Abstract
Fourier transform infrared (FT-IR) spectroscopy and chemometric techniques were used to discriminate five closely related Salmonella enterica serotype Enteritidis phage types, phage type 1 (PT1), PT1b, PT4b, PT6, and PT6a. Intact cells and outer membrane protein (OMP) extracts from bacterial cell membranes were subjected to FT-IR analysis in transmittance mode. Spectra were collected over a wavenumber range from 4,000 to 600 cm−1. Partial least-squares discriminant analysis (PLS-DA) was used to develop calibration models based on preprocessed FT-IR spectra. The analysis based on OMP extracts provided greater separation between the Salmonella Enteritidis PT1-PT1b, PT4b, and PT6-PT6a groups than the intact cell analysis. When these three phage type groups were considered, the method based on OMP extract FT-IR spectra was 100% accurate. Moreover, complementary local models that considered only the PT1-PT1b and PT6-PT6a groups were developed, and the level of discrimination increased. PT1 and PT1b isolates were differentiated successfully with the local model using the entire OMP extract spectrum (98.3% correct predictions), whereas the accuracy of discrimination between PT6 and PT6a isolates was 86.0%. Isolates belonging to different phage types (PT19, PT20, and PT21) were used with the model to test its robustness. For the first time it was demonstrated that FT-IR analysis of OMP extracts can be used for construction of robust models that allow fast and accurate discrimination of different Salmonella Enteritidis phage types.
Over the past 10 years there has been an increase in the incidence of gastrointestinal infections caused by Salmonella enterica serovar Enteritidis, which is now one of the leading S. enterica serotypes worldwide (21, 27). Poultry, poultry products, cattle, and dairy products are the predominant sources of Salmonella-contaminated food products that cause human salmonellosis (28). Large-scale infections continue to occur in developed countries (8). Unrestricted international movement of commercially prepared food and food ingredients and dissimilarities in government and industry food safety controls during the processing, distribution, and marketing of products have surely contributed to the increase in food-borne outbreaks. Salmonella is a tremendous challenge for the agricultural and food processing industries because of its ability to survive under adverse conditions, such as low levels of nutrients and suboptimal temperatures (4, 13).
Salmonella Enteritidis isolates can be categorized for epidemiological purposes by using a variety of typing tools (13). These tools include typing techniques such as serological and phage typing (29) and antibiotic resistance patterns (25). These methods are now supplemented by molecular genetics techniques, such as DNA fingerprinting (23), plasmid profiling (16), and pulsed-field gel electrophoresis (26). Phage typing has been used to diagnose Salmonella outbreaks, including S. enterica serovar Typhi and S. enterica serovar Typhimurium outbreaks (29). It is useful to evaluate whether isolates obtained from different sources at different times are similar or distinct in terms of their reactions with a specific collection of bacteriophages used for typing. The correlation between phage type and the source of an epidemic is high (22). Although very effective, existing classification methods are time-consuming, laborious, and expensive, and they often require special training of personnel and expertise, which can prevent a rapid response to the presence of pathogenic bacterial species.
Fourier transform infrared (FT-IR) spectroscopy has been successfully used for differentiation and classification of microorganisms at the species and subspecies levels (7, 9, 12, 15, 18, 19, 20). This technique has been shown to have high discriminatory power and allows identification of bacteria at distinct taxonomic levels based on differences in the infrared absorption patterns of microbial cells. FT-IR spectroscopy has been used to differentiate and characterize intact microbial cells based on outer membrane cell components, including lipopolysaccharides (LPS), lipoproteins, and phospholipids (24). Several studies in which S. enterica serotypes have been discriminated using multivariate data analysis and FT-IR spectroscopy have been performed (1, 2, 10, 11). Kim et al. (11) compared the FT-IR spectra of intact cells and the FT-IR spectra of outer membrane protein (OMP) extracts from S. enterica serotypes to discriminate serotypes. Analysis of spectra of OMP extracts in the 1,800- to 1,500-cm−1 region resulted in 100% correct classification of the serotypes investigated.
Previously, there have been no reports of differentiation of Salmonella Enteritidis phage types by FT-IR spectroscopy and chemometric methods. To discriminate closely related phage types of Salmonella Enteritidis in this study, intact cells and OMP extracts of bacterial cell membranes were subjected to FT-IR analysis. The isolates analyzed included isolates belonging to five of the phage types of Salmonella Enteritidis found most frequently in Portuguese hospitals in the period from 2004 to 2006, phage type 1 (PT1), PT1b, PT4b, PT6, and PT6a (5, 14). Chemometric models were used to discriminate between phage types based on infrared spectra.
