Abstract
Genotypic interpretation systems (GISs) for darunavir and tipranavir susceptibility are rarely tested by the use of independent data sets. The virtual phenotype (the phenotype determined by Virco [the “Vircotype”]) was used to interpret all genotypes in Québec, Canada, and phenotypes were determined for isolates predicted to be resistant to all protease inhibitors other than darunavir and tipranavir. We used multivariate analyses to predict relative phenotypic susceptibility to darunavir and tipranavir. We compared the performance characteristics of the Agence Nationale de Recherche sur le Sida scoring algorithm, the Stanford HIV database scoring algorithm (with separate analyses of the discrete and numerical scores), the Vircotype, and the darunavir and tipranavir manufacturers' scores for prediction of the phenotype. Of the 100 isolates whose phenotypes were determined, 89 and 72 were susceptible to darunavir and tipranavir, respectively. In multivariate analyses, the presence of I84V and V82T and the lack of L10F predicted that the isolates would be more susceptible to darunavir than tipranavir. The presence of I54L, V32I, and I47V predicted that the isolates would be more susceptible to tipranavir. All GISs except the system that provided the Stanford HIV database discrete score performed well in predicting the darunavir resistance phenotype (R2 = 0.61 to 0.69); the R2 value for the Stanford HIV database discrete scoring system was 0.38. Other than the system that provided the Vircotype (R2 = 0.80), all GISs performed poorly in predicting the tipranavir resistance phenotype (R2 = 0.00 to 0.31). In this independent cohort harboring highly protease inhibitor-resistant HIV isolates, reduced phenotypic susceptibility to darunavir and tipranavir was rare. Generally, GISs predict susceptibility to darunavir substantially better than they predict susceptibility to tipranavir.
HIV-infected patients harboring multidrug-resistant virus face limited protease inhibitor (PI) treatment options. Studies suggest that approximately 10% of HIV-infected patients initiating therapy experience triple-class treatment failure (18), although this rate may be declining as the treatment options improve (4). For those patients given therapy prior to the availability of highly active antiretroviral therapy, the rate of triple-class resistance exceeds 20% (25a).
Approved in June 2005, tipranavir is a nonpeptidic PI specifically developed for the management of patients harboring PI-resistant virus. In clinical trials, the use of tipranavir resulted in a virologic response superior to that achieved with the comparator PIs (11). However, tipranavir requires twice-daily dosing and coadministration with 200 mg of ritonavir and food and has many drug-drug interactions and an adverse side effect profile. Patients given tipranavir have increased rates of hepatotoxicity, hyperlipidemia, rash, and therapy discontinuation than patients receiving a comparator PI; and the use of tipranavir has been linked to intracranial hemorrhage (1).
In June 2006, the U.S. Food and Drug Administration approved twice-daily darunavir coadministered with 100 mg ritonavir for use by treatment-experienced adults. The use of darunavir resulted in a virologic response superior to that achieved with the comparator PIs in patients harboring PI-resistant virus (2, 16). In those pivotal studies, darunavir had a side effect profile similar to that of the comparator PIs, other than a lower incidence of diarrhea.
While darunavir and tipranavir are both active against isolates highly resistant to PIs, they have not been compared directly in clinical studies, although one analysis suggests that darunavir may have more activity (12). Given the lack of clinical data, the interpretation of genotypic resistance test results is often used to choose between darunavir and tipranavir.
Over 20 genotypic interpretation systems (GISs) have been developed to interpret the complex patterns of amino acid substitutions seen with PI-associated resistance (14, 15, 20), but their performance characteristics have rarely been compared. Given the complexity of PI-associated resistance, a phenotype can be determined to better characterize resistance, although clinical data in support of such a strategy are limited (17). For drugs for which limited clinical data are available, such as darunavir and tipranavir, the phenotype may provide a more reliable measure of the activity of a drug, as the phenotype measures in vitro susceptibility to specific drugs and the genotype analyzes the sequence information of the virus and infers drug resistance from the mutations present.
