Abstract
To examine the effect of pathogens on the diversity and structure of plant-associated bacterial communities, we carried out a molecular analysis using citrus and huanglongbing as a host-disease model. 16S rRNA gene clone library analysis of citrus roots revealed shifts in microbial diversity in response to pathogen infection. The clone library of the uninfected root samples has a majority of phylotypes showing similarity to well-known plant growth-promoting bacteria, including Caulobacter, Burkholderia, Lysobacter, Pantoea, Pseudomonas, Stenotrophomonas, Bacillus, and Paenibacillus. Infection by “Candidatus Liberibacter asiaticus” restructured the native microbial community associated with citrus roots and led to the loss of detection of most phylotypes while promoting the growth of bacteria such as Methylobacterium and Sphingobacterium. In pairwise comparisons, the clone library from uninfected roots contained significantly higher 16S rRNA gene diversity, as reflected in the higher Chao 1 richness estimation (P ≤ 0.01) of 237.13 versus 42.14 for the uninfected and infected clone libraries, respectively. Similarly, the Shannon index of the uninfected clone library (4.46) was significantly higher than that of the infected clone library (2.61). Comparison of the uninfected clone library with the infected clone library using LIBSHUFF statistics showed a significant difference (P ≤ 0.05). Quantitative PCR analysis revealed that the bacterial community changes not only qualitatively but also quantitatively. The relative proportions of different groups of bacteria changed significantly after infection with the pathogen. These data indicate that infection of citrus by “Ca. Liberibacter asiaticus” has a profound effect on the structure and composition of the bacterial community associated with citrus roots.
Plants synthesize an almost infinite range of carbonaceous compounds by capturing energy from sunlight and utilizing this for the reduction of carbon contained in atmospheric CO2. These photosynthates provide carbon, nitrogen, and energy sources and make plants excellent ecosystems for microorganisms, particularly bacteria (14). Plants themselves can be divided into different microenvironments, and conditions differ considerably between the highly variable aerial plant parts and the more-stable root system (51). Microbes interact with plant tissues and cells with different degrees of dependence and have developed several strategies for adapting to the plant environment. Plant-microbe interactions include competition, commensalism, mutualism, and parasitism (14, 34, 39). The microbial community structure of plant-associated bacteria changes in response to a variety of processes, and these variations have been suggested to affect ecosystem processes (e.g., nutrient recycling, decomposition) or the effectiveness of microbial invasion (e.g., growth of pathogens, release of plant growth-promoting rhizobacteria) (11, 30, 31).
Losses caused by plant pathogens have been and remain important constraints on efforts to increase plant production and productivity worldwide. Among the bacterial pathogens, the most intensively studied members belong to the phylum Proteobacteria (for example, Agrobacterium, Erwinia, Pseudomonas, Ralstonia, and Xanthomonas) (1). These pathogens are spread by wind, rain, insects, or cultivation practices and enter plant tissues either through wounds or natural openings. Plant-pathogenic bacteria of the phylum Proteobacteria cause diverse disease symptoms and cause host cell death in various plant parts. These symptoms affect both the yield and the quality of the plant produce and can have various effects on the economy and society (42). Only a few studies have examined the influence of phytopathogens on the microbial diversity of plant-associated bacteria (2, 28, 29, 33, 48).
Although extensively studied, pathogenic interactions represent only a fraction of the overall plant-microbe interactions. In fact, out of 5,806 known bacterial species in about 1,094 genera, plant-pathogenic bacteria are recorded only in 132 species in 29 genera (49). The majority of plant-microbe interactions are either commensalistic or mutualistic (3, 37, 43). Plants can benefit from these bacterial associations in terms of growth enhancement, nutrient uptake, and/or stress reduction (41). The diversity and stability of the plant-associated bacterial communities heavily influence soil and plant quality and ecosystem sustainability (20, 27, 31). Much of the basic information regarding the community structure of plant-associated bacteria, their principal functions, their relative ecological stability, and the organizing forces that govern their continuity is still lacking. Also, the interactions between plant-associated bacterial communities and phytopathogens are not well understood, and our knowledge of the intimacy and decisiveness of such associations with respect to the behavior and survival of participating organisms is still in its infancy.
Theoretically, plants interact simultaneously with different groups of bacteria via compounds exuded by the roots (3). However, it has been suggested that plants can specifically attract bacteria for their own ecological and evolutionary benefit (3, 39, 41). This selection process allows the recruitment of different groups of plant-associated bacteria possessing general plant growth-promoting traits. Once recruited, these bacteria undergo host-specific adaptations, the outcome of which is a highly specialized mutualism (24). Such mutualism may make plants better able to tolerate plant-associated bacteria without recognizing them as pathogens, while the bacteria, in turn, become more responsive to the plant's metabolism. We hypothesize that the introduction of pathogens to this finely tuned system will impact plant-associated microbes and can result in a shift in the structure of the microbial community. The pathogen can mediate this restructuring by various mechanisms, which include microbial cross talk, competition for nutrients and space, production of metabolites, and/or changes in the niche environment. Description of microbial diversity and its variation, or the assessment of the factors structuring the composition of the community in the plant, would provide insights into the ecological behavior of pathogenic bacteria in the context of the other microorganisms present in the same niches.
Because of its enormous economic importance, one aspect of plant-microbe interactions that has been extensively studied is the plant-pathogen interaction. Most of the studies in this field have been directed toward a better understanding of the molecular mechanisms of disease induction by the pathogen and defense responses by the plants (15, 19). However, very few studies have investigated the effect of plant pathogens on microbial diversity. The goal of this study is to attain a better understanding of how plant pathogens affect microbial diversity. In the current study, we have used citrus as the model, because citrus has a long life and is usually maintained for more than 20 years in the citrus grove. Thus, the microbial diversity in citrus is expected to be stable and mature. Also, the microbial diversity associated with citrus roots has not been reported in the literature.
To test our hypothesis, we have evaluated the effect of citrus huanglongbing (HLB, or citrus greening) in relation to changes in the native microbial community of citrus roots. HLB is associated with three species of phloem-limited, Gram-negative, fastidious alphaproteobacteria (5). A comprehensive study of bacterial diversity associated with HLB diseased trees has indicated that “Candidatus Liberibacter asiaticus” was the only pathogen responsible for HLB disease in Florida (38, 45). HLB was found in Florida in 2005. Because of its relatively recent entry in Florida, we postulate that exposure of citrus-associated bacteria to HLB is at a juvenile stage. This is in contrast to the situation for some other well-established pathogens to which plants have been exposed previously; in such cases, microbial diversity has been changed due to host response or other factors. To achieve our goal, we have undertaken community profiling of the plant-associated bacteria of citrus root samples, both those infected with the HLB pathogen and uninfected samples, by sequence analysis of 16S rRNA gene libraries. Bacterial community sizes were compared using taxon-specific quantitative real-time PCR (qPCR). This study will provide information about interspecies relationships that could be exploited through biotechnology for the management of plant diseases.
MATERIALS AND METHODS
Sample collection.