MATERIALS AND METHODS
Bacterial strains.
The isolates used in this study were obtained from strain collections of the Enterobacteriaceae Unity of the Bacteriology Center of the Portuguese National Health Institute Ricardo Jorge (Lisbon, Portugal) and originated from clinical and food samples. A total of 42 isolates of S. enterica serotype Enteritidis were analyzed (7 PT1 isolates, 13 PT1b isolates, 9 PT4b isolates, 6 PT6 isolates, and 7 PT6a isolates). Eight additional isolates belonging to phage types 19, 20, and 21 were also analyzed and used to test the robustness of the method used.
Phage typing: reference method.
The Salmonella isolates were analyzed at the Enterobacteriaceae Unity of the Bacteriology Center of the Portuguese National Health Institute Ricardo Jorge (Lisbon, Portugal) by using a serologic identification method, including agglutination with specific sera (29). Isolates of S. Enteritidis were inoculated into tubes containing nutrient broth, incubated at 37°C, and centrifuged at 10,000 × g for 60 min. The bacterial suspensions were inoculated onto plates containing nutrient agar to obtain confluent cultures. The medium surface was dried at 37°C for about 60 min. After this, a phage applicator was used to apply 16 phages specific for S. Enteritidis. The plates were incubated at 37°C for 16 to 18 h. The lysed areas were observed with oblique light. The results were interpreted on the basis of the lysis patterns, using the method of Ward et al. (29). The typing scheme for the Salmonella Enteritidis phage types used in this study is shown in Table 1.
TABLE 1.
Typing scheme for Salmonella Enteritidis phage types used in this study
| Phage type | Results for the following phages at routine test dilutiona: |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | H | F | O | Q | P | L | J1 | J2 | J3 | J4 | R | S | J5 | 15 | 16 | |
| 1b | OL | SCL | CL | <OL | CL | SCL | CL | OL | <OL | OL | CL | CL | CL | CL | OL | OL |
| 1 | OL | SCL | CL | <OL | CL | SCL | CL | OL | OL | OL | CL | CL | CL | CL | − | − |
| 4b | − | SCL | CL | <OL | CL | SCL | CL | OL | <OL | OL | CL | CL | CL | − | − | <SCL |
| 6 | − | SCL | − | SCL | − | SCL | − | OL | <OL | OL | − | − | − | − | − | − |
| 6a | − | SCL | − | SCL | − | SCL | − | − | <OL | − | − | − | − | − | − | − |
SCL, semiconfluent lysis; CL, confluent lysis; OL, opaque lysis; <SCL and <OL, intermediate levels of lysis.
Preparation of outer membrane protein extracts.
Isolates were cultured in tryptose soya agar (TSA) at 37°C for 16 to 18 h. The outer membrane proteins of S. Enteritidis isolates were individually extracted using the procedure proposed by Kim et al. (11), with modifications. Briefly, an overgrown culture was harvested by centrifugation (MBC centrifuge; Hawksley, England) at 10,000 × g for 10 min at 4°C. At the time of harvesting, the cells were at the same growth stage (end of exponential phase). The pellets were resuspended in 1 ml of 10 mM Tris-HCl buffer (pH 8.0), and the suspensions were sonicated three times for 10 s using a Vibracell Sonifier (Bioblock Scientific, Illkirch, France) and were then centrifuged (5402 centrifuge; Eppendorf, Engelsdorf, Germany) at 14,000 × g for 45 min at 4°C. Each pellet was resuspended in 1 ml of a solution containing 10 mM Tris-HCl buffer (pH 8.0) and N-lauroyl-sarcosine (Sigma-Aldrich GmbH, Germany). After incubation at room temperature for 20 min, the suspensions were centrifuged at 14,000 × g for 45 min at 4°C, and each pellet was suspended in 120 ml of distilled water.
Preparation of intact cells.
The strains were cultured on TSA at 37°C for 24 h. Individually selected colonies of each strain were carefully harvested from plates with a calibrated 1-μl loop. The colonies were suspended in 125 μl of sterile distilled water and homogenized with a vortex mixer.
Phenotypic typing (FT-IR method).