In the study described here we compared the ability of several of the most commonly used GISs (those of the Agence Nationale de Recherche sur le Sida [ANRS] and the Stanford HIV database, the Virco system for the determination of the virtual phenotype [Vircotype], and the darunavir and tipranavir manufacturers' scores) to predict the phenotypes for resistance to darunavir and tipranavir for a set of highly resistant clinical isolates and developed a model that may be used to predict the relative susceptibility to darunavir and tipranavir (4a, 21, 23; www.hivfrenchresistance.org/; Virco).
(This study was presented at the 5th International AIDS Society [IAS] Conference on HIV Pathogenesis, Treatment, and Prevention, 19 to 22 July 2009, Cape Town, South Africa [abstr. WEPEB202].)
MATERIALS AND METHODS
In the province of Québec, Canada, all genotypic resistance tests are performed centrally and the interpretation of the results is provided by Virco. These results are referred to as the Vircotypes. For establishment of these Vircotypes (the Virco HIV-1 resistance genotypes), the genotype-phenotype pairs available within the company's database are used to predict the phenotype on the basis of the patient's genotype. In Québec, per protocol, a phenotype (the antivirogram phenotype) is determined for isolates predicted to be resistant to all PIs other than darunavir and tipranavir on the basis of the Vircotype.
For the present study, we included isolates collected from January 2007 through July 2008 for which the genotype, Vircotype, and phenotype were available. For each isolate included in the study, a score was generated for each GIS of interest for darunavir and tipranavir. For the ANRS system, the score was categorized as 0 for susceptible, 1 for intermediate, and 2 for resistant. For the Stanford HIV database algorithm, we analyzed both the five-way discrete score and the numerical score (e.g., from 0 to 60+) produced by the algorithm. The discrete score was categorized as 0 for susceptible, 1 for potentially low-level resistance, 2 for low-level resistance, 3 for intermediate resistance, and 4 for high-level resistance. Both ANRS and the Stanford HIV database occasionally update their scoring systems. We used the versions publicly available in September 2008. The Vircotype was categorized as 0 for susceptible/maximal response, 1 for reduced response, and 2 for resistant/minimal response, on the basis of the cutoffs for darunavir and tipranavir established by Virco in September 2008 and applied to all the isolates. We used the current darunavir manufacturer's score, based on the number of the 11 mutations present in a given isolate, and the weighted tipranavir manufacturer's score, which includes 19 mutations of interest (4a, 23).
We plotted each score versus the natural log of the fold change in susceptibility, as this transformation to the fold change led to the best linear correlation between the two variables. We compared the predictive ability of each score with an R2 calculation.
For the real phenotype, we used the clinical cutoffs (CCOs) established by Virco (the darunavir CCO1 is a 10-fold change, the darunavir CCO2 is a 40-fold change, the tipranavir CCO1 is a 3-fold change, and the tipranavir CCO is a 10-fold change). We constructed models to predict relative phenotypic susceptibility to darunavir and tipranavir. We performed forward stepwise multivariate logistic regression with the Akaike information criterion (AIC), used for variable entry, to predict which isolates had increased susceptibility to darunavir compared to their susceptibility to tipranavir (and increased susceptibility to tipranavir compared to their susceptibility to darunavir), on the basis of the CCO for each drug. We performed 2,500 bootstrap replicates of this AIC model selection technique to estimate the proportion of times that the AIC would select given mutations (6, 26). We limited the number of mutations in each darunavir-versus-tipranavir and tipranavir-versus-darunavir comparison and model to 8 and 5, respectively, to limit overfitting.
RESULTS
During the study period, approximately 4,200 genotypic resistance tests were performed in Québec; among the isolates evaluated in those tests, 55% presented reduced susceptibility to at least one drug. One hundred isolates were resistant to all PIs, excluding darunavir and tipranavir, and phenotypic resistance testing was performed with those isolates.
Of those 100 isolates, the majority were susceptible to both drugs; 89 were susceptible to darunavir and 72 were susceptible to tipranavir (Table 1). Two isolates had intermediate susceptibility to darunavir and nine were fully resistant. For tipranavir, 20 isolates were reported to have intermediate susceptibility and 8 were fully resistant. Only two isolates were resistant to both darunavir and tipranavir.
TABLE 1.
