG-associated β-lactamase expression. In strain KJN2xylEΩ(pRK-NG), the induced β-lactamase activity was restored, whereas strain KJN2xylEΩ(pRK-G) retained a low uninducible β-lactamase activity (Table 3). To assure the ampG gene in plasmid pRK-G was indeed expressed, the ampG transcript of strains KJN2xylEΩ and KJN2xylEΩ(pRK-G) was quantified by qRT-PCR. The ampG transcript ratio of KJN2xylEΩ(pRK-G) to KJN2xylEΩ was 169. Accordingly, both AmpN and AmpG are essential for β-lactamase expression in S. maltophilia.
ampN-ampG operon displays a nondosage effect on β-lactamase activity.
Korfmann and Sanders (17) have shown that in the E. cloacae system, AmpG does not have a dosage effect on the expression of the cloned ampR-ampC module. Whether a similar phenomenon occurs in S. maltophilia is thus of interest. The basal and induced β-lactamase activities of strains KJ(pRK415) and KJ(pRK-NG) were determined, as shown in Table 3. The ampG transcript of strains KJ and KJ(pRK-NG) was quantified by qRT-PCR to ensure that the ampG gene in the complementation plasmid was indeed expressed. The expression of ampG was 8-fold higher in KJ(pRK-NG) than in KJ(pRK415). Table 3 shows that under both basal and induced conditions, the strains KJ(pRK415) and KJ(pRK-NG) exhibited equivalent β-lactamase activities. This indicates that the ampN-ampG operon of S. maltophilia displays a nondosage effect on β-lactamase activity, which is consistent with that of E. coli (17).
ampG of E. coli can partially complement the ampN polar mutant of S. maltophilia.
The functional interchangeability of the AmpG permease between E. coli and S. maltophilia was tested. The β-lactamases activity of KJN2xylEΩ was partially complemented by the introduction of the plasmid pRK-EcG (Table 3), indicating that E. coli AmpG retains its biological function in S. maltophilia. The induced β-lactamase activity of KJN2xylEΩ(pRK-EcG), differentially determined by the nitrocefin-EDTA method (12), consists of 12% L1 and 88% L2 activity. Consequently, the complementation of E. coli AmpG involves the L1 and L2 β-lactamases.
The AmpG proteins of S. maltophilia and E. coli share only approximately 33% identity, suggesting that only weak amino acid sequence constraints are placed on the AmpG permease. The substrate of the E. coli AmpG permease must contain the structural unit of disaccharide, N-acetylglucosaminyl-β-1,6-anhydro-N-acetylmuramic acid (GlcNAc-anhMurNAc). The length of the peptide side chain, which is linked to anhydro-N-acetylmuramic acid (anhMurNAc), is not critical for AmpG function (2). Therefore, the precursor of the ligand activating the expression of the L1 and L2 genes can be disaccharide, disaccharide-peptides, or derivatives. However, compared to KJN2xylEΩ(pRK-NG), KJN2xylEΩ(pRK-EcG) recovers only 60% β-lactamases inducibility (Table 3). Two explanations can account for this difference. (i) In the S. maltophilia biological system, E. coli AmpG is not expressed enough for the complete complementation. (ii) AmpG of S. maltophilia and AmpG of E. coli could have different substrate specificities.
Susceptibility tests of wild-type KJ and its derived mutants.
To understand the effect of ampN and ampG inactivation on β-lactam resistance, the β-lactam MICs of ampN and ampG mutants and their complemented derivatives were also evaluated. The results of susceptibility tests (Table 4) were in overall good agreement with those of β-lactamase activity and qRT-PCR.
TABLE 4.
MICs of β-lactam antibiotics for S. maltophilia KJ and its derivatives
| Strain | MIC of antibiotic (μg/ml)a |
||||||
|---|---|---|---|---|---|---|---|
| PIP | CAR | FOX | CXM | IPM | MEM | ATM | |
| KJ | 1,024 | 1,024 | 1,024 | >1,024 | >32 | >32 | >1,024 |
| KJN1xylEΩ | 32 | 8 | 32 | 32 | 4 | 0.5 | 32 |
| KJN2xylEΩ | 16 | 2 | 32 | 32 | 2 | 0.38 | 16 |
| KJGxylEΩ | 32 | 2 | 32 | 32 | 2 | 0.38 | 16 |
| KJΔN2 | 32 | 4 | 32 | 32 | 4 | 0.38 | 32 |
| KJΔN2GxylEΩ | 16 | 2 | 32 | 32 | 2 | 0.38 | 16 |
| KJ(pRK415) | 1,024 | 1,024 | 1,024 | >1,024 | >32 | >32 | >1,024 |
| KJ(pRK-NG) | 1,024 | 1,024 | 1,024 | >1,024 | >32 | >32 | >1,024 |
| KJN2xylEΩ(pRK415) | 16 | 4 | 32 | 32 | 1.5 | 0.5 | 16 |
| KJN2xylEΩ(pRK-G) | 32 | 4 | 64 | 64 | 3 | 0.75 | 32 |
| KJN2xylEΩ(pRK-NG) | 1,024 | 1,024 | 1,024 | >1,024 | >32 | >32 | >1,024 |
| KJN2xylEΩ(pRKEc-G) | 512 | 512 | 1,024 | >1,024 | >32 | >32 | >1,024 |
PIP, piperacillin; CAR, carbenicillin; FOX, cefoxitin; CXM, cefuroxime; IPM, imipenem; MEM, meropenem; ATM, aztreonam.