Aliquots (35 μl) of the bacterial suspensions and OMP extracts were evenly applied to wells in a silicium plate (an optical plate on which bacterial suspensions were dried to obtain a transparent film suitable for FT-IR measurement). The amounts of extracted OMP were equivalent for all samples, although the precise amounts were not determined. Prior to analysis, samples were oven dried at 44°C for 30 min. Each sample was measured in triplicate, and the entire procedure was repeated on two consecutive days (using two different bacterial suspensions). The procedure was repeated on 2 days with different suspensions to ensure that the method was reproducible. Spectra was collected with an FT-IR TENSOR spectrometer (Bruker Optik GmbH, Karlsruhe, Germany) in transmittance mode at wavenumbers from 4,000 to 600 cm−1 with a resolution of 4 cm−1 using OPUS software, version 5.0 (Bruker Optik GmbH, Karlsruhe, Germany). The OPUS spectral quality control test for the FT-IR spectra was used routinely with thresholds for the minimum absorbance (0.345) and maximum absorbance (1.245) for detector linearity, signal-to-noise (S/N) ratio, and water vapor. Spectra that did not pass the quality control test were not used for further analysis. If more than two spectra for one isolate did not pass the quality control test, the data for the isolate were not included in the data set. Therefore, despite the fact that the intact cell and OMP extract spectra were obtained for the same isolates, the numbers of isolates used in the analysis were not the same for the two sampling methods. A total of 45 isolates were analyzed (intact cells and OMP extracts). Forty-two of these isolates passed the quality control test for intact cells, and 38 of these isolates passed the quality control test for OMP extracts. Because the quality control test results were assessed several days after the spectra were collected (we used the ultimate thresholds described by Bruker), it was not practical to repeat the analysis with isolates that did not pass the quality control test. For each isolate that passed the quality control test, the mean spectrum was obtained and stored until it was analyzed. Two FT-IR spectra were therefore available for each isolate, corresponding to the two bacterial suspensions of the same isolate analyzed (referred to as replicates below). During routine analysis, when spectra for new samples were acquired, the operator had to verify that the quality control test was passed. If it was not, the spectrum acquisition procedure had to be repeated.
Data analysis.
The calculations for all models were carried out using Matlab (version 6.5, release 13; MathWorks, Natick, MA) and the PLS Toolbox (version 3.5 for Matlab; Eigenvector Research, Manson, WA). The Savitzky-Golay filter, multiplicative scatter correction, and standard normal variate were evaluated as preprocessing methods (3). The entire wavenumber region was considered in the analysis (4,000 to 600 cm−1) since no improvement was obtained if spectral regions were selected. The partial least-squares discriminant analysis (PLS-DA) algorithm was used to develop calibration models for discrimination of bacteria (6, 30). The models were based on the mean-centered preprocessed spectral data. PLS-DA requires appropriate validation to prevent overfitting. The procedure used was based on division of the original data set into a training set and a validation set (17); 70% of the data were selected for calibration, and the remaining 30% were used for validation. Isolates were selected randomly, but the following two rules were followed: (i) the proportion of phage types in the original isolates was maintained in both the calibration and validation sets (to ensure that there were correctly balanced calibration and validation data sets) and (ii) the two replicates of each isolate were always assigned to the calibration or validation set (to prevent overfitting). PLS-DA models that considered different numbers of latent variables were constructed based on the calibration data set and were tested with the validation set. The serologic identification method results were used as targets for the PLS-DA models. The PLS-DA model that yielded the lowest validation error was kept. However, the results could depend on a particular training-validation division. Therefore, the entire procedure was repeated 200 times. The final results were obtained by averaging the results of the 200 PLS-DA optimal models. The results were expressed in the form of confusion matrices. The confusion matrices compared the phage type of each isolate (obtained by the reference serologic identification method) with the corresponding FT-IR-based prediction. The results were expressed as percentages.
RESULTS AND DISCUSSION
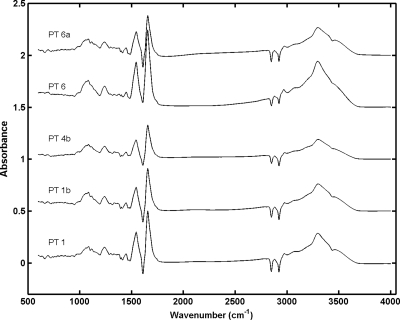
Normalized spectra of intact cells for the region from 600 to 4,000 cm−1 were compared with the corresponding spectra of OMP extracts (Fig. 1). Difference spectra were obtained by subtracting the normalized intact cell spectra from the normalized OMP extract spectra for one isolate belonging to each phage type analyzed (the standard normal variate normalization procedure was employed). Spectral differences were very important over the entire spectral range, as expected. The spectra of intact cells did not differ much for the phage types analyzed, whereas there were some visible dissimilarities between spectra of OMP extracts, especially in the lipid region (2,800 to 3,000 cm−1) and the phospholipid-DNA-RNA region (1,200 to 1,500 cm−1).
FIG. 1.
FT-IR difference spectra for intact cells and OMP extracts for one isolate of each S. Enteritidis phage type used in this study.
