Abstract
Listeria monocytogenes is a Gram-positive facultative intracellular pathogen which invades different cell types, including nonphagocytic cells, where it is able to replicate and survive. The different steps of the cellular infectious process have been well described and consist of bacterial entry, lysis of the endocytic vacuole, intracellular replication, and spreading to neighboring cells. To study the listerial infectious process, gentamicin survival assays, plaque formation, and direct microscopy observations are typically used; however, there are some caveats with each of these techniques. In this study we describe new single-cell techniques based on use of an array of integrative fluorescent plasmids (green, cyan, and yellow fluorescent proteins) to easily, rapidly, and quantitatively detect L. monocytogenes in vitro and in vivo. We describe construction of 13 integrative and multicopy plasmids which can be used for detecting intracellular bacteria, for measuring invasion, cell-to-cell spreading, and intracellular replication, for monitoring in vivo infections, and for generating transcriptional or translational reporters. Furthermore, we tested these plasmids in a variety of epifluorescence- and flow cytometry-based assays. We showed that we could (i) determine the expression of a particular promoter during the cell cycle, (ii) establish in one rapid experiment at which step in the cell cycle a particular mutant is defective, and (iii) easily measure the number of infected cells in vitro and in mouse organs. The plasmids that are described and the methods to detect them are new powerful tools to study host-Listeria interactions in a fast, robust, and high-throughput manner.
Listeria monocytogenes is a facultative intracellular pathogen and is responsible for human listeriosis. This bacterium is able to cross three tissue barriers, the intestinal barrier, the hemato-encephalic barrier, and the feto-placental barrier, by subverting cellular effectors and functions, thereby allowing bacterial internalization, replication, and survival within different types of cells, including nonphagocytic cells and macrophages (6, 15). Bacterial entry into nonphagocytic cells is induced by binding of two bacterial surface proteins, InlA and InlB, to their cognate receptors on the host cell, E-cadherin and the hepatocyte growth factor (HGF) receptor Met, respectively (2, 15, 34). After entry, Listeria is engulfed in a vacuole, which is rapidly lysed by the action of listeriolysin O (LLO). Following escape from the vacuole, L. monocytogenes replicates in the cytosol. Subsequently, the bacterium polymerizes host actin at the bacterial pole, forming actin tails, which provide the driving force for intracellular motility. The bacterial surface protein ActA is sufficient to promote actin recruitment and polymerization events (15). Using this actin-based motility system, L. monocytogenes can spread from cell to cell by forming protrusions that are engulfed by neighboring cells. After escape from the secondary vacuole that forms in the newly invaded cell, the bacterium may continue its intracellular life cycle (6). Although several aspects of L. monocytogenes virulence have been described, many aspects of this virulence remain unknown; the functions of at least 35% of the Listeria genes have not been identified, and the roles of these genes in virulence have not been determined (12). To address this issue, sensitive high-throughput assays are necessary.
The classical method used to assess the invasiveness of a bacterium is the gentamicin survival assay, which determines the number of bacteria that survive treatment with gentamicin after entry into cells. The two methods commonly used to measure cell-to-cell spread are the plaque assay, which relies on infection of a fibroblast monolayer, and direct microscopic observation of individual foci of infection initiated by uptake of a single bacterium in the cell monolayer in the presence of gentamicin. These techniques are widely used; however, as experiments are increasingly performed at the single-cell level and using high-throughput assays, methods for studying bacterial infections need to be improved.
The use of green fluorescent protein (GFP) as a tool to study host-pathogen interactions has been reported for Gram-negative (28, 32, 33) as well as Gram-positive (1, 10, 20, 22) bacteria. GFP, which was first identified in the jellyfish Aequorea victoria in the 1960s (16, 27), enables direct visualization of tagged bacteria. By introducing mutations into the gfp gene, many useful GFP variants have been developed, including the commonly used enhanced GFP (which contains a Ser65Thr substitution), as well as cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) (4). Additionally, a red fluorescent protein cloned from Discosoma coral (DsRed) and variants of this protein (mRFP1 and mCherry) have been generated and used in reporter systems (25). The emission wavelengths of the different fluorescent protein variants cover the visual spectrum, and these variants are useful for single-cell studies.
