Abstract
Symbiotic associations with midgut bacteria have been commonly found in diverse phytophagous heteropteran groups, where microbiological characterization of the symbiotic bacteria has been restricted to the stinkbug families Acanthosomatidae, Plataspidae, Pentatomidae, Alydidae, and Pyrrhocoridae. Here we investigated the midgut bacterial symbiont of Cantao ocellatus, a stinkbug of the family Scutelleridae. A specific gammaproteobacterium was consistently identified from the insects of different geographic origins. The bacterium was detected in all 116 insects collected from 9 natural host populations. Phylogenetic analyses revealed that the bacterium constitutes a distinct lineage in the Gammaproteobacteria, not closely related to gut symbionts of other stinkbugs. Diagnostic PCR and in situ hybridization demonstrated that the bacterium is extracellularly located in the midgut 4th section with crypts. Electron microscopy of the crypts revealed a peculiar histological configuration at the host-symbiont interface. Egg sterilization experiments confirmed that the bacterium is vertically transmitted to stinkbug nymphs via egg surface contamination. In addition to the gut symbiont, some individuals of C. ocellatus harbored another bacterial symbiont in their gonads, which was closely related to Sodalis glossinidius, the secondary endosymbiont of tsetse flies. Biological aspects of the primary gut symbiont and the secondary Sodalis-allied symbiont are discussed.
Insects are among the largest animal groups on the earth, embracing 750,000 to several millions of species (37, 52). Diverse insects are symbiotically associated with microorganisms, especially bacteria (5-7). In some insects, symbiotic bacteria are harbored in specialized host cells called bacteriocytes (or mycetocytes), constituting obligate mutualistic associations. For example, Buchnera aphidicola is harbored within bacteriocytes in the abdominal body cavity of almost all aphids and provides essential amino acids that are lacking in the phloem sap diet of the insects (9, 47). Wigglesworthia glossinidia is localized in a midgut-associated bacteriome of tsetse flies and plays pivotal roles in biosynthesis of B vitamins that are deficient in the vertebrate blood diet of the insects (2, 34). These obligate endocellular symbionts are often collectively referred to as “primary symbionts.”
In contrast, there are facultative endosymbiotic microorganisms not essential for their host insects, often collectively called “secondary symbionts.” For example, many aphids are known to harbor various facultative symbionts, which belong to distinct lineages in the Gamma- and Alphaproteobacteria (33, 43) and the Mollicutes (10). While the majority of those facultative bacteria are either parasitic or commensalistic for their hosts, some of them affect the host fitness beneficially in particular ecological contexts (29, 32, 36, 44, 51). In addition to the obligate primary symbiont Wigglesworthia, tsetse flies harbor the facultative secondary symbiont Sodalis glossinidius, whose biological function for the hosts is currently elusive (3, 8).
Members of the suborder Heteroptera, known as true bugs and consisting of over 38,000 described species, are characterized by their sucking mouthparts, half-membranous forewings, and incomplete metamorphosis (46). In the Heteroptera, symbiotic associations with bacteria are mainly found in phytophagous groups, especially in stinkbugs of the infraorder Pentatomomorpha. These stinkbugs generally possess many sacs or tubular outgrowths, called crypts or ceca, in a posterior region of the midgut, whose lumen is densely populated by a specific bacterial symbiont (7, 16). In some cases, experimental elimination of the symbiotic bacteria resulted in retarded growth and high mortality of the host insects (1, 13, 21, 26, 27, 39), indicating that these gut symbionts play important biological roles. Most of the gut symbionts are vertically transmitted through host generations by such mechanisms as egg surface contamination in the families Pentatomidae and Acanthosomatidae (1, 27, 39, 40, 42), coprophagy in the Cydnidae and Coreidae (22, 45), and capsule transmission in the Plataspidae (20), whereas a case of environmental acquisition has been reported from the Alydidae (26). Thus far, gut symbiotic bacteria of some members of the Acanthosomatidae, Plataspidae, Pentatomidae, Alydidae, and Pyrrhocoridae have been characterized using molecular techniques (21, 23, 25, 27, 38), while phylogenetic and biological aspects of gut symbiotic bacteria have been untouched in many other stinkbug groups.
These gut symbiotic bacteria are, despite their extracellular localization, regarded as “primary symbionts” of the stinkbugs. On the other hand, some stinkbugs may, in addition to the gut symbiotic bacteria, also be associated with facultative “secondary symbionts.” For example, Wolbachia infections have been detected from diverse stinkbugs, most of which are probably of parasitic or commensalistic nature (24). Besides Wolbachia, there has been no report on facultative, secondary symbionts from stinkbugs.
Members of the family Scutelleridae, often referred to as jewel bugs or shield-backed bugs, are stinkbugs characterized by their greatly enlarged convex scutellum that usually covers the entire abdomen. Some tropical species are also known for their vivid and beautiful body coloration (46). The family contains approximately 80 genera and 450 species, and in Japan, at least 7 genera and 9 species have been recorded (50). In the early 20th century, the presence of symbiotic bacteria was histologically described in midgut crypts of several scutellerid species (16, 31, 42). Since these pioneer works, however, no studies have been conducted on the symbiotic bacteria of scutellerid stinkbugs.
Here we investigated the midgut symbiont of Cantao ocellatus, a scutellerid stinkbug widely distributed in Asian countries, including Japan, and known to guard their eggs and newborn nymphs (Fig. 1A) (50). In addition to the gut symbiont, we also identified a Sodalis-allied facultative secondary symbiont from gonads of the insect.
FIG. 1.