Preprocessing of the spectra was required before the spectra were modeled. Spectra were preprocessed using multiplicative scatter correction to eliminate light scattering effects. A Savitzky-Golay filter was also applied to reduce noise, and the second derivative was considered. The Savitzky-Golay filter parameters were a 13.5-cm−1 filter window and a third-order polynomial and second derivative. Preprocessed spectra were also mean centered prior to PLS-DA modeling. Despite the fact that other preprocessing methods (e.g., standard normal variate or vector normalization) could be used for the same purpose, the processing methods used yielded the best modeling results. Use of the second derivative also maximized the accuracy of the method.
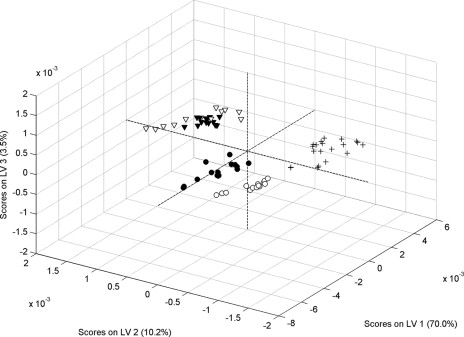
The discriminating features in the FT-IR spectra of intact cells and OMP extracts were evaluated by developing two PLS-DA classifications. Initially, the entire spectral range (600 to 4,000 cm−1) was considered in the analysis. The results of the two PLS-DA models are shown as confusion matrices in Table 2 for intact cells and OMP extracts. The two main PLS-DA model scores are also shown in Fig. 2 for intact cells and OMP extracts. Data in Table 2 demonstrate that using the intact cell spectra was not enough to identify the correct phage types of isolates. The results obtained for OMP show that it was possible to obtain a rate of correct predictions of 90.7%. Additionally, this model correctly identified all isolates belonging to PT4b. It must be emphasized that the results were obtained using the average for 200 different models (different training-validation sets). This means that 100% correct predictions for PT4b were obtained for each of the 200 models, indicating that the method's accuracy was excellent. The PLS-DA scores shown in Fig. 2 indicate that good separation was possible using the OMP spectra. The results also demonstrate that no PT1 or PT1b isolate was assigned to PT6 or PT6a and vice versa. There were incorrect predictions only within the PT1-PT1b and PT6-PT6a groups. Therefore, the OMP extract FT-IR spectra could be used to discriminate between the PT1-PT1b, PT4b, and PT6-PT6a groups. When these three phage type groups were considered, the method based on OMP extract FT-IR spectra was 100% accurate.
TABLE 2.
Confusion matrices for S. Enteritidis intact cell- and OMP extract-based PLS-DA discrimination models for phage types 1, 1b, 4b, 6, and 6a
| Model | FT-IR method | Reference methoda |
|||||
|---|---|---|---|---|---|---|---|
| PT1 | PT1b | PT4b | PT6 | PT6a | Total | ||
| Intact cell | PT1 | 10.0 | 1.1 | 1.8 | 0.9 | 0.4 | 14.2 |
| PT1b | 1.3 | 18.2 | 5.3 | 0.9 | 6.4 | 32.2 | |
| PT4b | 6.7 | 3.3 | 9.1 | 5.1 | 1.6 | 25.8 | |
| PT6 | 2.0 | 1.1 | 2.7 | 6.2 | 2.9 | 14.9 | |
| PT6a | 0.0 | 2.9 | 1.1 | 0.2 | 8.7 | 12.9 | |
| Total | 20.0 | 26.7 | 20.0 | 13.3 | 20.0 | 100.0 | |
| OMP extract | PT1 | 19.6 | 0.4 | 0.0 | 0.0 | 0.0 | 20.0 |
| PT1b | 1.8 | 21.1 | 0.0 | 0.0 | 0.0 | 22.9 | |
| PT4b | 0.0 | 0.0 | 21.4 | 0.0 | 0.0 | 21.4 | |
| PT6 | 0.0 | 0.0 | 0.0 | 19.3 | 5.0 | 24.3 | |
| PT6a | 0.0 | 0.0 | 0.0 | 2.1 | 9.3 | 11.4 | |
| Total | 21.4 | 21.4 | 21.4 | 21.4 | 14.3 | 100.0 | |
The values are percentages.
FIG. 2.
PLS-DA scores for the discrimination models for S. Enteritidis phage types 1, 1b, 4b, 6, and 6a using FT-IR spectra of OMP extracts. •, PT1; ○, PT1b; +, PT4b; ▾, PT6; ▿, PT6a. LV, latent variable.