L. monocytogenes strains harboring plasmids expressing GFP have been used in previous studies (10, 17, 36). However, these studies used multicopy plasmids bearing GFP and antibiotic selection genes for maintenance of the fluorescence plasmids over long periods of time. Furthermore, heterogeneity in the plasmid copy number throughout the cell population prevents use of these plasmids for quantitative single-cell studies. In this study, we constructed 9 integrative plasmids (and 4 multicopy plasmids) that allowed chromosomal labeling of L. monocytogenes with different fluorescent proteins (GFP, CFP, and YFP). Since these proteins were expressed from the chromosome, there was no need for antibiotic pressure, which allowed long-term experiments to be performed. Furthermore, a single copy resulted in homogeneous fluorescence; therefore, quantitative studies could be performed. Our plasmids either express the fluorescent proteins under various promoters (Phyper [a constitutive promoter], PinlC, and PactA) or can be used to generate transcriptional or translational fusions. The powerful methods described below could ultimately be used for high-throughput screens to identify either new host factors or new bacterial factors that are involved in cellular infection or virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cell lines.
Strains and plasmids used in this study are listed in Table 1. Listeria strains were grown at 37°C in brain heart infusion (BHI) (Difco Laboratories, Detroit, MI). Escherichia coli strains were grown in Luria-Bertani (LB) broth at 37°C. When required, chloramphenicol was used at a final concentration of 7 μg/ml for L. monocytogenes and at a final concentration of 35 μg/ml for E. coli, and kanamycin was used at a final concentration of 50 μg/ml for E. coli. The tissue culture cells used in this study were Caco-2 cells (human epithelial colon cells; ATCC HTB-37), Jeg-3 cells (human epithelial placental cells; ATCC HTB-36), HeLa cells (human epithelial cervix cells; ATCC CCL-2), and J774A.1 cells (BALB/c mouse macrophage cells; ATCC TIB-67). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 1 mM sodium pyruvate, 2 mM l-glutamine, 0.1 mM nonessential amino acids, and 10% (vol/vol) fetal calf serum (Biowest). Cells were grown at 37°C with 10% CO2.
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Characteristics | Collection no. | Antibiotic resistancea | Source or reference |
|---|---|---|---|---|
| Plasmids | ||||
| pCR-Blunt | Blunt-end PCR cloning vector | Km | Invitrogen | |
| pCR2.1-TOPO | PCR cloning vector | Ap | Invitrogen | |
| pmCherry | Vector harboring the mCherry-encoding gene | Ap | Clontech | |
| pAT18 | E. coli-L. monocytogenes shuttle cloning vector | 31 | ||
| pPL2 | L. monocytogenes site-specific phage integration vector | BUG 2176 | Cm | 18 |
| pH-hly gfp-PL3 | pHPL3 expressing constitutive GFP | BUG 2377 | Cm | 26 |