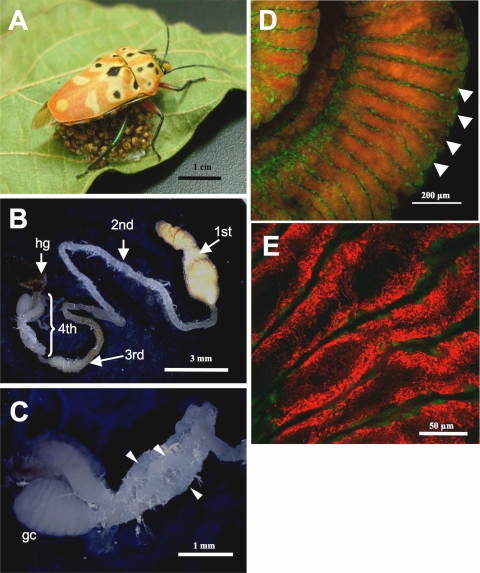
(A) Adult female of Cantao ocellatus, guarding hatchlings under her body. (B) Dissected midgut from an adult female of C. ocellatus. 1st, midgut 1st section; 2nd, midgut 2nd section; 3rd, midgut 3rd section; 4th, midgut 4th section with crypts; hg, hindgut. (C) Enlarged image of the midgut 4th section with crypts. Arrowheads indicate three rows of crypts, while a fourth row is hidden behind. Glandular crypts (gc) are developed in adult females specifically, which may be involved in egg surface contamination with the symbiont. (D) An in situ hybridization image of the midgut 4th section, in which red and green signals indicate the gut symbiont and the host nucleus, respectively. Each arrow shows a crypt. (E) An enlarged image of the symbiotic bacteria in the crypts.
MATERIALS AND METHODS
Insects.
Collection data for the adult C. ocellatus insects used in this study are summarized in Table 1. Insect samples were collected from the Japanese mallotus tree, Mallotus japonicus, at nine localities in Japan in 2008 and 2009.
TABLE 1.
Samples of Cantao ocellatus used in this study
| Collection locality | Collection date | Collector | Accession no. for: |
|
|---|---|---|---|---|
| Midgut symbiont | Sodalis-allied symbiont | |||
| Onna, Okinawa Island, Okinawa Prefecture, Japan | April 2008 | Masaaki Kimura | AB541001 (16S rRNA) | |
| AB541011 (groEL) | ||||
| Kumejima Island, Okinawa Prefecture, Japan | April 2008 | Takahiro Hosokawa | AB541002 (16S rRNA) | |
| Kumejima Island, Okinawa Prefecture, Japan | May 2008 | Suguru Ohno | ||
| Kunigami, Okinawa Island, Okinawa Prefecture, Japan | June 2008 | Takahiro Hosokawa | AB541003 (16S rRNA) | |
| Ishigaki Island, Okinawa Prefecture, Japan | July 2008 | Takahiro Hosokawa | AB541004 (16S rRNA) | AB541010 (16S rRNA) |
| AB541012 (groEL) | ||||
| Ishigaki Island, Okinawa Prefecture, Japan | September 2008 | Takahiro Hosokawa | ||
| Ishigaki Island, Okinawa Prefecture, Japan | September 2009 | Takahiro Hosokawa | ||
| Iriomote Island, Okinawa Prefecture, Japan | August 2009 | Hiromi Mukai | AB541005 (16S rRNA) | |
| Muroto, Kochi Prefecture, Japan | October 2008 | Katsura Ito | AB541006 (16S rRNA) | |
| Tosa-Shimizu, Kochi Prefecture, Japan | November 2008 | Mikio Takai | AB541007 (16S rRNA) | |
| Naze, Amami-Oshima Island, Kagoshima Prefecture, Japan | June 2008 | Yuki G. Baba | AB541008 (16S rRNA) | |
| Naze, Amami-Oshima Island, Kagoshima Prefecture, Japan | July 2009 | Takahiro Hosokawa | ||
| Nishino-Omote, Tanagashima Island, Kagoshima Prefecture, Japan | July 2009 | Takahiro Hosokawa | AB541009 (16S rRNA) | |
Dissection and DNA extraction.
Ovaries, testes, and midgut 1st, 2nd, 3rd, and 4th sections were dissected from adult insects using a pair of fine forceps under a binocular microscope in a plastic petri dish filled with phosphate-buffered saline (PBS; 137 mM NaCl, 8.1 mM Na2HPO4, 2.7 mM KCl, 1.5 mM KH2PO4 [pH 7.5]). These insect tissues were immediately subjected to DNA extraction using Nucleo Spin tissue kit (Macherey-Nagel).
DNA cloning and sequencing.
A 1.5-kb segment of bacterial 16S rRNA gene was amplified with the primers 16SA1 (5′-AGAGTTTGATCMTGGCTCAG-3′) and 16SB1 (5′-TACGGYTACCTTGTTACGACTT-3′) (12). A 1.7-kb fragment of bacterial groEL gene was amplified with the primers Gro-F1 (5′-ATGGCAGCWAAAGACGTAAATTYGG-3′) and Gro-R1 (5′-TTACATCATKCCGCCCATGC-3′). PCR was conducted with Ampli Taq DNA polymerase (Applied Biosystems) and the supplemented buffer system, under the following temperature profile: 95°C for 10 min, followed by 35 cycles consisting of 95°C for 30 s, 55°C for 1 min, and 72°C for 2 min. The PCR products were subjected to the cloning using TA cloning vector pT7Blue (Takara Bio) and Escherichia coli DH5α competent cells (Takara Bio) with a blue-white selection system, using ampicillin and 5-bromo-4-chloro-3-indolyl-d-galactopyranoside (X-Gal). In order to check the length of the inserted DNA fragment, white colonies, which were expected to contain inserted plasmids, were directly subjected to PCR with the primers Univ19 (5′-GTTTTCCCAGTCACGACGT-3′) and Rev20 (5′-AGCTATGACCATGATTACGC-3′). When a PCR product of expected size (1.5 kb and 1.7 kb, respectively) was obtained, the product was subjected to restriction fragment length polymorphism (RFLP) genotyping. Three or more clones for each of the RFLP genotypes were cultured at 37°C in LB liquid medium overnight and subjected to plasmid extraction using a QIAprep-Spin miniprep kit (Qiagen). The purified plasmids were subjected to DNA sequencing with the primers Univ19 and Rev20 and the internal primers Eub925r (5′-CCGYCAATTCCTTTGAGTTT-3′) and Eub1405r (5′-GACGGGCGGTGTGTRCA-3′) for the 16S rRNA gene, groE-GS (5′-TAATGCTGATGAAAGCGT-3′) for the groEL gene of the midgut symbiont, and groE-SD (5′-ACTATCTCCGCCAACTC-3′) for the groEL genes of the Sodalis-allied symbiont, under a temperature profile of 96°C for 1 min, followed by 26 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min, using a Genetic Analyzer (Applied Biosystems 3130x1).