Root samples were collected from Valencia orange (Citrus sinensis) trees (3 each) identified as HLB symptomatic or asymptomatic on the basis of visible symptoms in a heavily infected grove at Fort Pierce, Florida. Root segments were collected from the stem base with a shovel at a depth of 5 to 15 cm. The samples were brought to the lab in a cooler with ice in November 2008 and were processed immediately.
Sample processing.
Roots were washed with tap water to remove attached soil. Subsequently, the roots were immersed in 70% ethanol for 3 min, washed with fresh sodium hypochlorite solution (2.5% available Cl−) for 5 min, rinsed in 70% ethanol for 30 s, and finally washed five times with sterile distilled water. To assess surface sterility, 100-μl aliquots of the final rinse water were spread on tryptone yeast extract (TYA) agar plates. The plates were examined for bacterial growth after incubation at 28°C for 3 days. An additional 1-ml aliquot of the final wash water, boiled to release DNA, was assessed using eubacterial primers (799F and 1492R) the PCR procedure as outlined in the following sections. Samples that were not contaminated, as determined by both culture-dependent and PCR-based sterility tests, were used for further analysis.
Total-DNA extraction.
DNA was extracted from root samples using the Wizard genomic DNA purification kit (Promega Corp., Madison, WI) by following the protocol for isolating genomic DNA from plant tissue. The DNA pellet was dried in a Vacufuge (Eppendorf, Westbury, NY) for 15 min and was then dissolved in 100 μl of DNA rehydration solution (Promega).
Detection of “Ca. Liberibacter asiaticus.”
PCR using primers A2 and J5 was performed to confirm the presence of “Ca. Liberibacter asiaticus” in the samples (17) (Table 1). All PCRs in this study were performed in a DNA Engine Peltier thermal cycler (Bio-Rad Laboratories, Hercules, CA). Amplification of DNA was determined by electrophoresis on 1.2% agarose gels for about 30 to 45 min and was visualized by ethidium bromide staining.
TABLE 1.
PCR primers and thermal cycling conditions used for amplification of plant-associated bacteria, detection of “Candidatus Liberibacter asiaticus,” and quantification of the different phyla and classes
| Primer | Sequence (5′-3′) | Thermal conditions |
|---|---|---|
| Endophytic bacteria | ||
| 799F | AAC MGG ATT AGA TAC CCK G | 1 cycle at 94°C for 5 min; 30 cycles at |
| 1492R | GGY TAC CTT GTT ACG ACT T | 94°C for 1 min, 48-58°C (gradient = 48.0, 48.3, 48.9, 49.7, 50.8, 52.3, 54.0, 55.4, 56.5, 57.3, 57.8, 58.0°C) for 45 s, 72°C for 1 min; 1 cycle at 72°C for 8 min |
| Detection of “Candidatus Liberibacter asiaticus” | ||
| Conventional PCR | ||
| A2 | TAT AAA GGT TGA CCT TTC GAG TTT | 1 cycle at 94°C for 2 min; 35 cycles at 94°C for 10 s, 65°C for 10 s, 72°C for 1 min; 1 cycle at 72°C for 4 min |
| J5 | ACA AAA GCA GAA ATA GCA CGA ACA A | |
| qPCR | ||
| CQULA04F | TGG AGG TGT AAA AGT TGC CAA A | 1 cycle at 50°C for 2 min; 1 cycle at 95°C for 15 min; 45 cycles at 94°C for 15 s and 60°C for 1 min |
| CQULAP10P | ATC GTC TCG TCA AGA TTG CTA TCC GTG ATA CTA G | |
| CQULA04R | CCA ACG AAA AGA TCA GAT ATT CCT CTA | |
| Taxon-specific qPCR | ||
| Total bacteria | ||
| Eub338 | ACT CCT ACG GGA GGC AGC AG | 1 cycle at 95°C for 15 min; 40 cycles at 95°C for 1 min, 53°C for 30 s, 72°C for 1 min |
| Eub518 | ATT ACC GCG GCT GCT GG | |
| Acidobacteria | ||
| Acid31 | GAT CCT GGC TCA GAA TC | 1 cycle at 95°C for 15 min; 40 cycles at 95°C for 1 min, 50°C for 30 s, 72°C for 1 min |
| Eub518 | ATT ACC GCG GCT GCT GG | |
| Actinobacteria | ||
| Actino235 | CGC GGC CTA TCA GCT TGT TG | 1 cycle at 95°C for 15 min; 40 cycles at 95°C for 1 min, 60°C for 30 s, 72°C for 1 min |
| Eub518 | ATT ACC GCG GCT GCT GG | |
| Alphaproteobacteria | ||
| Eub338 | ACT CCT ACG GGA GGC AGC AG | 1 cycle at 95°C for 15 min; 40 cycles at 95°C for 1 min, 60°C for 30 s, 72°C for 1 min |
| Alfa685 | TCT ACG RAT TTC ACC YC TAC | |
| Bacteroidetes | ||
| Cfb319 | GTA CTG AGA CAC GGA CCA | 1 cycle at 95°C for 15 min; 40 cycles at 95°C for 1 min, 65°C for 30 s, 72°C for 1 min |
| Eub518 | ATT ACC GCG GCT GCT GG | |
| Betaproteobacteria | ||
| Eub338 | ACT CCT ACG GGA GGC AGC AG | 1 cycle at 95°C for 15 min; 40 cycles at 95°C for 1 min, 60°C for 30 s, 72°C for 1 min |
| Bet680 | TCA CTG CTA CAC GYG | |
| Firmicutes | ||
| Lgc353 | GCA GTA GGG AAT CTT CCG | 1 cycle at 95°C for 15 min; 40 cycles at 95°C for 1 min, 60°C for 30 s, 72°C for 1 min |
| Eub518 | ATT ACC GCG GCT GCT GG |
All qPCR assays were performed in a 96-well plate using an ABI Prism 7500 sequence detection system (Applied Biosystems, Foster City, CA). The CQULA04F-CQULAP10P-CQULA04R primer-probe set was used to target the β-operon region of “Ca. Liberibacter asiaticus,” and qPCRs were performed under conditions described previously (47) (Table 1). Each individual sample was replicated four times on a 96-well plate, and the whole reaction was repeated twice to verify the consistency of the method. The results were analyzed using ABI Prism software. Raw data were analyzed using the default settings (threshold, 0.2) of the software. The standard equation developed by Trivedi et al. (46) was used to convert the individual threshold cycle (CT) values into quantification of the bacterial population as cells per microgram of total tissue DNA.
Amplification of the bacterial 16S rRNA genes.
Bacterial primers 799F and 1492R (6) (Table 1) were used to amplify partial 16S rRNA genes from the samples. These primers can successfully differentiate between bacterial rRNA genes from chloroplast DNA and mitochondrial products. The 50-μl PCR mixture contained 100 ng of DNA extract, 1× Taq reaction buffer, 20 pmol of each primer, 200 μM each deoxynucleoside triphosphate (dNTP), and 1.5 U of Taq DNA polymerase (Promega). The DNA was amplified by using gradient PCR (Table 1), and products of all 12 temperatures on the gradient belonging to one sample type (e.g., the uninfected root of tree 1) were pooled before electrophoresis. The band of approximately 735 bp in the electrophoresis pattern was excised from an agarose gel and purified by the Wizard SV gel and PCR cleanup system (Promega) as described by the manufacturer.