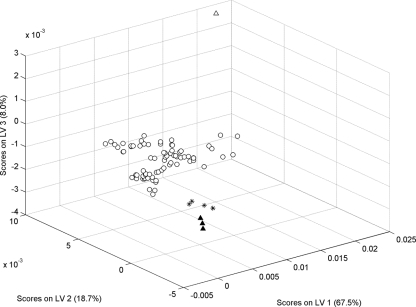
To assess the robustness of the proposed method, whether it was able to identify isolates that did not belong to the five phage types modeled had to be determined. We expected that when an isolate belonging to a different phage type was examined with this model, it would be identified as different or not belonging to any of the phage types included. Therefore, eight isolates belonging to phage types different than those used to calibrate the model were examined, and the results were analyzed. These isolates belong to PT19, PT20, and PT21. They were not used for development of the method because only a small number of isolates were available. However, they could be used to test the robustness of the model. Spectra of these isolates were examined with the PLS-DA model that was developed, and the resulting scores were compared with the scores obtained for the other phage types. It was observed (Fig. 3) that not only were these scores identified by the model as outliers (they fell outside the range covered by PT1, PT1b, PT4b, PT6, and PT6a, as shown in the score plot), but they also appeared to cluster according the corresponding phage type (a further indication of the ability of the method to discriminate between phage types). Because the number of isolates with these projected phage types was limited, we could not draw any conclusion regarding their classification, but the results clearly indicate that the method is able to detect isolates that do not belong to the phage types used for calibration, which is a guarantee of model robustness. In practice, the new samples should be considered outliers, and phage type predictions for these isolates would have no validity.
FIG. 3.
PLS-DA scores obtained when eight isolates belonging to PT19, PT20, and PT21 were projected onto the model. ○, PT1, PT1b, PT4b, PT6, and PT6a; ▴, PT19; ▵, PT20; *, PT21. LV, latent variable.
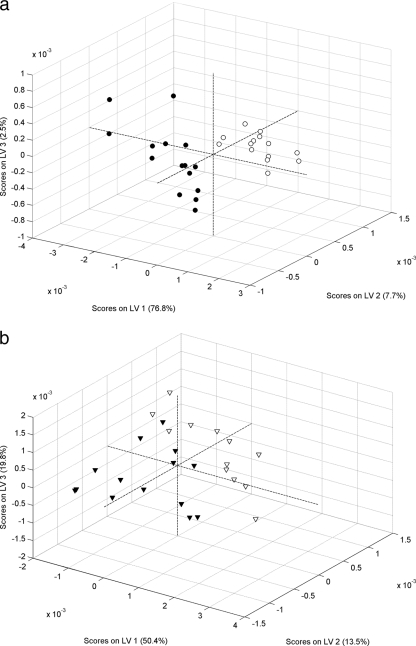
The analysis described above was performed using the entire wavenumber region. To better assess the importance of different spectral regions in the discrimination models, the analysis was repeated for intact cell and OMP extract FT-IR spectra by considering subsets of the entire spectral range. FT-IR spectra of bacteria contain information from all chemical structures of the cells. However, it is common to subdivide the spectral range according to some of the principal biological building blocks, including lipids (2,800 to 3,000 cm−1), proteins and amides I and II (1,500 to 1,700 cm−1), phospholipids, DNA, and RNA (1,200 to 1,500 cm−1), polysaccharides (900 to 1,200 cm−1), and the fingerprint region (600 to 900 cm−1). More information about the specific absorption in the infrared region of the main biochemical blocks is available elsewhere (19). To determine whether better discrimination could be obtained by considering each spectral window separately, five PLS-DA models were built using the intact cell spectra and five models were built using the OMP extract spectra. The rates of correct predictions were then compared to the rates obtained when the entire spectral range was used. The results obtained (not shown) showed that the best model was the model obtained when the entire spectral range was used both for intact cells and OMP extracts. Previous results suggested that the model that aimed at discriminating all phage types simultaneously could not successfully discriminate within the PT1-PT1b and PT6-PT6a groups. Additionally, the model based on OMP extracts performed better, especially for discriminating between PT1 and PT1b. Local models considering only the PT1-PT1b and PT6-PT6a groups could identify the variability within the phage type groups and thus complement the main model. In order to confirm the possible discrimination within the closely related PT 1-PT1b and PT 6-PT6a groups, two local PLS-DA models using the entire spectral range were developed on the basis of FT-IR spectra of OMP extracts; one of these local models considered only PT1 and PT1b isolates, and the other local model considered only PT6 and PT6a isolates. The results are shown in Table 3 in the form of confusion matrices. It was clear that the level of discrimination within the PT1-PT1b and PT6-PT6a groups increased when the local models were developed. The error rate obtained for discrimination between PT1 and PT1b was 1.7%, which is significantly lower than the 5% error rate obtained for the global model (Table 2). A similar result was obtained for discrimination between PT6 and PT6a isolates. The error rate for the OMP extract-based local model was 14%, which is lower than the 20% error rate obtained with the global model (Table 2). While successful discrimination between PT1 and PT1b isolates was obtained with the local model (98.3% correct predictions), the same was not true for discrimination between PT6 and PT6a isolates (only 86% correct predictions). The significant level of misclassification obtained for the latter isolates could not be overcome by using different spectrum preprocessing techniques or by restricting the wavenumber regions. As described above, for each model different spectral windows were used to determine if better predictions could be obtained. However, our conclusion was that the best model was obtained when the entire spectral range was considered. The main PLS-DA scores for the two models are shown in Fig. 4a and 4b. The analysis of the PLS-DA scores confirmed that a better model was obtained for the model discriminating PT1 and PT1b (better score separation). Nevertheless, the scores for the model discriminating PT6 and PT6a also appear to be separated, but the evidence was not as strong as the evidence for the model discriminating PT1 and PT1b.