Molecular phylogenetic analysis.
Bacterial 16S rRNA and groEL gene sequences were subjected to molecular phylogenetic analysis together with those of gammaproteobacterial representatives. Multiple alignments were generated using the program ClustalW (48). Aligned nucleotide sites containing a gap were removed from the data set using the program SeaView (15), and the final alignment was corrected manually. Phylogenetic analyses were conducted by maximum likelihood (ML) and maximum parsimony (MP) methods. The best-fit substitution models were selected by using the program MrModeltest v2.1 (J. A. Nylander, 2004; http://www.ebc.uu.se/systzoo/staff/nylander.html). ML phylogenies, with 100 bootstrap resamplings, were estimated using the program phyML 3.0 (18) with the GTR model for both 16S rRNA and groEL phylogenies. MP phylogenies, with 1,000 bootstrap resamplings, were estimated using the program phylo_win (15).
Relative rate test.
A relative rate test, based on genetic distances estimated under the Kimura's two-parameter model (28), was performed using the program RRTree (41). For 16S rRNA gene sequences, 1,426 unambiguously aligned nucleotide sites were subjected to the analysis. For groEL gene sequences, 1,040 unambiguously aligned nucleotide sites at the first- and second-codon positions were analyzed, while nucleotide sites at the third-codon positions were omitted from the analysis due to saturated nucleotide substitutions.
Diagnostic PCR.
The following specific primer sets were used for diagnostic PCR detection of 16S rRNA gene of the symbiotic bacteria: 16SA2 (5′-GTGCCAGCAGCCGCGGTAATAC-3′) and akagi16S780R (5′-GCTACGGAGGCTACTTATT-3′) for a 0.16-kb segment from the midgut symbiont; and sodalis370F (5′-CGRTRGCGTTAAYAGCGC-3′) and 16SB2 (5′-CGAGCTGACGACARCCATGC-3′) for a 0.62-kb segment from the Sodalis-allied symbiont. The temperature profile was 95°C for 10 min, followed by 35 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 1 min. To check the quality of the DNA samples, a 1.5-kb segment of the insect mitochondrial 16S rRNA gene was amplified with the primers MtrA1 (5′-AAWAAACTAGGATTAGATACCCTA-3′) and MtrB1 (5′-TCTTAATYCAACATCGAGGTCGCAA-3′) (11).
Whole-mount FISH.
The Alexa 555-labeled oligonucleotide probe Al555-AKG447 (5′-GTGTATTATTTTTCATCACC-3′) was used for whole-mount fluorescent in situ hybridization (FISH) targeting 16S rRNA of the midgut symbiont. Midgut sections with crypts were dissected from adult females and fixed in Carnoy's solution (6:3:1 ethanol-chloroform-acetic acid) overnight. The fixed samples were incubated in ethanolic 6% H2O2 solution for a week to quench the autofluorescence of the tissues (30). The insects were thoroughly washed and equilibrated with a hybridization buffer (20 mM Tris HCl [pH 8.0], 0.9 M NaCl, 0.01% sodium dodecyl sulfate, 30% formamide), to which the specific probe, SYTOX green and DAPI (4′,6-diamidino-2-phenylindole) were added at final concentrations of 0.1 μM, 0.5 μM, and 0.1 μg/ml, respectively. After an overnight incubation, the samples were thoroughly washed in PBSTx (PBS containing 0.3% Triton X-100) and observed under an epifluorescence microscope (Axiophoto; Carl Zeiss) and a laser confocal microscope (PASCAL5; Carl Zeiss). To confirm the specificity of the detection, the following control experiments were conducted: a no-probe control experiment, a no-SYTOX control experiment, and a competitive suppression control experiment with excess unlabeled probe.
Transmission electron microscopy.
The midgut crypts were dissected from adult females in 0.1 M phosphate buffer (pH 7.4) containing 2.5% glutaraldehyde, prefixed in the fixative at 4°C overnight, and postfixed in the phosphate buffer containing 2% osmium tetroxide at 4°C for 60 min. After dehydration through an ethanol series, the materials were embedded in Spurr resin (Nisshin-EM). Ultrathin sections (thickness, 80 nm) were made on an ultramicrotome (Ultracat-N; Leichert-Nissei), mounted on collodion-coated copper meshes, stained with uranyl acetate and lead citrate, and observed under a transmission electron microscope (model H-7000; Hitachi).
Egg sterilization experiments.
In total, 17 egg masses laid by different females collected at Ishigaki Island in 2008 and 2009 were subjected to the experiments. Nine randomly selected egg masses were treated with 70% ethanol for 5 min and 4% formalin for 15 min, rinsed with 70% ethanol, and dried in air. For a control, five egg masses were treated with water for 20 min instead of the surface sterilization. The other three egg masses were kept untreated. Each of the 17 egg masses was separately placed in a plastic petri dish (5 cm in diameter and 1 cm in depth) with a wet cotton ball at 25°C under a long-day condition (16 h of light and 8 h of dark [16L/8D]) until hatching. The hatchlings were fed with raw peanuts, dry soybeans, and distilled water at 25°C (16L/8D) until they reached the 3rd instar. Ten to 20 nymphs were randomly collected from each of the egg masses and preserved in acetone and were subjected to DNA extraction and diagnostic PCR.
RESULTS
General observation of digestive tract.
Digestive organs were dissected from adult females of C. ocellatus (Fig. 1B). The enlarged midgut 1st section was filled with liquid, followed by the elongated midgut 2nd and 3rd sections. The midgut 4th section bore a number of well-developed crypts, which were white, arranged in four rows, and fused to each other into a loose helical assemblage (Fig. 1C).
Bacterial gene sequences from midgut section with crypts.
Insects collected at nine localities in Japan were examined. One to three insect samples from each of the localities were dissected, and their midgut 4th sections were subjected to DNA extraction and PCR amplification of the bacterial 16S rRNA gene. The PCR products were cloned, and three to eight clones per insect were sequenced. All of the bacterial sequences obtained were identical without geographic and populational variations. The bacterial groEL gene was also amplified, cloned, and sequenced from an insect collected at Onna, Okinawa Island.
Phylogenetic placement of the gut symbiont.