Profile of phenotypic resistance to darunavir and tipranavir among 100 PI-resistant isolates
| Tipranavir susceptibilitya | No. of isolates with the following darunavir susceptibilityb |
||
|---|---|---|---|
| Sensitive | Intermediate | Resistant | |
| Sensitive | 67 | 2 | 3 |
| Intermediate | 16 | 0 | 4 |
| Resistant | 6 | 0 | 2 |
Tipranavir fold change cutoffs were <3 for sensitive, 3 to <10 for intermediate, and ≥10 for resistant.
Darunavir fold change cutoffs were <10 for sensitive, 10 to <40 for intermediate, and ≥40 for resistant.
As expected, the 100 isolates were highly PI resistant. They had a median of 12.0 (interquartile range [IQR] = 10.0, 14.0) PI-associated mutations and median numbers of 1.0 (IQR = 0.0, 2.0) and 4.0 (IQR = 3.0, 6.0) darunavir and tipranavir mutations, respectively, according to the IAS-USA mutation list (13). Figure 1 displays the frequency of amino acid changes at various positions in the protease sequences of the isolates. On the basis of the IAS-USA mutation list, the most frequent darunavir resistance-conferring mutations were I84V, L33F, and V32I, which occurred at frequencies of 42%, 32%, and 15%, respectively. The most frequent tipranavir resistance-conferring mutations were at L90M, M36I, and I54V, which occurred at frequencies of 67%, 62%, and 52%, respectively.
FIG. 1.
Frequency of mutations by protease position (n = 100). T, position of IAS-USA major tipranavir resistance-conferring mutation; D, position of IAS-USA major darunavir resistance-conferring mutation (13).
For darunavir, all GISs except the Stanford HIV database discrete score algorithm performed similarly well in predicting the darunavir resistance phenotype (R2 = 0.61 to 0.69; Fig. 2); the Stanford HIV database discrete score was less predictive (R2 = 0.38). The category “intermediate resistance” from the Stanford HIV database algorithm encompasses a wide range of numerical scores (from 30 to 59), and 46 of the 100 isolates were categorized as having intermediate resistance. Comparison of Fig. 2b and Fig. 2c shows that a substantial amount of discriminatory ability was lost by using the discrete category rather than the numerical score for this group, as the fold change appeared to increase with higher scores within the intermediate resistance group.
FIG. 2.
Correlation between genotypic interpretation scores and natural log of fold change for darunavir (DRV). (a) ANRS score versus natural log of the darunavir fold change; (b) Stanford HIV database (HIVDB) discrete score versus natural log of the darunavir fold change; (c) Stanford HIV database numerical score versus natural log of the darunavir fold change; (d) Vircotype versus natural log of the darunavir fold change; (e) darunavir score versus natural log of the darunavir fold change.
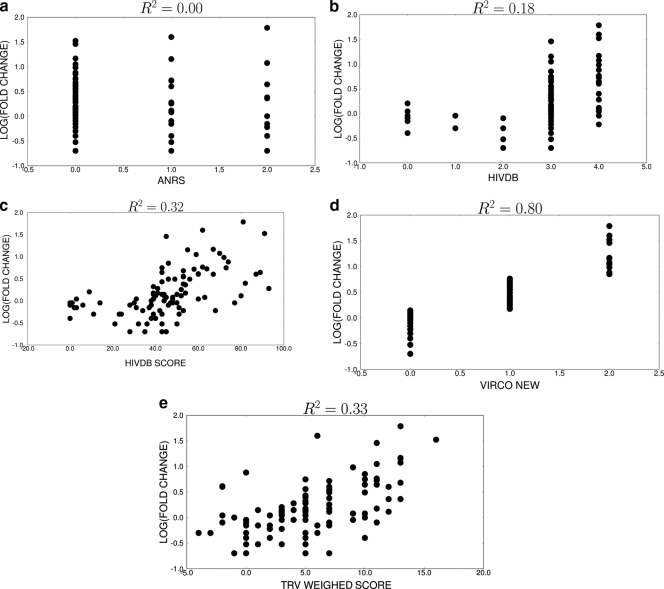
In general, the GISs for tipranavir performed poorly (Fig. 3). The interpretation produced by the ANRS system was not significantly associated with the natural log of the fold change (R2 = 0.00; P = 0.62). The Stanford HIV database scores (both the discrete and the numerical scores) and the tipranavir manufacturer's scores were minimally predictive of the fold change (R2 = 0.18 to 0.33). Interestingly, the Vircotype was unique in its ability to predict the tipranavir fold change (R2 = 0.80).