| pJEBAM2 | cfp gene under control of the L. monocytogenes Pdlt promoter | BUG 2461 | Ery | 1 |
| pJEBAM3 | yfp gene under control of the L. monocytogenes Pdlt promoter | BUG 2462 | Ery | 1 |
| Cloning plasmids | ||||
| pTOPO-Phyper | Terminator-Phyper cloned in pCR2.1-TOPO | BUG 2529 | Km | This study |
| pTOPO-PinlC | Terminator-PinlC cloned in pCR2.1-TOPO | BUG 2486 | Km | This study |
| pTOPO-PactA | Terminator-PactA cloned in pCR2.1-TOPO | BUG 2765 | Km | This study |
| pBlun-UTR-GFP | hly UTR-gfp cloned in pCR-Blunt | BUG 2526 | Km | This study |
| pBlunt-UTR-YFP | hly UTR-yfp cloned in pCR-Blunt | BUG 2768 | Km | This study |
| pBlunt-UTR-CFP | hly UTR-cfp cloned in pCR-Blunt | BUG 2769 | Km | This study |
| pBlunt-UTR-Cherry | hly UTR-mCherry cloned in pCR-Blunt | BUG 2770 | Km | This study |
| Plasmids used for constitutive labeling of Listeria | ||||
| pAD1-cGFP | pPL2-Phyper-GFP (constitutive) | BUG 2479 | Cm | This study |
| pAD1-cYFP | pPL2-Phyper-YFP (constitutive) | BUG 2771 | Cm | This study |
| pAD1-cCFP | pPL2-Phyper-CFP (constitutive) | BUG 2772 | Cm | This study |
| pAD1-cCherry | pPL2-Phyper-mCherry (constitutive) | BUG 2773 | Cm | This study |
| pAT18-cGFP | pAT18-Phyper-GFPmut2 (constitutive) | BUG 2533 | Ery | This study |
| pAT18-cYFP | pAT18-Phyper-YFP (constitutive) | BUG 2534 | Ery | This study |
| pAT18-cCFP | pAT18-Phyper-CFP (constitutive) | BUG 2535 | Ery | This study |
| pAT18-cCherry | pAT18-Phyper-mCherry (constitutive) | BUG 2818 | Ery | This study |
| Transcriptional fusions | ||||
| pAD2-PinlC-GFP | pPL2 expressing GFP under control of PinlC | BUG 2491 | Cm | This study |
| pAD2-PinlC-CFP | pPL2 expressing CFP under control of PinlC | BUG 2792 | Cm | This study |
| pAD3-PactA-GFP | pPL2 expressing GFP under control of PactA | BUG 2793 | Cm | This study |
| pAD3-PactA-CFP | pPL2 expressing CFP under control of PactA | BUG 2795 | Cm | This study |
| pAD3-PactA-YFP | pPL2 expressing YFP under control of PactA | BUG 2794 | Cm | This study |
| Plasmids encoding fluorescent proteins to generate translational fusions | ||||
| pTL-GFP | GFP cloned in pCR-Blunt | BUG 2799 | Km | This study |
| pTL-CFP | CFP cloned in pCR-Blunt | BUG 2484 | Km | This study |
| pTL-YFP | YFP cloned in pCR-Blunt | BUG 2485 | Km | This study |
| pTL-Cherry | mCherry cloned in pCR-Blunt | BUG 2800 | Km | This study |
| Escherichia coli strains | ||||
| TG1 λatt cat-gfp | TG1 strain λ-att gfpmut2-cat | BUG 2512 | Cm35 | J. M. Ghigo |
| XL1-Blue | ||||
| Listeria strains | ||||
| EGD | L. monocytogenes wild-type strain | BUG 600 | 13 | |
| EGD-e | L. monocytogenes wild-type strain | BUG 1600 | 12 | |
| ΔinlA EGD | L. monocytogenes EGD InlA deletion mutant | BUG 947 | 7 | |
| ΔinlB EGD | L. monocytogenes EGD InlB deletion mutant | BUG 1047 | 7 | |
| ΔinlAB EGD | L. monocytogenes EGD InlAB deletion mutant | BUG 949 | 7 | |