Construction of a 16S rRNA gene clone library.
The PCR products were immediately cloned into the pCR2.1-TOPO TA cloning vector and were then transformed into chemically competent Escherichia coli TOP10 cells (Invitrogen, Carlsbad, CA). Transformed cells were plated onto Luria-Bertani (LB) agar plates with ampicillin (100 μg liter−1) and kanamycin (10 μg liter−1). Plates were incubated overnight at 37°C and were then stored at 4°C for 24 h. Transformants were screened by blue/white colony selection, and individual white colonies were inoculated into 96-well culture blocks (Eppendorf) containing 1 ml of freezing medium (LB broth with 10% [vol/vol] anhydrous glycerol, 25 μg liter−1 ampicillin, and 12.5 μg liter−1 kanamycin) per well. The blocks were incubated at 37°C for 16 h on a rotary shaker at 200 rpm. A 150-μl aliquot of grown cultures was then transferred to sterile 96-well microtitration plates (Corning Inc., NY) for sequencing. Plates were sealed with aluminum seal tape (Eppendorf), and both the blocks and the plates were stored at −80°C. Sequencing was performed using the M13 forward (−20) primer at the sequencing facility of the Interdisciplinary Center for Biotechnology Research at the University of Florida.
Taxonomic assignment and phylogenetic analysis.
The presence of possible chimeric sequences was investigated by using the CHIMERA_CHECK program of Ribosomal Database Project II (RDP II) (7). The taxonomic hierarchy of the sequences in each library was determined using the RDP classifier tool with a confidence level of 80% (http://rdp.cme.msu.edu/). Based on this percentage of similarity, taxonomic assignments were then made. The similarity cutoff values were 75%, 85%, 91%, 92%, 95%, and 100% for phylum, class, order, family, genus, and species designations, respectively. Clone libraries were compared using the RDP Library Comparing tool. Comparison was performed on the phylum level (on both the phylum and class levels for Proteobacteria) with a confidence level of 80%. All the sequences obtained were then compared with the sequences in the GenBank/EMBL/DDBJ database by using the BLASTN search program (http://blast.ncbi.nlm.nih.gov). An operational taxonomic unit (OTU) was defined as a group with ≥97% identity in the rRNA gene sequences according to the conventional definition of a microbial “species” (36).
Species richness estimation and diversity.
All species richness and diversity index estimations were performed using the FASTGroupII program, with a default of 80% similarity in the sequence match (50) (http://biome.sdsu.edu/fastgroup/fg_tools.htm). Each sequence was treated as a separate sample. Poor-quality sequences and chimeras were removed from all groups. The diversity of the clone library was also investigated by rarefaction analysis. Rarefaction curves were calculated using the freeware program aRarefactWin (18). To examine the differences in the compositions of bacterial communities, principal coordinate analysis (PCA) using UniFrac (25) was performed. First, a phylogenetic tree was constructed for the 16S rRNA gene sequences using the neighbor-joining method. The phylogenetic tree and the environment file were subsequently uploaded to UniFrac (http://bmf2.colorado.edu/unifrac/index.psp). To determine the significance of differences between the infected and uninfected clone libraries, LIBSHUFF software was used (http://libshuff.mib.uga.edu/) (40). The matrix analyzed by LIBSHUFF was generated by the PRELIBSHUFF program (http://libshuff.mib.uga.edu/) and the DNA-DIST program of PHYLIP (http://evolution.genetics.washington.edu/phylip.html) using the Jukes-Cantor model for nucleotide substitution.
Taxon-specific qPCR.
Real-time PCR quantifications for Acidobacteria, Actinobacteria, Firmicutes, Alphaproteobacteria, Bacteroidetes, Betaproteobacteria, and total bacteria were performed using the primers and cycling conditions described by Fierer et al. (13) (Table 1). qPCRs were carried out on DNAs extracted from different root samples by using Absolute qPCR SYBR green mixtures (Qiagen Inc., CA) on an ABI Prism 7500 sequence detection system. Known template standards were made from plasmids containing previously characterized full-length 16S rRNA gene inserts. Standard curves were run in parallel, corresponding to a range of 108 to 101 copies per μl. Standard curve regression coefficients were consistently above 0.99, and melting curve analysis verified the presence of a single amplicon per reaction. Samples and standards were assessed in at least two different runs to confirm the reproducibility of the quantification. Target copy numbers for each reaction were calculated from the standard curve and were used to ascertain the number of copies per microgram of DNA. The relative fractional abundance for each of the groups was calculated by determining the copy numbers measured with each group-specific qPCR assay and with the “total-bacteria” assay (13). In the analysis of qPCR data, Student t tests were used to determine the significance of pairs of mean values.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of a single representative of each OTU have been deposited in GenBank with accession numbers GU166607 to GU166679.
RESULTS
Detection of “Ca. Liberibacter asiaticus.”
In order to avoid the false-positive or false-negative effect of conventional PCR or qPCR, we combined the two methods to detect “Ca. Liberibacter asiaticus” in the root samples. Conventional PCR using primers A2 and J5, which target the 16S rRNA genes of “Ca. Liberibacter asiaticus,” showed a band of approximately 703 bp in the infected root samples, which was not detected in uninfected samples (Fig. 1). qPCR results showed the absence of “Ca. Liberibacter asiaticus” in the root samples of uninfected citrus trees. The number of “Ca. Liberibacter asiaticus” organisms in infected trees ranged from 2.94 × 104 to 3.70 × 105 genome equivalents μg of total DNA−1.
FIG. 1.
PCR-based detection of “Ca. Liberibacter asiaticus” in the root samples of infected versus uninfected trees. The results of agarose gel electrophoresis of PCR products amplified using primers specific for “Ca. Liberibacter asiaticus” are shown. The 703-bp amplicon is indicative of the presence of “Ca. Liberibacter asiaticus” in samples. Lane M, DNA molecular weight markers; lanes 1 to 9, infected samples from 3 different trees with 3 subsamples each; lanes 10 to 18, uninfected samples from 3 different trees with 3 subsamples each.
Analysis of plant-associated bacterial communities by using clone libraries.