FIG. 3.
Correlation between genotypic interpretation scores and natural log of fold change for tipranavir (TPV). (a) ANRS score versus natural log of the tipranavir fold change; (b) Stanford HIV Database discrete score versus natural log of the tipranavir fold change; (c) Stanford HIV Database numerical score versus natural log of the tipranavir fold change; (d) Vircotype versus natural log of the tipranavir fold change; (e) tipranavir score versus natural log of tipranavir fold change.
In bootstrap analyses, the presence of I84V and V82T and the lack of L10F were the most robust predictors that isolates would be more susceptible to darunavir than tipranavir (Table 2). I84V was selected in 97% of the bootstrap models and had a mean odds ratio of 1.5 (95% confidence interval [CI], 1.2, 1.8) for relative darunavir susceptibility. That is, isolates with I84V were 50% more likely to be less resistant (on the basis of the phenotypic cutoffs) to darunavir than to tipranavir than the typical isolate within the cohort. V82T was selected in 86% of the models and had a mean odds ratio of 1.6 (95% CI, 1.3, 2.1). The lack of L10F was selected in 82% of the bootstrap models, with its lack being associated with a mean odds ratio of 1.4 (95% CI, 1.2, 1.6).
TABLE 2.
Mutations most strongly associated with relative darunavir susceptibility versus tipranavir susceptibility
| Mutation | Mean odds ratio (95% CI) | % of models | Mean rank in models |
|---|---|---|---|
| 82T | 1.6 (1.3, 2.1) | 86 | 2.5 |
| 84V | 1.5 (1.2, 1.8) | 97 | 1.9 |
| Lack of 10F | 1.4 (1.2, 1.6) | 82 | 3.3 |
The presence of I54L, V32I, and I47V predicted that the isolates would be more susceptible to tipranavir than darunavir (Table 3). I54L was selected in 99% of the models and had a mean odds ratio of 2.0 (95% CI, 1.5, 2.7) for predicting relative tipranavir susceptibility over relative darunavir susceptibility. V32I was selected in 94% of the models and had a mean odds ratio of 1.4 (95% CI, 1.1, 1.9), and I47V was selected in 88% of the models and had a mean odds ratio of 1.7 (95% CI, 1.2, 2.7).
TABLE 3.
Mutations most strongly associated with relative tipranavir susceptibility versus darunavir susceptibility
| Mutation | Mean odds ratio (95% CI) | % of models | Mean rank in models |
|---|---|---|---|
| 54L | 2.0 (1.5, 2.7) | 99 | 1.5 |
| 47V | 1.7 (1.2, 2.7) | 88 | 2.3 |
| 32I | 1.4 (1.1, 1.9) | 94 | 2.3 |
When these mutations occurred together, the combined presence of I84V and V82T and the lack of L10F resulted in an odds ratio of 2.8 (95% CI, 1.6, 5.0) for relatively greater darunavir susceptibility than tipranavir susceptibility. The presence of I54L, V32I, and I47V together predicted that the isolates would be more susceptible to tipranavir than to darunavir and had an odds ratio of 4.4 (95% CI, 2.7, 7.3).
DISCUSSION
Various studies have evaluated the performance characteristics of different GISs in predicting susceptibility to first-generation PIs by the use of independent data sets (8, 10, 19). However, the performance of GISs with an independent data set for darunavir and tipranavir has rarely been evaluated. Independent data sets are important for the evaluation of GISs, as a score created on the basis of a given data set will always perform well due to some degree of model overfitting. Even if a portion of the data set is used as a training data set and the remainder is used as a validation data set, the similarities of the treatment histories and other patient characteristics between the training and the validation data sets will likely also lead to the overly good performance of a given scoring system.