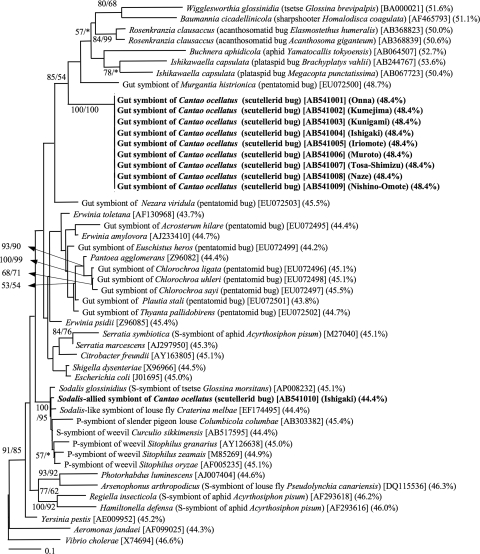
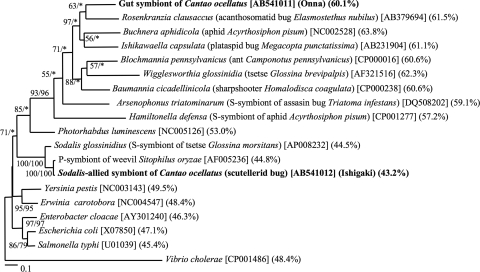
Phylogenetic analyses of the 16S rRNA gene sequences revealed that the gut symbiont constituted a distinct lineage in the Gammaproteobacteria, not closely related to gut symbionts of other stinkbugs. Meanwhile, the lineage was placed in a cluster of diverse insect symbionts, including endocellular symbionts of aphids, sharpshooters, and tsetse flies, and also gut symbionts of acanthosomatids, plataspids, and pentatomids, although the phylogenetic assemblage was not conclusive, with relatively low bootstrap supports (Fig. 2). Phylogenetic analyses on the basis of groEL gene sequences yielded a similar phylogenetic relationship: the gut symbiont was placed in a cluster of insect symbionts consisting of endocellular symbionts of aphids, sharpshooters, and tsetse flies, and also gut symbionts of acanthosomatids and plataspids (Fig. 3).
FIG. 2.
Phylogenetic placement of the symbiotic bacteria of C. ocellatus on the basis of 16S rRNA gene sequences. A maximum likelihood (ML) tree inferred from a total of 1,243 aligned nucleotide sites is shown, whereas a maximum parsimony (MP) analysis gave substantially the same topology (data not shown). Bootstrap values higher than 50% are indicated at the nodes in the order of ML/MP. Asterisks indicate support values lower than 50%. Sequence accession numbers and AT contents of the nucleotide sequences are in brackets and parentheses, respectively. As for insect endosymbionts, the name of the host insect is also indicated in parentheses. P-symbiont, primary symbiont; S-symbiont, secondary symbiont.
FIG. 3.
Phylogenetic placement of the symbiotic bacteria from C. ocellatus on the basis of groEL gene sequences. A total of 1,104 aligned nucleotide sites at first and second codon positions were subjected to the analysis. The third codon positions were not used because of saturated nucleotide substitutions. Analysis of deduced amino acid sequences gave substantially the same results (data not shown). A maximum likelihood tree is shown. Support values for the nodes are indicated as in Fig. 2. Sequence accession numbers and AT contents of the nucleotide sequences are in brackets and parentheses, respectively. Note that the AT content values are based on the data of all codon positions.
Localization and morphology of the gut symbiont.
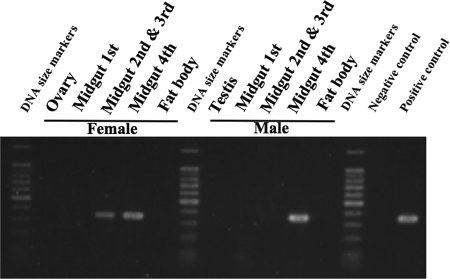
Diagnostic PCR analysis of dissected tissues detected the symbiont DNA sequence preferentially in the midgut 4th section with crypts, while the midgut 2nd and 3rd sections of females also exhibited the symbiont infection (Fig. 4). In situ hybridization targeting 16S rRNA of the gut symbiont identified strong and specific signals in the lumen of the crypts in the midgut 4th section (Fig. 1D). In each of the crypts, tubular symbiont cells were distributed not uniformly but concentrated near the epithelial cell layer (Fig. 1E).
FIG. 4.
Diagnostic PCR detection of the midgut symbiont in dissected tissues of adult C. ocellatus. Negative control, distilled water; positive control, DNA from midgut crypts of C. ocellatus; lane M, DNA size markers (from bottom to top) from 100 bp to 1,000 bp in 100-bp increments and 1,500 bp.
Fine structure of the midgut crypt and the gut symbiont.
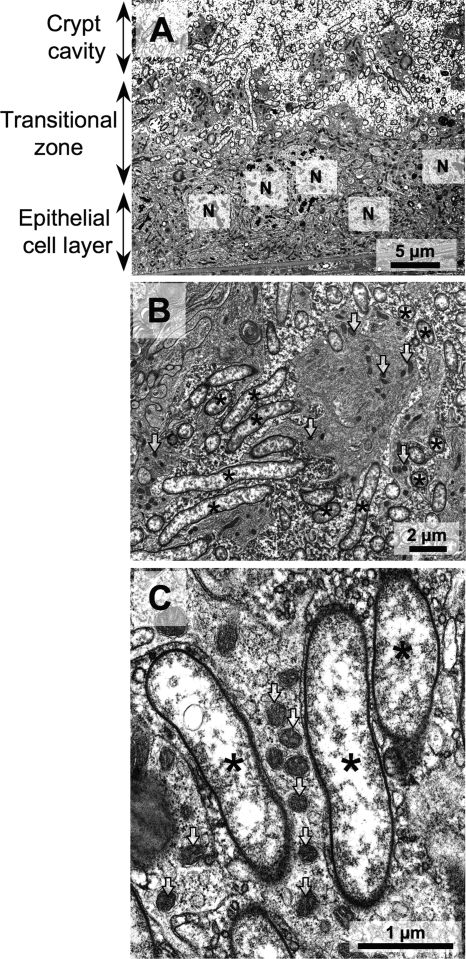
Electron microscopy revealed a peculiar histological configuration inside the midgut crypts. At the central region of the crypts, the symbiont cells were found in the extracellular space in a relatively dispersed manner (Fig. 5A). Meanwhile, at the peripheral zone of the crypts, the symbiont cells and the host epithelial cells were closely associated with each other in an intermingled manner (Fig. 5A and B). Some of the symbiont cells were seen in the epithelial cell layer, next to mitochondria and endoplasmic reticula, as if they were located in the host cytoplasm (Fig. 5C). However, close examination of a number of electron microscopic images, including those from serial sections, revealed that those symbiont cells were actually located in extracellular spaces. It was postulated that the symbiont cells were entrapped in narrow extracellular spaces constituted by convoluted projections from the epithelial cell layer. Of course, we cannot rule out the possibility that, while the majority of the symbiont cells were found extracellularly, a small population of the symbiont cells might be nearly or entirely engulfed by the gut epithelial cells.