Concluding remarks.
Regarding the role of ampN in ampG expression and β-lactamase induction, two conclusions can be reached based on the results of this study. (i) The ΔampN allele has a polar effect on the transcription of the downstream ampG gene. (ii) Both ampN and ampG are essential for β-lactamase induction. Furthermore, the possible function of ampN includes the following. (i) AmpN is expected to be a cytosolic protein and is likely to be involved in processing the AmpG-transported ligands to form the actual inducer for L1 and L2 induction. (ii) AmpN may assemble with AmpG to form a functional AmpN/AmpG permease. The transmembrane protein-associated operons of bacteria generally contain a gene encoding an accessory subunit for the major transmembrane protein, for example, the Nqo protein in Thermus thermophilus (30).
AmpN and AmpG, which are essential for L1 and L2 β-lactamases induction of S. maltophilia, have been identified in this study. Accordingly, in addition to the known ampR (19) and ampDI(36) genes, the L1 and L2 β-lactamases induction system of S. maltophilia can be expanded as the ampR-ampDI-ampN-ampG network.
Acknowledgments
This research was supported by grant NSC 98-2320-B-039-011-MY3 from the National Science Council and grant CMU98-S-33 from China Medical University.
Footnotes
Published ahead of print on 12 April 2010.
REFERENCES
- 1.Cheng, Q., H. Li, K. Merdek, and J. T. Park. 2000. Molecular characterization of the beta-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J. Bacteriol. 182:4836-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng, Q., and J. T. Park. 2002. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 184:6434-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing of bacteria; 14th informational supplement, M07-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.Crossman, L. C., V. C. Gould, J. M. Dow, G. S. Vernikos, A. Okazaki, M. Sebaihia, D. Saunders, C. Arrowsmith, T. Carver, N. Peters, E. Adlem, A. Kerhornou, A. Lord, L. Murphy, K. Seeger, R. Squares, S. Rutter, M. A. Quail, M. A. Rajandream, D. Harris, C. Churcher, S. D. Bentley, J. Parkhill, N. R. Thomson, and M. B. Avison. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Gen. Biol. 9:R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dietz, H., D. Pfeifle, and B. Wiedemann. 1996. Location of N-acetylmuramyl-L-alanyl-D-glutamylmesodiaminopimelic acid, presumed signal molecule for beta-lactamase induction in the bacterial cell. Antimicrob. Agents Chemother. 40:2173-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietz, H., D. Pfeifle, and B. Wiedemann. 1997. The signal molecule for beta-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob. Agents Chemother. 41:2113-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanson, N., and C. C. Sanders. 1999. Regulation of inducible AmpC beta-lactamase expression among Enterobacteriaceae. Curr. Pharm. Dis. 5:881-894. [PubMed] [Google Scholar]
- 8.Hill, C. V., and B. W. Harnish. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 78:7069-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 10.Holtje, J. V., U. Kopp, A. Ursinus, and B. Wiedemann. 1994. The negative regulator of beta-lactamase induction AmpD is a N-acetyl-anhydromuramyl-L-alanine amidase. FEMS Microbiol. Lett. 122:159-164. [DOI] [PubMed] [Google Scholar]
- 11.Honore, N., M. H. Nicolas, and S. T. Cole. 1986. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO. J. 5:3709-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu, R. M., K. H. Chiang, C. W. Lin, and T. C. Yang. 2008. Modified nitrocefin-EDTA method to differentially quantify the induced L1 and L2 β-lactamases in Stenotrophomonas maltophilia. Lett. Appl. Microbiol. 47:457-461. [DOI] [PubMed] [Google Scholar]
- 13.Hu, R. M., K. J. Huang., L. T. Wu, Y. J. Hsiao, and T. C. Yang. 2008. Induction of L1 and L2 β-lactamases of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 52:1198-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs, C., B. Joris, M. Jamin, K. Klarsov, J. Van Beeumen, D. Mengin-Lecreulx, J. van Heijenoort, J. T. Park, S. Normark, and J. M. Frère. 1995. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol. Microbiol. 15:553-559. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs, C., J. M. Frere, and S. Normark. 1997. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible beta-lactam resistance in gram-negative bacteria. Cell 88:823-832. [DOI] [PubMed] [Google Scholar]