16S rRNA gene clone libraries were constructed from “Ca. Liberibacter asiaticus”-infected and uninfected citrus root samples. Out of a total of 576 clones, 20 were identified as chimeric sequences and 38 showed similarity with cyanobacteria; these were excluded from further analysis. A total of 241 and 277 clones from “Ca. Liberibacter asiaticus”-infected and uninfected root samples, respectively, were analyzed. Using the RDP classifier, these sequences were placed in a taxonomic hierarchy. A confidence limit of 50% in the RDP classifier was used in a previous study to provide taxonomic affiliations and to perform library comparisons (21). We have chosen a more stringent confidence limit, 80%, in order to gain a better understanding of the prevailing levels of diversity in our clone libraries. The relative abundances of the main phyla, as determined with the 80% confidence level, are presented in Fig. 2. Members of 4 different phyla were observed in infected samples, while the clone library of the uninfected samples comprised members of 6 phyla. Clones representing Bacteroidetes were found only in the clone library of infected samples, whereas the phyla Acidobacteria, Deinococcus-Thermus, and Actinobacteria were represented only in uninfected samples. There were significant differences in the actual percentages of members of different phyla (different classes for Proteobacteria) between the two clone libraries. The dominant bacterial groups (constituting ≥10% of clones) in uninfected samples were Alphaproteobacteria (32.6%), Betaproteobacteria (21.0%), Gammaproteobacteria (16.7%), Firmicutes (10.1%), and Actinobacteria (9.8%). A majority of the clones in the library from infected samples belonged to the Alphaproteobacteria (77.4%). The other group making a significant contribution was Bacteroidetes, members of which formed 9.9% of total clones in the library of symptomatic root samples. Although the class Alphaproteobacteria was found to be the most dominant group in both clone libraries, there was a significant difference in the actual number of clones between the two clone libraries.
FIG. 2.
Composition of each 16S rRNA gene clone library at the phylum level (at the class level for Proteobacteria), determined by using the RDP Classifier tool with an 80% confidence level. The y axis represents the abundance (percentage) of each taxon within a given library. *, P ≤ 0.10.
The relatedness of representative clones (one for each taxon found) to their nearest relatives in the NCBI database is shown in Table 2. Most of the clones showed ≥98% similarity to the known reference strains. In some cases, clearly distinct groups within one genus were observed. The clone libraries of infected and uninfected root samples consisted of bacteria belonging to 22 and 65 OTUs, respectively. The sequences related to Proteobacteria made up the largest fraction of the clone libraries. The group Alphaproteobacteria, comprising 14 and 9 OTUs in the uninfected and infected samples, respectively, was the dominant subclass of Proteobacteria. One hundred seventeen clones in the infected clone library showed 99% similarity with “Ca. Liberibacter asiaticus.” This OTU was not represented in the uninfected clone library. Another major difference was in the abundance of clones showing similarity to the genus Methylobacterium. This group was represented by 6 OTUs with 51 clones in the infected library, while only 2 OTUs having 8 clones were found in the uninfected library. Prominent groups of the uninfected library were related to Caulobacter, Brevundimonas, and Mesorhizobium spp. These were not observed in the infected clone library.
TABLE 2.
Distribution of 16S rRNA gene clones of endophytic bacteria from root samples of “Ca. Liberibacter asiaticus”-infected and uninfected citrus trees
| Group | Closest NCBI match (accession no.) | % Identity | Uninfected trees |
Infected trees |
||||
|---|---|---|---|---|---|---|---|---|
| No. of OTUs | No. of clonesa | % of total clonesa | No. of OTUs | No. of clonesa | % of total clonesa | |||
| Alphaproteobacteria | 14 | 85 | 30.68 | 9 | 174 | 72.19 | ||
| Mesorhizobium sp. Aci124 (AB480767) | 100 | 7 | 2.52 | |||||
| Mesorhizobium sp. S23423 (D84623) | 97 | 5 | 1.80 | |||||
| Rhizobium sp. HGR13 (GQ483459) | 99 | 2 | 0.72 | 3 | 1.24 | |||
| Ochrobactrum sp. 1605 (DQ989292) | 98 | 6 | 2.16 | |||||
| Caulobacter sp. DSV3 M (FJ948826) | 100 | 15 | 5.41 | |||||
| Paracoccus sp. GN-N06-15.1 (EU518706) | 100 | 8 | 2.88 | |||||
| Methylosinus sp. LW2 (AF150786) | 97 | 9 | 3.24 | |||||
| Methylobacterium sp. AR5.1/5 (EU789486) | 100 | 5 | 1.80 | 3 | 1.24 | |||
| Methylobacterium hispanicum GN05-11d (DQ872462) | 100 | 3 | 1.08 | 4 | 1.64 | |||
| Brevundimonas sp. DSV1M (FJ948824) | 100 | 11 | 4.03 | |||||
| Sphingopyxis sp. Geo 48 (EU816422) | 99 | 5 | 1.80 | |||||
| Bosea thioxidans BI-42 (AF508803) | 100 | 4 | 1.44 | |||||
| Ochrobactrum anthropi (EU119263) | 99 | 1 | 0.36 | |||||
| Mesorhizobium sp. REG321 (EU703136) | 97 | 4 | 1.44 | |||||
| Methylobacterium sp. PR3/11 (EU789499) | 98 | 10 | 4.41 | |||||
| Methylobacterium sp. PR1/3 (EU789497) | 98 | 18 | 7.46 | |||||
| Methylobacteriaceae bacterium KVD-1894-12 (DQ490353) | 98 | 9 | 3.73 | |||||
| Methylobacterium sp. Evap_S_01 (DQ132876) | 97 | 7 | 2.90 | |||||
| “Candidatus Liberibacter” sp. clone DS500 (FJ388873) | 99 | 117 | 48.54 | |||||
| Sphingomonas yunnanensis 215 (EU730917) | 100 | 3 | 1.24 | |||||
| Betaproteobacteria | 12 | 52 | 18.77 | |||||
| Burkholderia vietnamiensis WBP (EU563934) | 99 | 14 | 5.05 | |||||
| Ralstonia mannitolilytica Pap In Ba8 (EU839656) | 99 | 2 | 0.72 | |||||
| Janthinobacterium sp. IC161 (AB196254) | 98 | 2 | 0.72 | |||||
| Delftia sp. Hq4-10 (EU304256) | 99 | 6 | 2.16 | |||||
| Oxalobacter formigenes (U49749) | 96 | 6 | 2.16 | |||||