FIG. 5.
Transmission electron microscopy of the midgut crypts in adult females of C. ocellatus. (A) Image of the interface between the crypt cavity and the epithelial cell layer; (B) enlarged image of the transitional zone; (C) enlarged image of the symbiont cells indicating an extracellular, rather than endocellular location. Asterisks and arrows indicate the symbiont cells and mitochondria, respectively. N, nucleus of the epithelial cell of the midgut crypt.
Detection and phylogenetic placement of the Sodalis-allied symbiont.
From ovarial DNA of an adult female collected at Ishigaki Island, bacterial 16S rRNA and groEL genes were detected. The PCR products were cloned, and eight clones were sequenced. All of the sequences were identical to each other. Phylogenetic analyses of the sequences revealed that the ovarial symbiont belongs to the Gammaproteobacteria, forming a well-supported clade with the secondary symbiont of tsetse flies Sodalis glossanidius, the bacteriome-associated symbiont of grain weevils, the symbiont of a louse fly, the bacteriocyte-associated symbiont of a pigeon louse, and the secondary symbiont of a Curculio weevil (Fig. 2 and 3).
AT richness of the gut symbiont genes.
The 16S rRNA gene sequence of the gut symbiont exhibited an AT content of 48.4%, which was slightly higher than the AT contents of free-living gammaproteobacteria around 45% (Fig. 2). The groEL gene sequence of the gut symbiont was 60.1% AT, which was drastically higher than the values of free-living gammaproteobacteria of around 45% (Fig. 3). On the other hand, the 16S rRNA gene sequence and the groEL gene sequence of the Sodalis-allied symbiont were not AT biased: 44.4% and 43.2% AT, respectively (Fig. 2 and 3).
Accelerated molecular evolution of the gut symbiont genes.
The evolutionary rates of the 16S rRNA and groEL gene sequences of the gut symbiont of C. ocellatus were significantly higher than those of free-living gammaproteobacteria, respectively. Meanwhile, the evolutionary rates in the Sodalis-allied symbiont of C. ocellatus were not significantly different from those of free-living gammaproteobacteria (Table 2).
TABLE 2.
Relative-rate tests for comparing the molecular evolutionary rates of 16S rRNA and groEL gene sequences between the lineages of the gut symbiont and the Sodalis-allied symbiont of Cantao ocellatus and their free-living relatives
| Gene | Lineage 1 | Lineage 2 | Outgroup | K1a | K2b | Difference of distancesc | Rate ratiod | P valuee |
|---|---|---|---|---|---|---|---|---|
| 16S rRNA | Gut symbiont of C. ocellatus (AB541001) | Escherichia coli (J01695) and Shigella dysenteriae (X96966) | Vibrio cholerae (X74694) | 0.052 | 0.027 | 0.025 | 1.9 | 0.0031* |
| groEL | Gut symbiont of C. ocellatus (AB541011) | Escherichia coli (X07850) and Salmonella enterica serovar Typhi (U01039) | Vibrio cholerae (CP001486) | 0.060 | 0.032 | 0.028 | 1.9 | 0.013* |
| 16S rRNA | Sodalis-allied symbiont of C. ocellatus (AB541010) | Escherichia coli (J01695) and Shigella dysenteriae (X96966) | Vibrio cholerae (X74694) | 0.030 | 0.026 | 0.004 | 1.2 | 0.53 |
| groEL | Sodalis-allied symbiont of C. ocellatus (AB541012) | Escherichia coli (X07850) and Salmonella enterica serovar Typhi (U01039) | Vibrio cholerae (CP001486) | 0.028 | 0.018 | 0.010 | 1.6 | 0.19 |
K1, estimated mean distance between lineage 1 and the last common ancestor of lineages 1 and 2.
K2, estimated mean distance between lineage 2 and the last common ancestor of lineages 1 and 2.
K1 − K2.
K1/K2 ratio.
P values were generated using the program RRTree (41). *, statistically significant difference.
Infection frequencies of the gut symbiont and the Sodalis-allied symbiont in natural host populations.
Table 3 shows infection frequencies of the gut symbiont and the Sodalis-allied symbiont in natural populations of C. ocellatus. The gut symbiont was detected from all 116 insects representing 9 populations examined. Meanwhile, the Sodalis-allied symbiont was detected only from 6 of 144 insects examined. All of the infected insects were collected at Ishigaki Island, where the infection frequency with the Sodalis-allied symbiont was 11% (6/53).
TABLE 3.