| ΔactA EGD | L. monocytogenes EGD ActA deletion mutant | BUG 2140 | 19 | |
| L. innocua | L. innocua wild-type strain | BUG 499 | 7 | |
| Listeria strains constitutively labeled with fluorescent proteins | ||||
| EGD-cGFP | pAD1-cGFP chromosomally integrated in EGD | BUG 2539 | Cm7 | This study |
| EGD-cYFP | pAD1-cYFP chromosomally integrated in EGD | BUG 2541 | Cm7 | This study |
| EGD-cCFP | pAD1-cCFP chromosomally integrated in EGD | BUG 2543 | Cm7 | This study |
| EGDe-cGFP | pAD1-cGFP chromosomally integrated in EGD-e | BUG 2538 | Cm7 | This study |
| EGDe-cYFP | pAD1-cYFP chromosomally integrated in EGD-e | BUG 2540 | Cm7 | This study |
| EGDe-cCFP | pAD1-cCFP chromosomally integrated in EGD-e | BUG 2542 | Cm7 | This study |
| ΔinlA-cGFP | pAD1-cGFP chromosomally integrated in ΔinlA EGD | BUG 2774 | Cm7 | This study |
| ΔinlA-cYFP | pAD1-cYFP chromosomally integrated in ΔinlA EGD | BUG 2775 | Cm7 | This study |
| ΔinlA-cCFP | pAD1-cCFP chromosomally integrated in ΔinlA EGD | BUG 2776 | Cm7 | This study |
| ΔinlB-cGFP | pAD1-cGFP chromosomally integrated in ΔinlB EGD | BUG 2553 | Cm7 | This study |
| ΔinlB-cYFP | pAD1-cYFP chromosomally integrated in ΔinlB EGD | BUG 2554 | Cm7 | This study |
| ΔinlB-cCFP | pAD1-cCFP chromosomally integrated in ΔinlB EGD | BUG 2555 | Cm7 | This study |
| ΔinlAB-cGFP | pAD1-cGFP chromosomally integrated in ΔinlAB EGD | BUG 2777 | Cm7 | This study |
| ΔinlAB-cYFP | pAD1-cYFP chromosomally integrated in ΔinlAB EGD | BUG 2778 | Cm7 | This study |
| ΔinlAB-cCFP | pAD1-cCFP chromosomally integrated in ΔinlAB EGD | BUG 2779 | Cm7 | This study |
| ΔactA-cGFP | pAD1-cGFP chromosomally integrated in ΔactA EGD | BUG 2783 | Cm7 | This study |
| ΔactA-cYFP | pAD1-cYFP chromosomally integrated in ΔactA EGD | BUG 2784 | Cm7 | This study |
| ΔactA-cCFP | pAD1-cCFP chromosomally integrated in ΔactA EGD | BUG 2785 | Cm7 | This study |
| EGDe-(pAT18-cGFP) | L. monocytogenes EGD-e harboring pAT18-cGFP | BUG 2546 | Ery5 | This study |
| EGDe-(pAT18-cYFP) | L. monocytogenes EGD-e harboring pAT18-cYFP | BUG 2547 | Ery5 | This study |
| EGDe-(pAT18-cCFP) | L. monocytogenes EGD-e harboring pAT18-cCFP | BUG 2548 | Ery5 | This study |
| L. innocua cGFP | pAD1-cGFP chromosomally integrated in L. innocua | BUG 2646 | Cm7 | This study |
| Listeria strains harboring transcriptional fusions | ||||
| EGD-PinlC-GFP | pAD2-PinlC-GFP chromosomally integrated in EGD | BUG 2545 | Cm7 | This study |
| EGD-PinlC-CFP | pAD2-PinlC-CFP chromosomally integrated in EGD | BUG 2853 | Cm7 | This study |
| EGD-PactA-GFP | pAD3-PactA-GFP chromosomally integrated in EGD | BUG 2797 | Cm7 | This study |
| EGD-PactA-CFP | pAD3-PactA-CFP chromosomally integrated in EGD | BUG 2798 | Cm7 | This study |
Km, kanamycin; Ap, ampicillin; Cm, chloramphenicol; Ery, erythromycin; Cm35, 35 μg/ml chloramphenicol; Cm7, 7 μg/ml chloramphenicol; Ery5, 5 μg/ml erythromycin.
Cloning techniques.