Within our collection of highly PI-resistant HIV isolates, the GISs performed well in predicting phenotypic susceptibility to darunavir. However, the Stanford HIV database discrete score performed less well, as it appeared that the intermediate resistance category was too broad and included isolates with a wide range of fold changes. The suggestion that there was a misclassification in the intermediate group is supported by the findings presented in Fig. 2b, in which there is a wide range of phenotypes in the intermediate category, and also by the fact that the numerical score performed quite well and had an R2 value of 0.68, consistent with the R2 values of the other GISs tested. The Stanford HIV database numerical score is readily available on the website when one enters a genotype into the web-based system, but clinicians frequently focus only on the discrete score when interpreting a genotype. Given these results, we would encourage clinicians to be sure to review the numerical score when deciding on the use of darunavir. Additionally, as a result of the findings of this study and others, the authors of the Stanford HIV database have revised their scores for darunavir to attempt to better predict darunavir susceptibility.
We found that the ability of the GISs to interpret tipranavir susceptibility was generally poor. GISs make qualitative judgments of susceptibility on the basis of the available clinical and laboratory data. As new results become available, the system is updated to incorporate the new data. The relative lack of publicly available clinical information on tipranavir may help explain the poor performance of rules-based GISs. Another possible explanation is that the development of genotypic resistance to tipranavir may be inherently more complex than the development of genotypic resistance to darunavir because several diverse pathways can lead to resistance to tipranavir. For instance, IAS-USA lists 21 resistance-conferring mutations for tipranavir and 11 for darunavir (13). With this complexity, it may be more difficult to create a rules-based algorithm for tipranavir. Our finding is consistent with the findings of others, who have also found that GISs do not perform well in evaluating tipranavir susceptibility (22).
The superior performance of the Vircotype for predicting phenotypic resistance to tipranavir was surprising. In previous studies, the virtual phenotype has not outperformed rules-based GISs in predicting the virologic response or phenotypic resistance (9, 25). However, Virco's access to a large pool of proprietary data for tipranavir may have allowed the Vircotype to have superior performance. In addition, the algorithm that Virco uses to correlate genotypic resistance to phenotypic resistance may be superior to rules-based algorithms in predicting phenotypic resistance for a drug with as complex a pattern of resistance as tipranavir.
We found the presence of I84V and V82T and the lack of L10F to be associated with relative susceptibility to darunavir compared to the level of susceptibility to tipranavir. The importance of V82T as an important predictor of relative darunavir susceptibility is not surprising, as V82T is a signature mutation for resistance to tipranavir and has not been associated with decreased darunavir susceptibility (7). On the basis of the tipranavir manufacturer's score, the presence of V82T alone is sufficient for reduced tipranavir susceptibility (10). I84V has been associated with decreased susceptibility to both darunavir and tipranavir in clinical studies (4a, 23), but in a number of scoring systems, I84V is given more weight in tipranavir resistance scores than in darunavir resistance scores (4a, 21, 23). I84V was also shown to be one of the first mutations to emerge during in vitro passage experiments with tipranavir (4a). L10F is a relatively common PI-associated mutation and has not been considered an important mutation conferring resistance to darunavir or tipranavir. The lack of the L10F mutation was unexpectedly associated with relative darunavir susceptibility, and its relevance should be confirmed by additional studies. It may be that the L10F is a proxy for other resistance-associated mutations that together affected the relative susceptibilities of these two new-generation PIs.
Not surprisingly, the presence of I54L was an important predictor of relative tipranavir susceptibility. It has been associated with an improved virologic response to tipranavir and has been given a negative weighting (the inverse of resistance) within the tipranavir manufacturer's score, while it not considered an important mutation for darunavir resistance (4a, 23). The emergence of V32I as an important mutation predicting relative tipranavir susceptibility is also consistent with the findings of previous work (4a, 13, 23). On the other hand, I47V has been associated with decreased responses to both darunavir and tipranavir, and its validity as a predictor of relative tipranavir susceptibility should be explored within other independent data sets. Again, this mutation may simply be a marker for other associated mutations that are present within our cohort.
The validation of GISs can be performed through genotype-clinical outcome correlation studies or with correlations with phenotypic resistance testing, as was done in this study. We did not have access to clinical data, which was a limitation of the study. Another limitation of the study is the limited number of clinical isolates that were available, which may not have allowed us to detect some important mutations for darunavir and tipranavir resistance.