| Alcaligenes sp. adx-4 (FJ169469) | 99 | 3 | 1.08 | |||||
| Achromobacter sp. MT-E3 (EU727196) | 98 | 3 | 1.08 | |||||
| Burkholderia cepacia MSMB16 (F114403) | 100 | 4 | 1.44 | |||||
| Achromobacter xylosoxidans S18 (GQ889256) | 98 | 5 | 1.80 | |||||
| Comamonas aquatica 634 (EU841530) | 99 | 3 | 1.08 | |||||
| Herbaspirillum seropedicae (AJ238361) | 97 | 2 | 0.72 | |||||
| Diaphorobacter sp. GS-1 (FJ158841) | 97 | 2 | 0.72 | |||||
| Gammaproteobacteria | 13 | 46 | 16.60 | 4 | 7 | 2.90 | ||
| Stenotrophomonas sp. P1A (AJ495804) | 98 | 2 | 0.72 | |||||
| Pseudomonas putida AD-21 (EU258552) | 99 | 5 | 1.80 | 1 | 0.41 | |||
| Lysobacter antibioticus 156 (FN398326) | 100 | 9 | 3.24 | |||||
| Pantoea ananatis ESS29 (EF602556) | 100 | 7 | 2.52 | |||||
| Acinetobacter sp. 12524 (GQ475503) | 100 | 2 | 0.72 | |||||
| Acinetobacter lwoffii GN-M06-04.1 (EU518693) | 100 | 3 | 1.08 | |||||
| Enterobacter sp. xw 16S (EF592491) | 100 | 2 | 0.72 | |||||
| Xanthomonas perforans BC2923 (GQ461740) | 100 | 2 | 0.72 | |||||
| Erwinia tasmaniensis ET1/99 (AM292081.1) | 98 | 1 | 0.36 | 4 | 1.65 | |||
| Acinetobacter calcoaceticus TMPSB-D12 (EU513394.1) | 100 | 6 | 2.16 | |||||
| Pseudomonas sp. Zj5 (GQ859170) | 99 | 1 | 0.36 | |||||
| Pseudomonas stutzeri DQgbc15 (GQ470400) | 100 | 3 | 1.08 | |||||
| Stenotrophomonas maltophilia VUN 10 (AF068009) | 99 | 3 | 1.08 | |||||
| Pseudomonas sp. BSw10041N (FJ416144) | 99 | 2 | 0.82 | |||||
| Deltaproteobacteria | 1 | 4 | 1.44 | |||||
| Bdellovibrio bacteriovorus 100 (AF084850.1) | 95 | 4 | 1.44 | |||||
| Acidobacteria | 1 | 5 | 1.80 | |||||
| Acidobacteriaceae bacterium Gsoil 149 (AB245339) | 98 | 5 | 1.80 | |||||
| Actinobacteria | 7 | 28 | 10.10 | |||||
| Brachybacterium sp. GN0406-11.4.3 (DQ890505) | 99 | 4 | 1.44 | |||||
| Friedmanniella antarctica AA-1042 (NR_026536) | 99 | 1 | 0.36 | |||||
| Microbacterium paraoxydans (FN257489) | 100 | 2 | 0.72 | |||||
| Streptomyces alni D65 (DQ460470) | 98 | 3 | 1.08 | |||||
| Curtobacterium sp. C01 (EF411134) | 99 | 12 | 4.33 | |||||
| Microbacterium paraoxydans SS18 (FN257489) | 100 | 4 | 1.44 | |||||
| Kocuria kristinae GN-NO6-23.3 (EU518711) | 100 | 2 | 0.72 | |||||
| Firmicutes | 10 | 30 | 10.83 | 3 | 6 | 2.46 | ||
| Bacillus sp. B1(2007) (EU281627) | 99 | 2 | 0.72 | 2 | 0.82 | |||
| Bacillus pumilus JK-SX001 (GQ169785) | 98 | 8 | 2.88 | |||||
| Bacillus cereus S45 (GQ462533) | 98 | 3 | 1.08 | |||||
| Bacillus subtilis HJ5 (GQ249662) | 100 | 2 | 0.72 | 2 | 0.82 | |||
| Paenibacillus lentimorbus (AB110988) | 98 | 5 | 1.80 | |||||
| Cohnella soli strain 5GH36-9 (EF368009) | 97 | 1 | 0.36 | |||||
| Paenibacillus sp. GT-H3 (GQ355277) | 99 | 3 | 1.08 | |||||
| Brevibacterium sp. BN53-1 (AB066340) | 99 | 1 | 0.36 | |||||
| Bacillus firmus 26-18 (FJ607047) | 99 | 4 | 1.44 | |||||
| Aerococcus sp. 4103 (FJ405327) | 98 | 1 | 0.36 | |||||
| Bacillus oshimensis 1P09AA (EU977653) | 99 | 2 | 0.82 | |||||
| Deinococcus-Thermus | 1 | 4 | 1.44 | |||||
| Deinococcus sp. X-121sp (EU718060) | 99 | 4 | 1.44 | |||||
| Bacteroidetes | 1 | 22 | 9.12 | |||||
| Sphingobacterium daejeonense (AB249372) | 98 | 1 | 22 | 9.12 | ||||
| Uncultured | 6 | 23 | 8.30 | 5 | 32 | 13.27 | ||
| Uncultured bacterium clone SN27 (EU735655) | 97 | 7 | 2.52 | 6 | 2.48 | |||
| Uncultured soil bacterium clone M16_Pitesti (DQ378236) | 99 | 6 | 2.16 | 6 | 2.48 | |||
| Uncultured bacterium clone 1959a-21 (AY917654) | 96 | 1 | 0.36 | 6 | 2.48 | |||
| Uncultured Simkania sp. clone DA81 (FJ388266) | 97 | 1 | 0.36 | 7 | 2.90 | |||
| Uncultured Brevundimonas sp. clone DA155 (FJ388340) | 98 | 1 | 0.36 | 7 | 2.90 | |||
| Kordiimonas sp. clone DA162 (FJ388347) | 97 | 7 | 2.52 | |||||
Blank entries indicate that phylotypes were not detected.
The sequences related to Betaproteobacteria comprised 12 OTUs in the uninfected clone library, while none were found in the infected library. Two OTUs, including 18 clones, were identified as Burkholderia spp. Other prominent OTUs have clones showing high similarity to bacteria belonging to Delftia, Oxalobacter, and Achromobacter spp. The Gammaproteobacteria were represented by 13 (46 clones) and 4 (7 clones) OTUs in the uninfected and infected libraries, respectively. Clones related to the genus Pseudomonas were found in both clone libraries. A substantial number of clones showing 100% similarity to Lysobacter antibioticus, Pantoea ananatis, and Acinetobacter spp. were found in the uninfected clone library. The levels of phylogenetic diversity of Deltaproteobacteria, Acidobacteria, and Deinococcus-Thermus were much lower; each of these groups was affiliated with only 1 OTU in the uninfected clone library. Within the group Bacteriodetes, only 1 OTU, showing high similarity with Sphingobacterium daejeonense, was observed in the clone library from infected samples. However, this OTU consisted of a total of 22 clones and was the second most dominant group. Seven OTUs belonging to the group Actinobacteria were observed exclusively in the uninfected clone library. Within this group, there was one predominant OTU having 12 clones, and it showed similarity to a Curtobacterium sp. The uninfected clone library included 10 OTUs for the group Firmicutes, compared to only 3 in the infected clone library. Clones belonging to the genus Bacillus were observed in both clone libraries, while Paenibacillus, Cohnella, Brevibacterium, and Aerococcus were present only in the uninfected clone library. Thirty-two and 23 clones in the infected and uninfected libraries, respectively, could not be assigned to any bacterial taxon on the basis of 16S rRNA gene sequence similarity. Most of these clones showed similarity to uncultured bacterial clones from different soils or to endophytes from “Ca. Liberibacter asiaticus”-infected citrus leaves (38).
Species richness estimation and diversity.