Primers used in this study are listed in Table 2. Standard techniques for DNA manipulation were used (24). PCRs were carried out using the Phusion high-fidelity PCR system (Finnzymes OY) according to the manufacturer's recommendations. A QIAquick PCR purification kit (Qiagen) and a QIAquick gel extraction kit (Qiagen) were used for purification of DNA fragments. Restriction endonucleases and DNA-modifying enzymes were obtained from New England Biolabs and used according to the manufacturer's instructions. Plasmid DNA was prepared using a Qiagen minispin prep kit (Qiagen). Transformation of E. coli XL1-Blue and L. monocytogenes was accomplished by performing electroporation with a 0.1-cm cuvette using a GenePulser apparatus (Bio-Rad) set to 25 μF, 400 Ω, and 1.25 kV.
TABLE 2.
Primers used in this study
| Primer | Sequence | Restriction site(s) |
|---|---|---|
| Generation of fragments containing transcriptional terminators and promoters of interest | ||
| Phyper-Term-Rv-EagI | GAGTCACGGCCGACACACATTATGCCACACCTTGTAGATAAAGTCAACAACTTTTTGCAAAATTAGGCCCTTTCGTCTTCAAGAA | EagI |
| PinlC-Term-Rv-EagI | GAGTCACGGCCGGGATCCTTATATGTTAGCAAAAATAAGAGATGTTTAAATTAACAAGCGTTAATAATCCCGGGCCCTTTCGTCTTCAAGAA | EagI |
| PactA-Term-Rv-EagI | GAGTCACGGCCGGGATCCTTTTAAGAATATCACTTGGAGAATTAATTTTTCTCTAACATTTGTTAATCAGTTAACCCCGGGCCCTTTCGTCTTCAAGAA | EagI |
| Term-Fw-SacI | GAGTCAGAGCTCGAATTCCGATCCCCAATTCCT | SacI |
| Generation of the hly 5′ UTR fragment | ||
| UTRhly-Fw-EagI | GAGTCACGGCCGATAAAGCAAGCATATAATA | EagI |
| UTRhly-Rv | GGGTTTCACTCTCCTTCTACA | |
| UTRhly+gfp-Fw | GGTTAAAAAATGTAGAAGGAGAGTGAAACCCATGCGTAAAGGAGAAGAACTTT | |
| UTRhly+cfp-yfp-Fw | GGTTAAAAAATGTAGAAGGAGAGTGAAACCCATGGCTAGCAAAGGAGAAGAACTTT | |
| UTRhly+mCherry-Fw | GGTTAAAAAATGTAGAAGGAGAGTGAAACCCATGGTGAGCAAGGGCGAGGAGG | |
| Generation of the fluorescent protein-encoding fragment | ||
| gfp-Rv-SalI | GAGTCAGTCGACTTATTTGTATAGTTCATCCATGCC | SalI |
| CFP-int-Rv | GTTGAGAGGTAATGGTTGTCTGGTAA | |
| CFP-int-Fw | CCAGACAACCATTACCTCTCAACACAATCTGCCCTTTCGAAA | |
| Y/CFP-Rv-SalI | GAGTCAGTCGACTTATTTGTAGAGTTCATCCATGCCACGTGTA | SalI |
| mCherry-Rv-SalI | GAGTCAGTCGACTTACTTGTACAGCTCGTCCATG | SalI |
| cfp-yfp-Fw-PstI | GAGTCACTGCAGGCTAGCAAAGGAGAAGAACTTT | PstI |
| mCherry-Fw-NsiI-BamHI | GAGTCAATGCATGGATCCGTGAGCAAGGGCGAGGAGG | NsiI/BamHI |
| Sequencing insert in pAD-based plasmid | ||
| pPL2-Fw | TTCGACCCGGTCGTCGGTTC | |
| pPL2-Rv | CTTAGACGTCATTAACCCTCAC | |
| Sequencing insert in pCR-Blunt | ||
| M13 | TGTAAAACGACGGCCAGT | |
| Rev | CAGGAAACAGCTATGACC | |
| Verification of pAD integration in the Listeria chromosome | ||
| NC16 | GTCAAAACATACGCTCTTATC | |
| PL95 | ACATAATCAGTCCAAAGTAGATGC |
Construction of plasmids encoding fluorescent proteins.