TABLE 3.
Confusion matrices for the S. Enteritidis OMP extract-based PLS-DA discrimination model for phage types 1 and 1b and phage types 6 and 6a
| FT-IR method | Reference methoda |
|||||
|---|---|---|---|---|---|---|
| PT1 and PT1b |
PT6 and PT6a |
|||||
| PT1 | PT1b | Total | PT6 | PT6a | Total | |
| PT1 | 50.0 | 1.7 | 51.7 | 54.0 | 8.0 | 62.0 |
| PT1b | 0.0 | 48.3 | 48.3 | 6.0 | 32.0 | 38.0 |
| Total | 50.0 | 50.0 | 100.0 | 60.0 | 40.0 | 100.0 |
The values are percentages of correct assignments.
FIG. 4.
PLS-DA scores for the discrimination models for phage types 1 and 1b (a) and phage types 6 and 6a (b) obtained using FT-IR spectra of S. Enteritidis OMP extracts. •, PT1; ○, PT1b; ▾, PT6; ▿, PT6a. LV, latent variable.
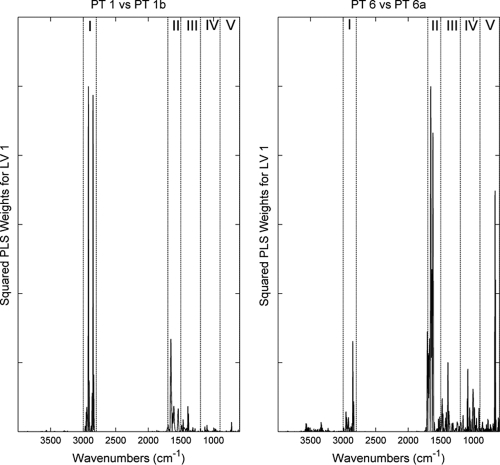
The two models were analyzed to determine the importance of each wavenumber for the discrimination obtained. For this analysis, the PLS-DA model weights for the first latent variable (the model component representing the major variance in the data) were analyzed (Fig. 5). For simplicity, the squares of the weights were used since the aim was to analyze absolute values. The first conclusion was that the region between 2,800 and 3,000 cm−1 (region I) was very important only for the OMP extract-based models. This was true for both models using OMP extract spectra. This very important region allowed increased model performance compared with the accuracy observed with intact cell spectra. The amide I and II region (region II) was also significant for both models.
FIG. 5.
Squared weights for the first latent variable (LV 1) for PLS-DA models for phage types 1 and 1b (left panel) and phage types 6 and 6a (right panel) obtained using S. Enteritidis OMP extracts. The main biochemical spectral regions examined were lipids (I), proteins (II), phospholipids, DNA, and RNA (III), polysaccharides (IV), and the fingerprint region (V).
Conclusions.
FT-IR spectroscopy together with chemometric methods was found to have considerable potential for effectively fulfilling the current requirements for rapid and correct differentiation of closely related Salmonella Enteritidis phage types. In this work, six phage types were used to assess this method's validity. However, if a representative number of isolates is available, isolates belonging to other phage types may also be used for calibration of the method and therefore improve its applicability. Developing optimal spectral preprocessing strategies to compensate for variations in spectra (baseline drift, noise, light scattering) is essential for development of robust models and successful discrimination of bacteria. The results showed that there was a significant difference between the performance of a discrimination model using intact cells and the performance of a discrimination model using OMP extracts from bacterial cell membranes. The analysis with OMP extracts proved to be superior. The analysis using OMP extracts provided greater separation between the Salmonella Enteritidis PT1-PT1b, PT4b, and PT6-PT6a groups than the intact cell analysis. The results demonstrated that when the OMP extract FT-IR spectra were used, the percentage of correct predictions was 90.7%. Moreover, when these three phage type groups were considered, the method based on OMP extract FT-IR spectra was 100% accurate. This method was tested with isolates belonging to different phage types, and the results showed that these isolates did not conform with the model and thus were outliers and the model was not suitable for obtaining a phage type estimate, as expected.