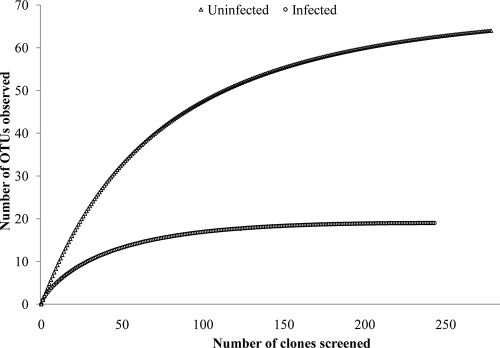
Rarefaction curves of 16S rRNA gene clone libraries of root-associated bacteria from “Ca. Liberibacter asiaticus”-infected and uninfected citrus tree roots are presented in Fig. 3. The rarefaction curves of both the clone libraries tended to plateau. This indicates that these two libraries are large enough to reflect the bacterial diversity of the respective samples. In pairwise comparisons, the uninfected clone library contained significantly higher 16S rRNA gene diversity, as reflected in the higher Chao 1 richness estimations (P ≤ 0.01). Chao 1 richness estimations were 237.13 and 42.14 for the uninfected and infected clone libraries, respectively. Similarly, the Shannon-Weiner diversity index of the uninfected clone library (4.46) was significantly higher than that of the infected clone library (2.61). PCA revealed a clear separation between the two sets of libraries. Both the groups could be observed along the first axis of the ordination plot, which explains 69.21% of the observed variation. Comparison of the infected clone library with the uninfected clone library using LIBSHUFF statistics revealed that most of the OTU diversity in the infected samples was represented in uninfected samples and that the infected samples were not significantly different from the uninfected samples (P ≤ 0.62). In contrast, comparison of the uninfected with the infected clone libraries showed that uninfected samples were significantly different (P ≤ 0.05), in that they contained additional sequences that did not occur in the infected samples.
FIG. 3.
Rarefaction curves of 16S rRNA gene clone libraries of plant-associated bacteria from “Ca. Liberibacter asiaticus”-infected and uninfected citrus roots.
Phylotype analysis by qPCR.
There was no significant difference in the total bacterial population, which ranged from 1.3 × 105 to 2.9 × 105 genome equivalents μg of DNA−1 in “Ca. Liberibacter asiaticus”-infected root samples and from 1.9 × 105 to 3.1 × 105 genome equivalents μg of DNA−1 in uninfected root samples. To determine the changes in different groups of bacteria, we have presented our results as fractional copy numbers, which provide a more accurate index of target abundances than the actual copy numbers, since they neutralize the bias produced by phylotypes with multiple copies of small-subunit (SSU) rRNA genes (4, 13). Clear differences in the fractional abundances of all the groups of bacteria were observed between “Ca. Liberibacter asiaticus”-infected and uninfected samples (Fig. 4). The proportion of Alphaproteobacteria was significantly higher (P ≤ 0.01) in “Ca. Liberibacter asiaticus”-infected root samples than in uninfected root samples. This is not surprising, since the HLB pathogen belongs to this group, and its infection might be the reason for the increase in the relative abundance of Alphaproteobacteria in the infected samples. The other group whose levels were higher in infected samples belonged to the phylum Bacteroidetes. The abundances of all other groups of bacteria were significantly higher in the uninfected root samples. In “Ca. Liberibacter asiaticus”-infected roots, a nearly 2-fold decrease in relative abundance was observed for Firmicutes and Betaproteobacteria (significant differences [P ≤ 0.1]). The relative abundances of bacteria belonging to the Acidobacteria and Actinobacteria were nearly four times greater in uninfected than in “Ca. Liberibacter asiaticus”-infected samples, a significant difference (P ≤ 0.01).
FIG. 4.
Relative abundances of the six bacterial groups in “Ca. Liberibacter asiaticus”-infected and uninfected citrus roots, as estimated using the qPCR assays. Error bars represent the standard errors of the means for three replicates. *, P ≤ 0.10; **, P ≤ 0.05; ***, P ≤ 0.01.
DISCUSSION
Plants form a complex ecosystem in which many factors affect the structure and species composition of the associated bacterial communities (41). The interactions between plants and bacteria are dynamic and can range from mutualism through commensalism to parasitism in a continuous manner (16, 35). Plant-associated bacterial communities play an important role in plant growth and health, but only a few reports have addressed the effect of an invading pathogen on the stability of native microbial communities (2, 28, 29, 33, 48). Therefore, this study was undertaken to provide insights into the nature and composition of bacteria associated with healthy plants and their shifts in relation to pathogen attack and infestation.
In accordance with our assumption that plants function as true filters of soil organisms and select mostly the mutualists as their associates, we observed various beneficial phylotypes in the clone library from uninfected root samples. These phylotypes belonged to the genera Caulobacter, Burkholderia, Lysobacter, Pantoea, Pseudomonas, Stenotrophomonas, Bacillus, and Paenibacillus, and the representative OTUs of these genera found in the uninfected clone library have been studied in detail for various biocontrol and plant growth promotion traits (8, 12, 26, 35, 44). Representatives of the phylum Actinobacteria, with the highest number of clones belonging to Curtobacterium spp., were observed only in the clone library from uninfected samples. Interestingly, Curtobacterium flaccumfaciens has been isolated frequently from asymptomatic sweet orange trees with citrus variegated chlorosis (CVC; caused by Xylella fastidiosa). It effectively controls CVC through the colonization of the same ecological niche and the production of three bacteriocins active against X. fastidiosa (2). We have not yet been able to demonstrate clearly whether the communities associated with healthy roots in fact promote disease suppression or whether their presence is simply a direct consequence of the absence of the pathogen. Nevertheless, the clear demonstration of differences in the bacterial communities associated with the roots of “Ca. Liberibacter asiaticus”-infected versus uninfected citrus trees certainly represents a good starting point for further analyses.
In the clone library from infected root samples, a majority of the clones showed similarity to “Ca. Liberibacter asiaticus,” while this was not observed in uninfected root samples. We have previously used clone library and PhyloChip-based analyses to demonstrate the association of “Ca. Liberibacter asiaticus” with HLB in Florida (38). Another major difference was in the abundance of bacteria belonging to the genus Methylobacterium, which was significantly greater in infected root samples. While studying the bacterial diversity of Citrus sinensis cultivars infected with CVC, Araújo et al. (2) observed a relationship between CVC symptoms and the frequency of isolation of species of Methylobacterium, which were frequently isolated from symptomatic plants. However, this group was not observed in the diversity analysis of “Ca. Liberibacter asiaticus”-infected leaf samples (38). Another prominent group in infected samples that was not observed in uninfected samples matched with Sphingobacterium daejeonense. PhyloChip analysis has shown that representatives of this group were more abundant in HLB symptomatic leaves than in asymptomatic leaves (38). The results of our study show that the disease could play a role in the establishment of a few groups of bacteria in the host plant. McSpadden Gardener and Weller (29) have also reported higher abundances of selected bacterial groups in the rhizosphere of wheat plants with take-all disease than in healthy plants.