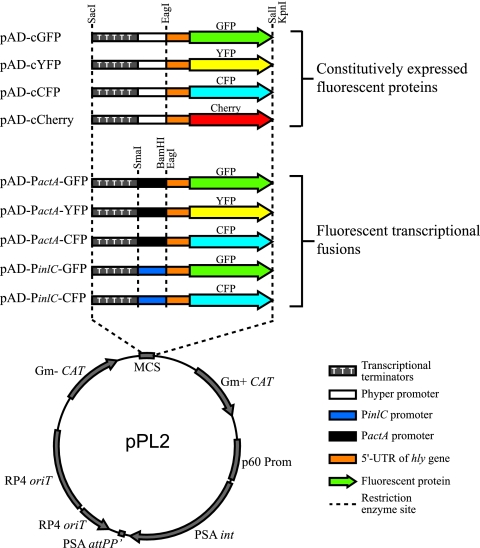
Site-specific pAD integration vectors were constructed using the pPL2 backbone (18). Similar to plasmid pH-hly gfp-PL3 (26), our pAD plasmids, which contain convenient restriction sites (Fig. 1), are composed of tandem copies of the rrnB T1 transcription terminator upstream of the Hyper-SPO1 (Phyper) constitutive promoter (23). Specifically, the rrnB T1 terminator region of the vector pH-hly gfp-PL3 was amplified with primers Term-Fw-SacI and Phyper-Term-Rv-EagI (Table 2). The Phyper promoter sequence was present in the Phyper-Term-Rv-EagI primer rather than amplified from the pH-hly gfp-PL3 plasmid. The resulting PCR product, containing the transcriptional terminators upstream of the Phyper promoter, was cloned into the pCR2.1-TOPO vector (Invitrogen) and was verified by sequencing using primers M13 and REV. Primers UTRhly-Fw-EagI and UTRhly-Rv were used to amplify the hly 5′ untranslated region (UTR) PCR fragment from the pH-hly gfp-PL3 plasmid, and primers UTRhly-gfp-Fw and gfp-Rv-SalI were used to amplify the gfpmut2 gene from genomic DNA of E. coli TG1 λatt cat-gfp (a kind gift from J. M. Ghigo). To fuse the hly 5′ UTR sequence to a fluorescence-encoding gene, the resulting PCR products were gel purified and then used as templates in an splicing-by-overlap-extension (SOE) PCR (35) with the flanking primers UTRhly-Fw-EagI and gfp-Rv-SalI. The fragments composed of the mCherry-, YFP-, or CFP-encoding gene fused to the hly 5′ UTR were generated similarly. The yfp and cherry genes were amplified from the pJEBAM3 (1) and pmCherry (Clontech) plasmids, respectively, using primer pairs UTRhly+cfp-yfp-Fw/Y/CFP-Rv-SalI and UTRhly+mCherry-Fw/mCherry-Rv-SalI. The cfp gene was amplified in two steps in order to suppress the SacI site present in the original plasmid, pJEBAM2 (1). The 5′ and 3′ parts of cfp were amplified from pJEBAM2 using primers UTRhly+cfp-yfp-Fw and CFP-int-Rv and primers CFP-int-Fw and Y/CFP-Rv-SalI, respectively. The two fragments were then coligated by SOE PCR using primers UTRhly+cfp-yfp-Fw and Y/CFP-Rv-SalI. The final SOE PCR products, containing the entire hly 5′ UTR sequence fused to the initiating codon of the fluorescent protein-encoding genes, were then cloned in pCR-Blunt (Invitrogen) and verified by sequencing using primers M13 and REV. Plasmids containing the transcriptional terminators were digested with SacI and EagI, while plasmids harboring the fluorescent protein-encoding genes were digested with EagI and SalI. Restriction fragments were gel purified and then coligated with the pPL2 vector (18) digested with SacI and SalI to generate plasmids pAD-cGFP, pAD-cYFP, and pAD-cCFP.