Complementary local models that considered only the PT1-PT1b and PT6-PT6a groups were developed, and the level of discrimination increased. Differentiation between PT1 and PT1b isolates was achieved with the local model using OMP extract spectra (98.3% correct predictions), and the accuracy of discrimination between PT6 and PT6a isolates was 90.4% when only the polysaccharide region (900 to 1,200 cm−1) of intact cell spectra was considered.
Reliable discrimination of bacteria at the phage type level by infrared methodology combined with multivariate analysis suggests that this technique may be useful in diagnostic laboratories or for efficient assessment of food safety. Application of different discrimination techniques to a single group of organisms unavoidably invites comparisons of the results. In order to draw reliable conclusions, examination of many strains would be very helpful. Thus, further work should focus on increasing the number of bacterial isolates used for investigation and identification of characteristic vibrational peaks that are important for FT-IR-based phage type identification.
Acknowledgments
We express our gratitude to Patrícia Malcato and Andreia Soares for their help with preparation of outer membrane protein extracts and phenotypic typing.
Ornella Preisner gratefully acknowledges the financial support provided by the Portuguese Foundation for Science and Technology (Ph.D. grant SFRH/BD/15218/2004).
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Baldauf, N. A., L. A. Rodriguez-Romo, A. E. Yousef, and L. E. Rodriguez-Saona. 2006. Differentiation of selected Salmonella enterica serovars by Fourier transform mid-infrared spectroscopy. Appl. Spectrosc. 60:592-598. [DOI] [PubMed] [Google Scholar]
- 2.Baldauf, N. A., L. A. Rodriguez-Romo, A. Männig, A. E. Yousef, and L. E. Rodriguez-Saona. 2007. Effect of selective growth media on the differentiation of Salmonella enterica serovars by Fourier-transform mid-infrared spectroscopy. J. Microbiol. Methods 68:106-114. [DOI] [PubMed] [Google Scholar]
- 3.Brereton, R. G. 2003. Chemometrics: data analysis for the laboratory and chemical plant. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 4.D'Aoust, J.-Y. 2001. Salmonella, p. 163-191. In R. G. Labbe, and S. Garcia (ed.), Guide to foodborne pathogens. Wiley-Interscience, New York, NY.
- 5.Fisher, I. S., S. Meakins, and Enter-net participants. 2006. Surveillance of enteric pathogens in Europe and beyond: Enter-net annual report for 2004. Euro Surveill. 11:3032.. [DOI] [PubMed] [Google Scholar]
- 6.Geladi, P., and B. R. Kowalski. 1986. Partial least-squares regression: a tutorial. Anal. Chim. Acta 185:1-17. [Google Scholar]
- 7.Helm, D., H. Labischinski, G. Schallehn, and D. Naumann. 1991. Classification and identification of bacteria by Fourier transform infrared spectroscopy. J. Gen. Microbiol. 137:69-79. [DOI] [PubMed] [Google Scholar]
- 8.Hickman-Brenner, F. W., A. D. Stubbs, and J. J. Farmer III. 1991. Phage typing of Salmonella enteritidis in the United States. J. Clin. Microbiol. 29:2817-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horbach, I., D. Naumann, and F. J. Fehrenbach. 1988. Simultaneous infections with different serogroups of Legionella pneumophila investigated by routine methods and Fourier transform infrared spectroscopy. J. Clin. Microbiol. 26:1106-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, S., B. L. Reuhs, and L. J. Mauer. 2005. Use of Fourier transform infrared spectra of crude bacterial lipopolysaccharides and chemometrics for differentiation of Salmonella enterica serotypes. J. Appl. Microbiol. 99:411-417. [DOI] [PubMed] [Google Scholar]
- 11.Kim, S., H. Kim, B. L. Reuhs, and L. J. Mauer. 2006. Differentiation of outer membrane proteins from Salmonella enterica serotypes using Fourier transform infrared spectroscopy and chemometrics. Lett. Appl. Microbiol. 42:229-234. [DOI] [PubMed] [Google Scholar]
- 12.Kirschner, C., K. Maquelin, P. Pina, N. A. Ngo Thi, L.-P. Choo-Smith, G. D. Sockalingum, C. Sandt, D. Ami, F. Orsini, S. M. Doglia, P. Allouch, M. Manfait, G. J. Puppels, and D. Naumann. 2001. Classification and identification of enterococci: a comparative phenotypic, genotypic, and vibrational spectroscopic study. J. Clin. Microbiol. 39:1763-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Minor, L. 1991. The genus Salmonella, p. 2760-2774. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 3. Springer-Verlag, New York, NY. [Google Scholar]
- 14.Machado, J., R. Vieira, and P. Themudo. 2004. Informações—vigilância epidemiológica laboratorial de Enterobacteriaceae. Instituto Nacional de Saúde Dr. Ricardo Jorge, Lisbon, Portugal.