Infection frequencies of the gut symbiont and the Sodalis-allied symbiont in natural populations of Cantao ocellatus
| Locality | Infection frequency (%)a |
|||||
|---|---|---|---|---|---|---|
| Gut symbiontb |
Sodalis-allied symbiontc |
|||||
| Male | Female | Total | Male | Female | Total | |
| Onna | 100 (3/3) | 100 (3/3) | 100 (3/3) | 0 (0/3) | 0 (0/3) | 0 (0/6) |
| Kumejima | 100 (4/4) | 100 (8/8) | 100 (12/12) | 0 (0/8) | 0 (0/5) | 0 (0/13) |
| Kunigami | 100 (7/7) | 100 (12/12) | 100 (19/19) | 0 (0/3) | 0 (0/1) | 0 (0/4) |
| Ishigaki | 100 (9/9) | 100 (2/2) | 100 (11/11) | 17 (2/12) | 10 (4/41) | 11 (6/53) |
| Iriomote | 100 (9/9) | No data | 100 (9/9) | 0 (0/9) | No data | 0 (0/9) |
| Naze | 100 (5/5) | 100 (8/8) | 100 (13/13) | 0 (0/3) | 0 (0/10) | 0 (0/13) |
| Muroto | 100 (1/1) | 100 (1/1) | 100 (2/2) | 0 (0/1) | 0 (0/1) | 0 (0/2) |
| Tosa-Shimizu | 100 (15/15) | 100 (15/15) | 100 (30/30) | 0 (0/15) | 0 (0/15) | 0 (0/30) |
| Nishino-Omote | 100 (5/5) | 100 (9/9) | 100 (14/14) | 0 (0/5) | 0 (0/9) | 0 (0/14) |
| Total | 100 (58/58) | 100 (58/58) | 100 (116/116) | 3 (2/59) | 5 (4/85) | 4 (6/144) |
Values in parentheses represent the number infected/number tested.
Dissected midgut 4th section with crypts was subjected to DNA extraction and diagnostic PCR.
Dissected testis/ovary was subjected to DNA extraction and diagnostic PCR.
Egg surface sterilization blocks vertical transmission of the gut symbiont.
Table 4 summarizes the results of the egg surface sterilization experiments. The formalin and ethanol sterilization procedure did not affect the hatching rates of the treated eggs, indicating no detrimental effects of the treatment on the embryonic development. All of the nymphs from the sterilized eggs were free of the gut symbiont, whereas most of the nymphs from the water-treated eggs and untreated eggs were infected with the gut symbiont.
TABLE 4.
Effects of egg surface sterilization on vertical transmission of the gut symbiont in Cantao ocellatus
| Treatment group and egg mass no. | Clutch size | Hatching rate (%) | Nymphal infection rate (%)a |
|---|---|---|---|
| Formalin and ethanol treatmentb | |||
| FE1 | 162 | 98 (159/162) | 0 (0/20) |
| FE2 | 82 | 97 (80/82) | 0 (0/14) |
| FE3 | 164 | 98 (161/164) | 0 (0/20) |
| FE4 | 196 | 86 (169/196) | 0 (0/11) |
| FE5 | 150 | 84 (126/150) | 0 (0/20) |
| FE6 | 175 | 97 (169/175) | 0 (0/18) |
| FE7 | 120 | 95 (114/120) | 0 (0/20) |
| FE8 | 98 | 99 (97/98) | 0 (0/20) |
| FE9 | 133 | 98 (130/133) | 0 (0/20) |
| Total | 1,280 | 94 (1,205/1,280) | 0 (0/163) |
| Water treatment controlc | |||
| WT1 | 209 | 99 (207/209) | 100 (14/14) |
| WT2 | 172 | 91 (157/172) | 100 (15/15) |
| WT3 | 148 | 100 (148/148) | 100 (10/10) |
| WT4 | 55 | 24 (13/55) | 80 (8/10) |
| WT5 | 192 | 98 (189/192) | 100 (13/13) |
| Total | 776 | 92 (714/776) | 97 (60/62) |
| No-treatment controld | |||
| UT1 | 110 | 90 (99/110) | 100 (14/14) |
| UT2 | 128 | 98 (125/128) | 100 (16/16) |
| UT3 | 100 | 96 (96/100) | 95 (19/20) |
| Total | 338 | 95 (320/338) | 98 (49/50) |
Inspected at 3rd instar.
Egg masses were treated with 70% ethanol for 5 min and 4% formalin for 15 min and rinsed with 70% ethanol for 10 s.
Egg masses were treated with water for 20 min.
Egg masses were untreated.
DISCUSSION
Here we identified a specific gammaproteobacterial midgut symbiont from the scutellerid stinkbug C. ocellatus. Previous studies have identified gammaproteobacterial midgut symbionts from stinkbugs of the families Acanthosomatidae, Plataspidae, and Pentatomidae. Although these symbionts are commonly harbored in the same symbiotic organ, midgut 4th section with crypts, irrespective of the host systematics, their microbiological affiliations are different: Rosenkranzia clausaccus with acanthosomatids, Ishikawaella capsulata with plataspids, and unnamed gammaproteobacterial lineages with pentatomids (21, 27, 38). Here we demonstrated that the gut symbiont of C. ocellatus is phylogenetically distinct from those stinkbug symbionts, indicating that the evolutionary origin of the gut symbiont in the Scutelleridae is independent of those of the gut symbionts in the Acanthosomatidae, Plataspidae, and Pentatomidae.
In the Acanthosomatidae and Plataspidae, respectively, the gut symbionts are monophyletic, cospeciate with the host insects, and exhibit AT-biased nucleotide composition and accelerated molecular evolution, indicating strict host-symbiont coevolution within each of the families over evolutionary time (21, 27). In the Pentatomidae, in contrast, the gut symbionts are polyphyletic: the gut symbionts of Acrosternum hilare, Murgantia histrionica, and Nezara viridula, the gut symbionts of Chlorochroa spp., the gut symbiont of Euschistus heros, and the gut symbionts of Plautia stali and Thyanta pallidovirens constitute distinct lineages in the Gammaproteobacteria, respectively (38). Their 16S rRNA gene sequences generally exhibit no remarkable AT bias (Fig. 2). Hence, although the gut symbionts are vertically transmitted and play biological roles in the Pentatomidae (1, 39, 40), host-symbiont coevolution is not so strict but occasional lateral transfers and/or symbiont replacements must have taken place. Since this study is the first to characterize a scutellerid gut symbiont, it is currently obscure whether the host-symbiont relationship in the Scutelleridae is similar to the situation in the Acanthosomatidae/Plataspidae or the situation in the Pentatomidae. AT-biased nucleotide composition (Fig. 2 and 3) and accelerated molecular evolution (Table 2) in the gut symbiont seem suggestive of an intimate host-symbiont relationship in C. ocellatus.