FIG. 1.
Schematic diagrams of the plasmids constructed in this study. The pAD plasmids are based on integrative plasmid pPL2. They contain four main elements: (i) tandem transcriptional terminators before the promoter to avoid any residual transcription from any upstream promoter, (ii) a constitutive promoter (Phyper) or a promoter of interest (PinlC and PactA), (iii) the hly 5′ UTR sequence, which was described as a stabilizer of the transcript, and (iv) a fluorescence-encoding gene (gfp, yfp, cfp, or cherry).
Plasmids pAD-PactA-XFP and pAD-PinlC-XFP were constructed as follows using the pAD-cXFP plasmid as the template. The inlC and actA promoters were incorporated into primers PinlC-Term-Rv-EagI and PactA-Term-Rv-EagI, respectively. Each of these primers was used with primer Term-Fw-SacI to amplify the transcriptional terminators from pH-hly gfp-PL3 as described above. PCR fragments were cloned in pCR-Blunt and verified by sequencing using primers M13 and REV. The SacI-EagI restriction fragments containing the promoters were gel purified and cloned into the corresponding SacI-EagI-digested pAD-cXFP plasmid. As a result, the Phyper promoter was replaced by the PinlC or PactA promoter. All pAD-based plasmids were verified by sequencing using primers pPL2-Rv and pPL2-Fw and were transformed into L. monocytogenes by electroporation. Integration into the chromosome was verified by PCR amplification using primers NC16 and PL95 (16).
For translational fusion, the GFP-, CFP-, YFP-, and mCherry-encoding sequences were amplified from pAD-cGFP, pAD-cCFP, pAD-cYFP, and pAD-cCherry, respectively, using primers pairs cfp-yfp-Fw-PstI/gfp-Rv-SalI, cfp-yfp-Fw-PstI/Y/CFP-Rv-SalI, cfp-yfp-Fw-PstI/Y/CFP-Rv-SalI, and mCherry-Fw-NsiI-BamHI/mCherry-Rv-SalI. PCR fragments were cloned in pCR-Blunt and could be used to perform translational fusion using the PstI or BamHI sites at the 5′ end of the fluorescence-encoding gene.
To construct the pAT18-based plasmids, the SacI/SalI restriction fragments of the pAD-based plasmids formed by the transcriptional terminators, the promoter, the hly UTR, and the color genes were cloned in SacI/SalI-digested plasmid pAT18.
Invasion assay.
Caco-2 cell suspensions obtained from confluent monolayers were seeded at a concentration of 1.5 × 105 cells per well in six-well tissue culture plates (Nalgene) and grown for 24 h in an antibiotic-free medium. Briefly, the Listeria strains were grown to an optical density at 600 nm (OD600) of 0.8, washed, and diluted in DMEM. Bacterial suspensions were added to the Caco-2 cells at a multiplicity of infection (MOI) of approximately 50 bacteria per cell and incubated for 1 h. The cells were then washed, and noninvasive bacteria were neutralized by adding complete medium containing 25 μg/ml of gentamicin. After incubation for the appropriate time, the cells were detached with trypsin and resuspended in phosphate-buffered saline (PBS). One volume of Cyto-chex (Streck) was added to preserve the cells. Sampling was done in triplicate, and the experiments were performed at least three times. For cytochalasin D treatment, cells were incubated in the presence of 5 mM cytochalasin D for 30 min prior to infection.
Analysis of cell-to-cell spread.