Clone libraries are useful for identifying and characterizing the dominant bacterial types in environmental samples. However, to accurately describe the microbial diversity within a sample, clone libraries usually need to be quite large. There are, as yet, few studies in which the representativeness issue has been satisfactorily resolved (14). Statistical techniques such as rarefaction analysis are therefore used to evaluate whether the clone library is sufficient to adequately cover the overall diversity (9, 32). Rarefaction analysis showed that the number of clones screened in this study was sufficient to yield a realistic picture of diversity. Clone library analysis of plant-associated bacteria in “Ca. Liberibacter asiaticus”-infected and uninfected citrus tree roots revealed differences in the structures of their microbial communities. Diversity indices and LIBSHUFF statistics clearly showed higher bacterial diversity in uninfected samples. In general, pathogens induce a cascade of reactions in plants, leading to the synthesis of stress metabolites, including H2O2, phytoalexins, and stress signals such as abscicic acid, jasmonic acid, and salicylic acid (23). The altered conditions after the pathogen attack could have variable effects on the survival and proliferation of different groups of bacteria. Microarray analysis has indicated that “Ca. Liberibacter asiaticus” infection significantly affected the expression of 624 genes, most of which were related to sugar metabolism, plant defense, phytohormone metabolism, and cell wall metabolism (22). Our results suggest that infection by the HLB pathogen promoted the growth of only a few phylotypes and caused a shift in microbial diversity.
Only a few studies have dealt directly with changes in the diversity of plant-associated bacteria in response to pathogen infection. In accordance with previous studies, we also noticed shifts in the bacterial community in diseased plants, but there were large differences in the extent of these changes. We observed negative effects of the pathogen on the stability of the plant-associated bacterial community, in contrast to earlier studies, which have reported increases in bacterial diversity in pathogen-infested plants (33, 48). Those investigators observed changes in the bacterial diversity of potato and avocado plants after infection by Erwinia carotovora and Phytopthora cinnamomi, respectively. Both of these pathogens cause rot, and the major virulence factor for both is the cell wall-degrading enzyme. The infection of host plants by these pathogens results in the degradation of the cell wall, which can allow the entry of various other bacteria into the plant system. “Ca. Liberibacter asiaticus,” in accordance with its intracellular nature, lacks genes for the production of extracellular lytic enzymes (10). Therefore, the reason for the differences in the microbial community could lie in the nature of the pathogen and in differences in the virulence mechanism involved in disease development.
Pathogens affect microbial diversity not only qualitatively but also quantitatively. To provide a more in-depth examination of a microbial community than can be done with clone libraries alone, we have used qPCR as a complementary molecular technique for the determination of the relative abundances of the dominant groups of bacteria in the samples. The qPCR approach adopted in the present study was at the coarsest level of taxonomic resolution, which would mask the diversity within each taxon chosen (13). Despite this, there were clear differences in the bacterial community structure between “Ca. Liberibacter asiaticus”-infected and uninfected samples. The results show that although the total number of bacteria remained the same, the relative proportions of certain groups changed significantly after “Ca. Liberibacter asiaticus” infection. The results of qPCR analysis support the trends observed in the clone libraries. The qPCR results also tended to show larger proportions of Alphaproteobacteria and Bacteroidetes in the infected root samples, while a reverse trend was observed for the other groups. Along with the clone library analysis, the qPCR results validate the postulation that competitive interactions between a pathogen and plant-associated bacteria shape the composition of the microbial community in plants.
The data from our study of bacterial diversity in “Ca. Liberibacter asiaticus”-infected and uninfected root samples are consistent with our initial hypothesis that microbial communities undergoing stress from invading pathogens tend to change their structure and diversity. In general, it appears that “Ca. Liberibacter asiaticus” infection restructures the microbial community: many of the species show reduced levels or are not detected and are replaced by other indigenous populations, which can better tolerate or adapt to the stress condition. These data indicate that infection of citrus by “Ca. Liberibacter asiaticus” has a profound effect on the structure and composition of the bacterial community associated with citrus tree roots. Examination of such interactions will help us to understand natural phenomena in the plant-microbe interactions and could lead us to applications resulting in sustainable resources, less impact on the environment, and disease management.
Acknowledgments
This work was supported by the Florida Citrus Production Research Advisory Council (FCPRAC).
Footnotes
Published ahead of print on 9 April 2010.
REFERENCES
- 1.Abramovitch, R. B., J. C. Anderson, and G. B. Martin. 2006. Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol. 7:601-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araújo, W. L., J. Marcon, W. Maccheroni, J. D. van Elsas, J. W. L. van Vuurde, and J. L. Azevedo. 2002. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl. Environ. Microbiol. 68:4906-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bais, H. P., T. L. Weir, L. G. Perry, S. Gilroy, and J. M. Vivanco. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57:233-266. [DOI] [PubMed] [Google Scholar]
- 4.Baxter, J., and S. P. Cummings. 2008. The degradation of the herbicide bromoxynil and its impact on bacterial diversity in a top soil. J. Appl. Microbiol. 104:1605-1616. [DOI] [PubMed] [Google Scholar]
- 5.Bové, J. M. 2006. Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 88:7-37. [Google Scholar]
- 6.Chelius, M. K., and E. W. Triplett. 2001. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 41:252-263. [DOI] [PubMed] [Google Scholar]
- 7.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Compant, S., B. Duffy, J. Nowak, C. Clément, and E. A. Barka. 2005. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Eviron. Microbiol. 71:4951-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. U. S. A. 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan, Y., L. Zhou, D. G. Hall, W. Li, H. Doddapaneni, H. Lin, L. Liu, D. Gabriel, C. M. Vahling, K. Williams, A. Dickerman, Y. Sun, and T. R. Gottwald. 2009. Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plant Microbe Interact. 22:1011-1020. [DOI] [PubMed] [Google Scholar]
- 11.Dunfield, K. E., and J. J. Germida. 2003. Seasonal changes in the rhizosphere microbial communities associated with field-grown genetically modified canola (Brassica napus). Appl. Environ. Microbiol. 69:7310-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emmert, E. A. B., and J. Handelsman. 1999. Biocontrol of plant disease: a (Gram-) positive perspective. FEMS Microbiol. Lett. 171:1-9. [DOI] [PubMed] [Google Scholar]
- 13.Fierer, N., J. A. Jackson, R. Vilgalys, and R. B. Jackson. 2005. The assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71:4117-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garbeva, P., J. A. V. van Veen, and J. D. van Elsas. 2004. Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 42:243-270. [DOI] [PubMed] [Google Scholar]
- 15.Glazebrook, J. 2005. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43:205-227. [DOI] [PubMed] [Google Scholar]
- 16.Hardoim, P. R., L. S. van Overbeek, and J. D. van Elsas. 2008. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16:463-471. [DOI] [PubMed] [Google Scholar]
- 17.Hocquellet, A., J. M. Bové, and M. Garnier. 1999. Isolation of DNA from the uncultured “Candidatus Liberobacter” species associated with citrus huanglongbing by RAPD. Curr. Microbiol. 38:176-182. [DOI] [PubMed] [Google Scholar]
- 18.Holland, S. 1998. aRarefactWin. University of Georgia, Athens, GA. http://www.uga.edu/strata/software/anRareReadme.html.