Genotypic resistance testing does not routinely sequence gag, while the Antivirogram phenotype assay incorporates only a portion of the C terminus of gag. Mutations in gag not only have been associated with restored replicative capacity but also have been independently associated with protease resistance (3). The clinical utility of evaluating gag during resistance testing has not been established, and since the currently available interpretation algorithms do not include gag mutations, we cannot speculate how the results of our study would have varied had these mutations been included. Nonetheless, our results do reflect the results that can be obtained by using the interpretation tools currently available to the clinician.
In patients harboring virus with extensive resistance to PIs, the choice of the PI to be used for salvage therapy is usually darunavir or tipranavir. By generating rules for predicting relative darunavir susceptibility versus relative tipranavir susceptibility, we attempt to provide guidance to clinicians for when one drug may be more active than the other, although additional studies are needed to validate the mutations selected by our models. Additionally, the activities of other drugs in the background regimen (especially etravirine, which cannot be coadministered with tipranavir) and the tolerability of the drugs will also be a factor in the selection of the appropriate PI (www.accessdata.fda.gov/drugsatfda_docs/label/2009/022187s002lbl.pdf). Furthermore, with the availability of a new class of agents, for some, the requirement for PIs in salvage therapy is less obvious (24).
In conclusion, for the set of isolates with high-level resistance to PIs evaluated in the present study, it was reassuring that the majority retained susceptibility to both darunavir and tipranavir. GISs effectively predict susceptibility to darunavir. However, other than use of the Vircotype, GISs cannot be relied upon to predict phenotypic susceptibility to tipranavir, but specific mutations may predict which isolates have relative tipranavir susceptibility over relative darunavir susceptibility. However, due to darunavir's ease of use and the fact that it retains activity even against highly resistant isolates, the role of tipranavir in salvage therapy will remain limited and tipranavir might be considered for use only in patients harboring the subgroup of isolates with the pattern of mutations that favor the use of tipranavir over darunavir.
Acknowledgments
Michel Roger has received grant support from the Fonds de la Recherche en Santé du Québec (FRSQ).
Footnotes
Published ahead of print on 5 April 2010.
REFERENCES
- 1.Chan-Tack, K. M., K. A. Struble, and D. B. Birnkrant. 2008. Intracranial hemorrhage and liver-associated deaths associated with tipranavir/ritonavir: review of cases from the FDA's Adverse Event Reporting System. AIDS Patient Care STDs 22:843-850. [DOI] [PubMed] [Google Scholar]
- 2.Clotet, B., N. Bellos, J. M. Molina, D. Cooper, J. C. Goffard, A. Lazzarin, A. Wöhrmann, C. Katlama, T. Wilkin, R. Haubrich, C. Cohen, C. Farthing, D. Jayaweera, M. Markowitz, P. Ruane, S. Spinosa-Guzman, and E. Lefebvre. 2007. Efficacy and safety of darunavir-ritonavir at week 48 in treatment-experienced patients with HIV-1 infection in POWER 1 and 2: a pooled subgroup analysis of data from two randomized trials. Lancet 369:1169-1178. [DOI] [PubMed] [Google Scholar]
- 3.Dam, E., R. Quercia, B. Glass, D. Descamps, O. Launay, X. Duval, H. G. Kräusslich, A. J. Hance, F. Clavel, and ANRS 109 Study Group. 2009. Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Pathog. 5:e1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deeks, S. G., S. J. Gange, M. M. Kitahata, M. S. Saag, A. C. Justice, R. S. Hogg, J. J. Eron, J. T. Brooks, S. B. Rourke, M. J. Gill, R. J. Bosch, C. A. Benson, A. C. Collier, J. N. Martin, M. B. Klein, L. P. Jacobson, B. Rodriguez, T. R. Sterling, G. D. Kirk, S. Napravnik, A. R. Rachlis, L. M. Calzavara, M. A. Horberg, M. J. Silverberg, K. A. Gebo, M. B. Kushel, J. J. Goedert, R. G. McKaig, and R. D. Moore. 2009. Trends in multidrug treatment failure and subsequent mortality among antiretroviral therapy-experienced patients with HIV-infection in North America. Clin. Infect. Dis. 49:1582-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.De Meyer, S., I. Dierynck, and E. Lathouwers. 2008. Abstr. 6th Eur. HIV Drug Resist. Workshop, abstr. 54.