- 16.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 17.Korfmann, G., and C. C. Sanders. 1989. ampG is essential for high-level expression of AmpC β-lactamases in Enterobacter cloacae. Antimicrob. Agents Chemother. 33:1946-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, C. W., C. S. Chiou, Y. C. Chang, and T. C. Yang. 2008. Comparison of pulsed-field gel electrophoresis and three rep-PCR methods for evaluating the genetic relatedness of Stenotrophomonas maltophilia isolates. Lett. Appl. Microbiol. 47:393-398. [DOI] [PubMed] [Google Scholar]
- 19.Lin, C. W., Y. W. Huang, R. M. Hu, K. H. Chiang, and T. C. Yang. 2009. The role of AmpR in the regulation of L1 and L2 β-lactamases in Stenotrophomonas maltophilia. Res. Microbiol. 160:152-158. [DOI] [PubMed] [Google Scholar]
- 20.Lindberg, F., L. Westman, and S. Normark. 1985. Regulatory components in Citrobacter freundii ampC beta-lactamase induction. Proc. Natl. Acad. Sci. U. S. A. 82:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindquist, S., K. Weston-Hafer, H. Schmidt, C. Pul, G. Kortmann, J. Erickson, C. Sanders, H. H. Martin, and S. Normark. 1993. AmpG, a signal transducer in chromosomal β-lactamase induction. Mol. Microbiol. 9:703-715. [DOI] [PubMed] [Google Scholar]
- 22.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(delta delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 23.Lodge, J., S. Busby, and L. Piddock. 1993. Investigation of the Pseudomonas aeruginosa ampR gene and its role at the chromosomal ampC beta-lactamase promoter. FEMS. Microbiol. Lett. 111:315-320. [DOI] [PubMed] [Google Scholar]
- 24.Lodge, J. M., S. D. Minchin, L. J. V. Piddock, and S. J. W. Busby. 1990. Cloning, sequencing and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal ampC β-lactamase. Biochem. J. 272:627-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Looney, W. J., M. Natrit, and K. Muhlemann. 2009. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect. Dis. 9:312-323. [DOI] [PubMed] [Google Scholar]
- 26.Moya, B., A. Dotsch, C. Juan, J. Blazquez, L. Zamorano, S. Haussler, and A. Oliver. 2009. Beta-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog. 5(3):e1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Normark, S. 1995. Beta-lactamase induction in gram-negative bacteria is intimately linked to peptidoglycan recycling. Microb. Drug Resist. 1:111-114. [DOI] [PubMed] [Google Scholar]
- 28.Okazaki, A., and M. B. Avison. 2008. Induction of L1 and L2 beta-lactamase production in Stenotrophomonas maltophilia is dependent on an AmpR-type regulator. Antimicrob. Agents Chemother. 52:1525-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, J. T., and T. Uehara. 2008. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72:211-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sazanov, L. A., and P. Hinchiffe. 2006. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311:1430-1436. [DOI] [PubMed] [Google Scholar]
- 31.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 32.Simon, R., M. O'Connell, M. Labes, and A. Puhler. 1986. Plasmid vectors for the genetic analysis and manipulation of Rhizobia and other Gram-negative bacteria. Methods Enzymol. 118:640-659. [DOI] [PubMed] [Google Scholar]
- 33.Trepanier, S., A. Prince, and A. Huletsky. 1997. Characterization of the penA and penR genes of Burkholderia cepacia 249 which encode the chromosomal class A penicillinase and its LysR-type transcriptional regulator. Antimicrob. Agents Chemother. 41:2399-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uehara, U., and J. T. Park. 2002. Role of the murein precursor UDP-N-acetylmuramyl-L-Ala-gamma-D-Glu-meso-disminopimelic acid-D-Ala-D-Ala in repression of beta-lactamase induction in cell division mutants. J. Bacteriol. 184:4233-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Votsch, W., and M. F. Templin. 2000. Characterization of a beta-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and beta-lactamase induction. J. Biol. Chem. 275:39032-39038. [DOI] [PubMed] [Google Scholar]
- 36.Yang, T. C., Y. W. Huang, R. M. Hu, S. C. Huang, and Y. T. Lin. 2009. AmpDI is involved in expression of the chromosomal L1 and L2 β-lactamases of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 53:2902-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]