- 15.Mariey, L., J. P. Signolle, C. Amiel, and J. Travert. 2001. Discrimination, classification, identification of microorganisms using FTIR spectroscopy and chemometrics. Vibration Spectrosc. 26:151-159. [Google Scholar]
- 16.Millemann, Y., M. C. Lesage, E. Chaslus-Dancla, and J. P. Lafont. 1995. Value of plasmid profiling, ribotyping, and detection of IS200 for tracing avian isolates of Salmonella typhimurium and S. enteritidis. J. Clin. Microbiol. 33(1):173-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Næs, T., T. Isaksson, T. Fearn, and T. Davies. 2002. A user-friendly guide to multivariate calibration and classification. NIR Publications, Chichester, United Kingdom.
- 18.Naumann, D., D. Helm, and H. Labischinski. 1991. Microbiological characterizations by FT-IR spectroscopy. Nature 351:81-82. [DOI] [PubMed] [Google Scholar]
- 19.Naumann, D. 2000. Infrared spectroscopy in microbiology, p. 102-131. In R. A. Meyers (ed.), Encyclopedia of analytical chemistry. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 20.Preisner, O., J. A. Lopes, R. Guiomar, J. Machado, and J. C. Menezes. 2007. Fourier transform infrared (FT-IR) spectroscopy in bacteriology: towards a reference method for bacteria discrimination. Anal. Bioanal. Chem. 387:1739-1748. [DOI] [PubMed] [Google Scholar]
- 21.Rodrigue, D. C., R. V. Tauxe, and B. Rowe. 1990. International increase in Salmonella enteritidis: a new pandemic? Epidemiol. Infect. 105:21-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rychlik, I., A. Svestkova, and R. Karpiskova. 2000. Subdivision of Salmonella enterica serovar enteritidis phage types PT14b and PT21 by plasmid profiling. Vet. Microbiol. 74:217-225. [DOI] [PubMed] [Google Scholar]
- 23.Stanley, J., and N. Baquar. 1994. Phylogenetics of Salmonella enteritidis. Int. J. Food Microbiol. 21:79-87. [DOI] [PubMed] [Google Scholar]
- 24.Stuart, B. H. 2004. Infrared spectroscopy: fundamentals and applications. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 25.Stubbs, A. D., F. W. Hickman-Brenner, D. N. Cameron, and J. J. Farmer III. 1994. Differentiation of Salmonella enteritidis phage type 8 strains: evaluation of three additional phage typing systems, plasmid profiles, antibiotic susceptibility patterns, and biotyping. J. Clin. Microbiol. 32:199-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki, Y., M. Ishihara, M. Matsumoto, S. Arakawa, M. Saito, N. Ishikawa, and T. Yokochi. 1995. Molecular epidemiology of Salmonella enteritidis. An outbreak and sporadic cases studied by means of pulsed-field gel electrophoresis. J. Infect. 31:211-217. [DOI] [PubMed] [Google Scholar]
- 27.Thong, K.-L., Y.-F. Ngeow, M. Altwegg, P. Navaratnam, and T. Pang. 1995. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J. Clin. Microbiol. 33:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tietjen, M., and D. Y. Fung. 1995. Salmonellae and food safety. Crit. Rev. Microbiol. 21:53-83. [DOI] [PubMed] [Google Scholar]
- 29.Ward, L. R., J. De Sa, and B. Rowe. 1987. A phage-typing scheme for Salmonella enteritidis. Epidemiol. Infect. 99:291-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wold, S., C. Albano, W. J. Dunn III, U. Edlund, K. H. Esbensen, P. Geladi, S. Hellberg, E. Johansson, W. Lindberg, and M. Sjöström. 1984. Multivariate data analysis in chemistry, p. 17-95. In B. R. Kowalski (ed.), Chemometrics. Mathematics and statistics in chemistry. Reidel Publishing Company, Dordrecht, Netherlands.