- 5.Reference deleted.
- 6.Efron, B., and R. Tibshirani. 1993. Introduction to the bootstrap. Chapman & Hall/CRC Press, Boca Raton, FL.
- 7.Elston, R. C., D. R. Kuritzkes, and R. Bethell. 2007. An investigation into the influence of the tipranavir-associated V82L/T mutations on the susceptibility to darunavir and brecanavir. Abstr. 14th Conf. Retrovir. Opportunistic Infect., abstr. 602.
- 8.Fox, Z. V., A. M. Geretti, J. Kjaer, U. B. Dragsted, A. N. Phillips, J. Gerstoft, S. Staszewski, B. Clotet, V. von Wyl, and J. D. Lundgren. 2007. The ability of four genotypic interpretation systems to predict virological response to ritonavir-boosted protease inhibitors. AIDS 21:2033-2042. [DOI] [PubMed] [Google Scholar]
- 9.Gallego, O., L. Martin-Carbonero, C. Aguero, J. de Mendoza, A. Corral, and V. Soriano. 2004. Correlation between rules-based interpretation and virtual phenotype interpretation of HIV-1 genotypes for predicting drug resistance in HIV-infected individuals. J. Virol. Methods 121:115-118. [DOI] [PubMed] [Google Scholar]
- 10.Garrido, C., E. Poveda, A. Corral, C. Mendoza, and V. Soriano. 2008. Low rate of tipranavir and darunavir resistance mutations in PI-experienced patients using the new interpretation scores—implications for PI sequencing. Antivir. Ther. 13(Suppl.):A107. [Google Scholar]
- 11.Hicks, C. B., P. Cahn, D. A. Cooper, S. L. Walmsley, C. Katlama, B. Clotet, A. Lazzarin, M. A. Johnson, D. Neubacher, D. Mayers, H. Valdez, and the RESIST Investigator Group. 2006. Durable efficacy of tipranavir-ritonavir in combination with an optimised background regimen of antiretroviral drugs for treatment-experimented HIV-1 infected patients at 48 weeks in the randomized evaluation of strategic intervention in drug resistant patients with tipranavir (RESIST) studies: an analysis of combined data from two open-label trials. Lancet 368:466-475. [DOI] [PubMed] [Google Scholar]
- 12.Hill, A., and G. Moyle. 2007. Relative antiviral efficacy of ritonavir-boosted darunavir and ritonavir-boosted tipranavir vs control protease inhibitor in the POWER and RESIST trials. HIV Med. 8:259-264. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, V. A., F. Vézina-Brun, B. Clotet, H. F. Gunthard, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2008. Update of the drug resistance mutations in HIV-1. Top. HIV Med. 16:138-145. [PubMed] [Google Scholar]
- 14.Kijak, G. H., A. E. Rubio, S. E. Pampuro, C. Zala, P. Cahn, R. Galli, J. S. Montaner, and H. Salomón. 2003. Discrepant results in the interpretation of HIV-1 drug-resistance genotypic data among widely used algorithms. HIV Med. 4:72-78. [DOI] [PubMed] [Google Scholar]
- 15.Luca, A., and C. F. Perno. 2003. Impact of different HIV resistance interpretations by distinct systems on clinical utility of resistance testing. Curr. Opin. Infect. Dis. 16:573-580. [DOI] [PubMed] [Google Scholar]
- 16.Madruga, J. V., D. Berger, M. McMurchie, F. Suter, D. Banhegyi, K. Ruxrungtham, D. Norris, E. Lefebvre, M. P. de Béthune, F. Tomaka, M. De Pauw, T. Vangeneugden, and S. Spinosa-Guzman. 2007. Efficacy and safety of darunavir-ritonavir compared with that of lopinavir-ritonavir at 48 weeks in treatment-experienced, HIV-infected patients in TITAN: a randomised controlled phase III trial. Lancet 370:49-58. [DOI] [PubMed] [Google Scholar]