In the midgut crypts of C. ocellatus, the gut symbiont was not distributed uniformly but was concentrated in the peripheral zone (Fig. 1E). Electron microscopy revealed a peculiar histological configuration at the host-symbiont interface: convoluted projections from the epithelial cell layer form an intermingled structure, where the symbiont cells are entrapped in narrow extracellular spaces in close association with the host cells (Fig. 5). In acanthosomatids and plataspids, electron microscopy detected no such cellular configuration: their midgut crypts were lined with thin and flat epithelial cells, and the crypt cavities were densely packed with symbiont cells (20, 27). The structure is probably the reason why the symbiont cells are concentrated near the epithelial cell layer in the midgut crypts (Fig. 1E).
The biological roles of the gut symbiont for C. ocellatus are currently unknown. Considering the essential roles of the gut symbionts in other stinkbug groups (1, 13, 21, 26, 27, 39) and the 100% infection frequencies in natural host populations (Table 3), it appears plausible that the gut symbiont plays some important roles for C. ocellatus. As is the case of scutellerid species, C. ocellatus is large with beautiful coloration (Fig. 1A). The gut symbiont might be involved in formation of these costly host traits, possibly via nutritional provisioning or other metabolic supplementation. The pericarp of Mallotus fruits, the main food source for C. ocellatus nymphs, contains rottlerin-like phloroglucinol derivatives and other cytotoxic compounds (4). Although speculative, the gut symbiont might be involved in detoxification of such plant defense substances.
In addition to the gut symbiont, we identified another bacterial associate of C. ocellatus. The bacterium was phylogenetically related to Sodalis glossinidius, the facultative symbiont of tsetse flies (Fig. 2 and 3), and detected only from a local host population at an infection frequency around 10% (Table 3). In this study, because of infrequent occurrences in our samples, we could not inspect biological aspects of the Sodalis-allied symbiont such as vertical transmission mechanism, effects on host fitness, etc. For a long time, Sodalis and allied symbionts have been known only from tsetse flies and grain weevils (8, 19). Recently, however, Sodalis-allied symbionts have been detected from a louse fly (35), a pigeon louse (14), a chestnut weevil (49), and a longicorn beetle (17). It has turned out that the Sodalis-allied symbionts are associated with a wider array of insects than previously envisioned.
In conventional model systems for insect symbiosis studies, like aphids and tsetse flies, the obligate primary symbiont is harbored in specialized host cells called bacteriocytes, while facultative secondary symbionts reside in either different cells, the body cavity, or the alimentary tract (5-7). The case of C. ocellatus is unique in that the obligate primary symbiont is harbored extracellularly in the midgut cavity, whereas the facultative secondary symbiont is associated with gonads of the host insect. How the extracellular primary symbiont and the secondary endosymbiont interact in the same host body is of interest, deserving future studies.
Acknowledgments
We thank K. Ito, M. Kimura, H. Mukai, S. Ohno, M. Takai, and Y. G. Baba for insect samples.
This study was funded by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN).
Footnotes
Published ahead of print on 16 April 2010.
REFERENCES
- 1.Abe, Y., K. Mishiro, and M. Takanashi. 1995. Symbiont of brown-winged green bug, Plautia stali Scott. Jpn. J. Appl. Entomol. Zool. 39:109-115. [Google Scholar]
- 2.Akman, L., A. Yamashita, H. Watanabe, K. Oshima, T. Shiba, M. Hattori, and S. Aksoy. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glosssinidia. Nat. Genet. 32:402-407. [DOI] [PubMed] [Google Scholar]
- 3.Aksoy, S., and V. M. R. Rio. 2005. Interactions among multiple genomes: tsetse, its symbionts and trypanosomes. Insect Biochem. Mol. Biol. 35:691-698. [DOI] [PubMed] [Google Scholar]
- 4.Arisawa, S. 2003. Constituents of the pericarps of Mallotus japonicus (Euphorbiaceae). Yakugaku Zasshi 123:217-224. (In Japanese with English abstract.) [DOI] [PubMed] [Google Scholar]
- 5.Bourtzis, K., and T. A. Miller. 2003. Insect symbiosis. CRC Press, Boca Raton, FL.
- 6.Bourtzis, K., and T. A. Miller. 2006. Insect symbiosis II. CRC Press, Boca Raton, FL.
- 7.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience, Troy, NY.
- 8.Dale, C., and I. Maudlin. 1999. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int. J. Syst. Bacteriol. 49:267-275. [DOI] [PubMed] [Google Scholar]
- 9.Douglas, A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17-37. [DOI] [PubMed] [Google Scholar]
- 10.Fukatsu, T., T. Tsuchida, N. Nikoh, and R. Koga. 2001. Spiroplasma symbiont of the pea aphid Acyrthosiphon pisum (Insecta: Homoptera). Appl. Environ. Microbiol. 67:1284-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukatsu, T., H. Shibao, N. Nikoh, and S. Aoki. 2001. Genetically distinct populations in an Asian soldier-producing aphid, Pseudoregma bambucicola (Homoptera: Aphididae), identified by DNA fingerprinting and molecular phylogenetic analysis. Mol. Phylogenet. Evol. 18:423-433. [DOI] [PubMed] [Google Scholar]
- 12.Fukatsu, T., and N. Nikoh. 1998. Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Appl. Environ. Microbiol. 64:3599-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukatsu, T., and T. Hosokawa. 2002. Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima. Appl. Environ. Microbiol. 68:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukatsu, T., R. Koga, W. A. Smith, K. Tanaka, N. Nikoh, K. Sasaki-Fukatsu, K. Yoshizawa, C. Dale, and D. H. Clayton. 2007. Bacterial endosymbiont of the slender pigeon louse, Columbicola columbae, allied to endosymbionts of grain weevils and tsetse flies. Appl. Environ. Microbiol. 73:6660-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHILO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 16.Glasgow, H. 1914. The gastric caeca and the caecal bacteria of the Heteroptera. Biol. Bull. 3:101-171. [Google Scholar]