- 19.Jones, J. D. G., and J. L. Dangl. 2006. The plant immune system. Nature 444:323-329. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy, A. C., and K. L. Smith. 1995. Soil microbial diversity and the sustainability of agricultural soils. Plant Soil 170:75-86. [Google Scholar]
- 21.Kielak, A., A. S. Pijl, J. A. van Veen, and G. A. Kowalchuk. 2008. Differences in vegetation composition and plant species identity lead to only minor changes in soil-borne microbial communities in a former arable field. FEMS Microbiol. Ecol. 63:372-382. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J., U. S. Sagaram, J. K. Burns, and N. Wang. 2009. Response of sweet orange (Citrus sinensis) to Candidatus Liberibacter asiaticus infection: microscopy and microarray analyses. Phytopathology 99:50-57. [DOI] [PubMed] [Google Scholar]
- 23.Lichtenthaler, H. K. 1998. The stress concept in plants: an introduction. Ann. N. Y. Acad. Sci. 851:187-198. [DOI] [PubMed] [Google Scholar]
- 24.Long, H. H., D. D. Schmidt, and I. T. Baldwin. 2008. Native bacterial endophytes promote host growth in a species-specific manner; phytohormone manipulations do not result in common growth responses. PLoS One 3(7):e2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lugtenberg, B., and F. Kamilova. 2009. Plant-growth-promoting Rhizobacteria. Annu. Rev. Microbiol. 63:541-556. [DOI] [PubMed] [Google Scholar]
- 27.Lukow, T., P. F. Dunfield, and W. Liesack. 2000. Use of T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32:241-247. [DOI] [PubMed] [Google Scholar]
- 28.Mazzola, M., and R. J. Cook. 1991. Effects of fungal pathogens on the population dynamics of biocontrol strains of fluorescent pseudomonads in the wheat rhizosphere. Appl. Environ. Microbiol. 57:2171-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McSpadden Gardener, B. B., and D. M. Weller. 2001. Changes in population of rhizosphere bacteria associated with take-all disease of wheat. Appl. Environ. Microbiol. 67:4414-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miethling, R., G. Wieland, H. Backhaus, and C. C. Tebbe. 2000. Variation of microbial rhizosphere communities in response to crop species, soil origin, and inoculation with Sinorhizobium meliloti L33. Microb. Ecol. 40:43-56. [DOI] [PubMed] [Google Scholar]
- 31.Nannipieri, P., J. Ascher, M. T. Ceccherini, L. Landi, G. Pietramellara, and G. Renella. 2003. Microbial diversity and soil functions. Eur. J. Soil Sci. 54:655-670. [Google Scholar]
- 32.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Aman. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiter, B., U. Pfeifer, H. Schwab, and A. Sessitsch. 2002. Response of endophytic bacterial communities in potato plants to infection with Erwinia carotovora subsp. atroseptica. Appl. Environ. Microbiol. 68:2261-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds, H. L., A. Packer, J. D. Bever, and K. Clay. 2003. Grassroots ecology: plant-microbe-soil interactions as drivers of plant community structure and dynamics. Ecology 84:2281-2291. [Google Scholar]
- 35.Rosenblueth, M., and E. Martínez-Romero. 2006. Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 19:827-837. [DOI] [PubMed] [Google Scholar]
- 36.Rosselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 37.Ryan, R. P., K. Germaine, A. Franks, D. J. Ryan, and D. N. Dowling. 2008. Bacterial endophytes: recent developments and applications. FEMS Microbiol. Lett. 278:1-9. [DOI] [PubMed] [Google Scholar]
- 38.Sagaram, U., K. M. DeAngelis, P. Trivedi, G. L. Andersen, S. E. Lu, and N. Wang. 2009. Bacterial diversity analysis of Huanglongbing pathogen-infected citrus, using PhyloChip arrays and 16S rRNA gene clone library sequencing. Appl. Environ. Microbiol. 75:1566-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schulz, B., and C. Boyle. 2006. What are endophytes? p. 1-13. In B. J. E. Schulz, C. J. C. Boyle, and T. N. Sieber (ed.), Microbial root endophytes. Springer-Verlag, Berlin, Germany.
- 40.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorensen, J., and A. Sessitsch. 2007. Plant-associated bacteria—lifestyle and molecular interactions, p. 211-236. In J. D. van Elsas, J. K. Jansson, and J. T. Trevors (ed.), Modern soil microbiology. CRC Press, Boca Raton, FL.
- 42.Strange, R. N., and P. R. Scott. 2005. Plant disease: a threat to global food security. Annu. Rev. Phytopathol. 43:83-116. [DOI] [PubMed] [Google Scholar]
- 43.Sturz, A. V., B. R. Christie, and J. Nowak. 2000. Bacterial endophytes: potential role in developing sustainable systems of crop production. Crit. Rev. Plant Sci. 19:1-30. [Google Scholar]
- 44.Sun, L., F. Qiu, X. Zhang, X. Dai, X. Dong, and W. Song. 2008. Endophytic bacteria diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microb. Ecol. 55:415-424. [DOI] [PubMed] [Google Scholar]
- 45.Tatineni, S., U. S. Sagaram, S. Gowda, C. J. Robertson, W. O. Dawson, T. Iwanami, and N. Wang. 2008. In planta distribution of ‘Candidatus Liberibacter asiaticus’ as revealed by polymerase chain reaction (PCR) and real-time PCR. Phytopathology 98:592-599. [DOI] [PubMed] [Google Scholar]
- 46.Trivedi, P., U. S. Sagaram, R. H. Brlansky, M. Rogers, L. L. Stelinski, C. Oswalt, J. S. Kim, and N. Wang. 2009. Quantification of viable Candidatus Liberibacter asiaticus in hosts using quantitative PCR with the aid of ethidium monoazide (EMA). Eur. J. Plant Pathol. 124:553-563. [Google Scholar]
- 47.Wang, Z., Y. Yin, H. Hu, Q. Yuan, G. Peng, and Y. Xia. 2006. Development and application of molecular-based diagnosis for ‘Candidatus Liberibacter asiaticus’, the causal pathogen of citrus huanglongbing. Plant Pathol. 55:630-638. [Google Scholar]
- 48.Yang, C., D. E. Crowley, and J. A. Menge. 2001. 16S rDNA fingerprinting of rhizosphere bacterial communities associated with healthy and Phytophthora infected avocado roots. FEMS Microbiol. Ecol. 35:129-136. [DOI] [PubMed] [Google Scholar]
- 49.Young, J. M. 2004. Classification and nomenclature of plant pathogenic bacteria, p. 286-289. In R. M. Goodman (ed.), Encyclopedia of plant and crop science. Marcel Dekker, New York, NY.
- 50.Yu, Y., M. Breitbart, P. McNairnie, and F. Rohwer. 2006. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinformatics 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zinniel, D. K., P. Lambrecht, N. B. Harris, Z. Feng, D. Kuczmarski, P. Higley, C. Ishimaru, A. Arunakumari, R. G. Barletta, and A. K. Vidaver. 2002. Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl. Environ. Microbiol. 68:2198-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]