- 17.Meynard, J. L., M. Vray, L. Morand-Joubert, E. Race, D. Descamps, G. Peytavin, S. Matheron, C. Lamotte, S. Guiramand, D. Costagliola, F. Brun-Vézinet, F. Clavel, and P. M. Girard. 2002. Phenotypic or genotypic resistance testing for choosing antiretroviral therapy after treatment failure: a randomized trial. AIDS 16:727-736. [DOI] [PubMed] [Google Scholar]
- 18.Phillips, A. N., C. Leen, A. Wilson, J. Anderson, D. Dunn, A. Schwenk, C. Orkin, T. Hill, M. Fisher, J. Walsh, D. Pillay, L. Bansi, B. Gazzard, P. Easterbrook, R. Gilson, M. Johnson, and C. A. Sabin. 2007. Risk of extensive virological failure to the three original antiretroviral drug classes over long-term follow-up from the start of therapy in patients with HIV infection: an observational cohort study. Lancet 370:1923-1928. [DOI] [PubMed] [Google Scholar]
- 19.Poveda, E., C. de Mendoza, L. Martin-Carbonero, A. Corral, V. Briz, J. González-Lahoz, and V. Soriano. 2007. Prevalence of darunavir resistance mutations in HIV-1 infected patients failing other protease inhibitors. J. Antimicrob. Chemother. 60:885-888. [DOI] [PubMed] [Google Scholar]
- 20.Ravela, J., B. J. Betts, F. Brun-Vézinet, A. M. Vandamme, D. Descamps, K. van Laethem, K. Smith, J. M. Schapiro, D. L. Winslow, C. Reid, and R. W. Shafer. 2003. HIV-1 protease and reverse transcriptase mutation patterns responsible for discordances between genotypic drug resistance interpretation algorithms. J. Acquir. Immune Defic. Syndr. 33:8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee, S., M. Gonzales, R. Kantor, B. J. Betts, J. Ravela, and R. W. Shafer. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saracino, A., L. Monno, A. Tartaglia, C. Tinelli, E. Seminari, F. Maggiolo, S. Bonora, S. Rusconi, V. Micheli, S. Lo Caputo, L. Lazzaroni, S. Ferrara, N. Ladisa, P. Nasta, G. Parruti, R. Bellagamba, F. Forbici, and G. Angarano. 2009. Clinical validation and applicability of different tipranavir/ritonavir genotypic scores in HIV-1 protease inhibitor-experienced patients. Curr. HIV Res. 7:425-433. [DOI] [PubMed] [Google Scholar]
- 23.Scherer, J., C. Boucher, J. D. Baxter, J. M. Schapiro, V. Kohlbrenner, and D. Hall. 2007. Improving the prediction of virologic response to tipranavir: the development of a tipranavir weighted score. Abstr. 11th Eur. AIDS Conf., abstr. P3.4/07.
- 24.Skiest, D., C. Cohen, B. Barker, M. Gottlieb, K. Mounzer, P. Bellman, E. Dejesus, H. Khanlou, B. Rashbaum, C. B. Hsiao, P. Ruane, K. Abriola, N. Bellos, D. Ward, J. Lalezari, F. Santiago, J. Garb, and A. Habel. 2009. Raltegravir without a protease inhibitor is highly efficacious in heavily pre-treated individuals, abstr. MOPEB072. Abstr. 5th Conf. HIV Pathog. Treatment Prev.
- 25.Torti, C., E. Quiros-Roldan, W. Keulen, L. Scudeller, S. Lo Caputo, C. Boucher, F. Castelli, F. Mazzotta, P. Pierotti, A. M. Been-Tiktak, G. Buccoliero, M. De Gennaro, G. Carosi, and C. Tinelli. 2003. Comparison between rules-based human immunodeficiency virus type 1 genotype interpretations and real or virtual phenotype: concordance analysis and correlation with clinical outcome in heavily treated patients. J. Infect. Dis. 188:194-201. [DOI] [PubMed] [Google Scholar]
- 25a.Zaccarelli, M., P. Lorenzini, and V. Tozzi. 2008. Abstr. 6th Eur. HIV Drug Resist. Workshop, abstr. 42.
- 26.Zolopa, A. R., R. W. Shafer, A. Warford, J. G. Montoya, P. Hsu, D. Katzenstein, T. C. Merigan, and B. Efron. 1999. HIV-1 genotypic resistance patterns predict response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann. Intern. Med. 131:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]