- 17.Grünwald, S., M. Pilhofer, and W. Höll. 2010. Microbial associations in gut systems of wood- and bark-inhabiting longhorned beetles [Coleptera: Cerambycidae]. Syst. Appl. Microbiol. 33:25-34. [DOI] [PubMed] [Google Scholar]
- 18.Guindon, S., F. Lethiec, P. Duroux, and O. Gascuel. 2005. PHYML Online: a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557-W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heddi, A., and P. Nardon. 2005. Sitophilus oryzae L.: a model for intracellular symbiosis in the Dryophthoridae weevils (Coleoptera). Symbiosis 39:1-11. [Google Scholar]
- 20.Hosokawa, T., Y. Kikuchi, X. Y. Meng, and T. Fukatsu. 2005. The making of symbiont capsule in the plataspid stinkbug Megacopta punctatissima. FEMS Microbiol. Ecol. 54:471-477. [DOI] [PubMed] [Google Scholar]
- 21.Hosokawa, T., Y. Kikuchi, N. Nikoh, M. Shimada, and T. Fukatsu. 2006. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 4:e337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber-Schneider, L. 1957. Morphologische und physiologische Untersuchungen an der Wanze Mesocerus marginatus L. und ihren Symbionten (Heteroptera). Z. Morphol. Ökol. Tiere 46:433-480. [Google Scholar]
- 23.Kaltenpoth, M., S. Winter, and A. Kleinhammer. 2009. Localization and transmission route of Coriobacterium glomerans, the endosymbiont of pyrrhocorid bugs. FEMS Microbiol. Ecol. 69:373-383. [DOI] [PubMed] [Google Scholar]
- 24.Kikuchi, Y., and T. Fukatsu. 2003. Diversity of Wolbachia endosymbionts in heteropteran bugs. Appl. Environ. Microbiol. 69:6082-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kikuchi, Y., X. Y. Meng, and T. Fukatsu. 2005. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl. Environ. Microbiol. 71:4035-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kikuchi, Y., T. Hosokawa, and T. Fukatsu. 2007. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial symbiont from the environment every generation. Appl. Environ. Microbiol. 73:4308-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kikuchi, Y., T. Hosokawa, N. Nikoh, X. Y. Meng, Y. Kamagata, and T. Fukatsu. 2009. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 29.Koga, R., T. Tsuchida, and T. Fukatsu. 2003. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc. R. Soc. Lond. B Biol. Sci. 270:2543-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koga, R., T. Tsuchida, and T. Fukatsu. 2009. Quenching autofluorescence of insect tissues for in situ detection of endosymbionts. Appl. Entomol. Zool. 44:281-291. [Google Scholar]
- 31.Kuskop, M. 1923. Bakteriensymbiosen bei Wanzen. Arch. Protistenkd. 47:350-385. [Google Scholar]
- 32.Montllor, C. B., A. Maxmen, and A. H. Purcell. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27:189-195. [Google Scholar]
- 33.Moran, N. A., J. A. Russell, R. Koga, and T. Fukatsu. 2005. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 71:3302-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nogge, G. 1981. Significance of symbionts for the maintenance of an optional nutritional state for successful reproduction in hematophagous arthropods. Parasitology 82:101-104. [Google Scholar]
- 35.Novakova, E., and V. Hypsa. 2007. A new Sodalis lineage from blood sucking fly Craterina melbae (Diptera, Hippoboscoidea) originated independently of tsetse flies symbiont Sodalis glossinidius. FEMS Microbiol. Lett. 269:131-135. [DOI] [PubMed] [Google Scholar]
- 36.Oliver, K. M., J. A. Russell, N. A. Moran, and M. S. Hunter. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. U. S. A. 100:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pimm, S. I., G. J. Russel, J. I. Gittleman, and T. M. Brooks. 1995. Future of biodiversity. Science 269:347-350. [DOI] [PubMed] [Google Scholar]
- 38.Prado, S. S., and R. P. P. Almeida. 2009. Phylogenetic placement of pentatomid stink bug gut symbionts. Curr. Microbiol. 58:64-69. [DOI] [PubMed] [Google Scholar]
- 39.Prado, S. S., and R. P. P. Almeida. 2009. Role of symbiotic gut bacteria in the development of Acrosternum hilare and Murgantia histrionica. Entomol. Exp. Appl. 132:21-29. [Google Scholar]
- 40.Prado, S. S., D. Rubinoff, and R. P. P. Almeida. 2006. Vertical transmission of a pentatomid caeca-associated symbiont. Ann. Entomol. Soc. Am. 99:577-585. [Google Scholar]
- 41.Robinson-Rechavi, M., and D. Huchon. 2000. RRTree: relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics 16:296-297. [DOI] [PubMed] [Google Scholar]
- 42.Rosenkranz, W. 1939. Die Symbiose der Pentatomiden (Hemiptera Heteroptera). Z. Morphol. Ökol. Tiere 36:279-309. [Google Scholar]
- 43.Sakurai, M., R. Koga, T. Tsuchida, X.-Y. Meng, and T. Fukatsu. 2005. Rickettsia symbiont of the pea aphid Acyrthosiphon pisum: novel cellular tropism, effect on the host fitness, and interaction with the essential symbiont Buchnera. Appl. Environ. Microbiol. 71:4069-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scarborough, C. L., J. Ferrari, and H. C. J. Godfray. 2005. Aphid protected from pathogen by endosymbiont. Science 310:1781. [DOI] [PubMed] [Google Scholar]
- 45.Schorr, H. 1957. Zur Verhaltensbiologie und Symbiose von Brachypelta aterrima Först (Cydnidae, Heteroptera). Z. Morphol. Ökol. Tiere 45:561-602. [Google Scholar]
- 46.Schuh, R. T., and J. A. Slater. 1995. True bugs of the world (Hemiptera: Heteroptera). Cornell University Press, Ithaca, NY.
- 47.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and J. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toju, H., T. Hosokawa, R. Koga, N. Nikoh, X. Y. Meng, N. Kimura, and T. Fukatsu. 2010. “Candidatus Curculioniphilus buchneri,” a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Appl. Environ. Microbiol. 76:275-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomokuni, M., T. Yasunaga, M. Kawamura, M. Takai, T. Kawasawa, and I. Yamashita. 1993. A field guide to Japanese bugs. Zenkoku Nouson Kyouiku Kyoukai, Tokyo, Japan. (In Japanese.)
- 51.Tsuchida, T., R. Koga, and T. Fukatsu. 2004. Host plant specialization governed by facultative symbiont. Science 303:1989. [DOI] [PubMed] [Google Scholar]
- 52.Wilson, E. O. 1992. The diversity of life. W. W. Norton & Company, New York, NY.