HeLa cells were grown on six-well plates, transfected with Cy3-labeled negative control 1 small interfering RNA (siRNA) (Ambion) using oligofectamine transfectant (Invitrogen) as recommended by the manufacturer, and incubated for 48 h. Mouse J774A.1 macrophages grown in 75-cm2 flasks were infected with L. monocytogenes GFP strains at an MOI of 20 for 45 min. Cells were then washed twice with PBS and were incubated for 3.5 h in complete medium containing 25 μg/ml of gentamicin. Cells were then washed, removed by scraping, and resuspended in 10 ml of complete medium containing gentamicin (25 μg/ml). The number of viable cells was determined using trypan blue. After the incubation period, the L. monocytogenes-infected J774A.1 cells were plated on HeLa cells at a ratio of 1 macrophage per 5 HeLa cells. The mixed cultures were incubated for 18 h to allow spreading of L. monocytogenes from the infected macrophages to the HeLa cells. Cells were detached with trypsin, washed in PBS, and fixed with CytoFix (BD) for 20 min at 4°C. Data were collected with a FACSCalibur (BD) equipped with a 15-mW argon laser (emission wavelength, 488 nm).
Epifluorescence analysis.
Infected Jeg-3 cells were fixed with a paraformaldehyde solution (4% in PBS) for 20 min at room temperature and permeabilized (0.4% Triton X-100 for 5 min in PBS). Cells were then rinsed five times in PBS, incubated with Alexa Fluor 488, 546, or 647 phalloïdin for 1 h at room temperature, rinsed five times in PBS, and mounted on glass coverslips using Fluoromount mounting medium (Electron Microscopy Sciences). Samples were analyzed with a Zeiss Axiovert 135 epifluorescence microscope (Carl Zeiss) connected to a charge-coupled device (CCD) camera. Images were acquired with a ×63 oil immersion objective, and images were processed with MetaMorph software (Universal Imaging).
In vivo and ex vivo infections.
Animal experiments were performed in accordance with the Institut Pasteur guidelines for laboratory animal husbandry. Bacterial virulence in mice was studied by injecting 8-week-old female BALB/c mice (Charles River) intravenously with a sublethal bacterial inoculum (105 CFU per mouse). At 72 h after infection, spleens were aseptically removed, single-cell spleen suspensions were prepared by mechanical homogenization using a 100-μm cell strainer (BD Biosciences) in cold fluorescence-activated cell sorting (FACS) buffer (PBS supplemented with 2.5 mM EDTA and 1% fetal calf serum), and erythrocytes were lysed with BD FACS lysing solution (BD Biosciences). Cells were then resuspended in cold FACS buffer.
Blood was collected from hearts of 8-week-old female BALB/c mice (Charles River) using a 21-gauge needle and a 2-ml syringe. The blood was immediately transferred to tubes containing heparin as an anticoagulant. Infection was performed using 109 bacteria/ml of blood for 1 h. Erythrocytes were lysed with BD FACS lysing solution (BD Biosciences), and white blood cells were resuspended in cold FACS buffer. All samples were fixed with CytoFix (BD Biosciences) for 20 min at 4°C and then analyzed by flow cytometry as described above.
RESULTS
Construction of fluorescent protein-encoding plasmids.
The plasmids that we generated in this study were derived from the pPL2 plasmid, which inserts in the Listeria chromosome at the tRNAArg-attBB site (18), thereby avoiding the requirement for antibiotic pressure to maintain the plasmid and heterogeneity of the fluorescence intensity due to variation in the plasmid copy number. A chloramphenicol antibiotic resistance cassette is present on the pPL2 plasmid, which also is integrated. Therefore, the presence of the construct can be verified by selection on chloramphenicol-containing medium and by PCR amplification (16).
Plasmids used for constitutive fluorescent labeling of L. monocytogenes.
We first generated integrative plasmids that constitutively express fluorescent proteins. Our plasmid constructs were derived from the L. monocytogenes integrative pH-hly gfp-PL3 plasmid (14, 18), which harbors the Hyper-SPO1 constitutive promoter (Phyper) fused to the hly 5′ UTR, as well as the gfp gene (26). In our plasmid constructs, we maintained most of the elements present in the pH-hly gfp-PL3 plasmid, but we engineered new, more convenient enzyme restriction sites that allowed easy excision and insertion of the different elements (Fig. 1). Specifically, we conserved from the pH-hly gfp-PL3 plasmid the tandem rrnB T1 transcription terminators, the Phyper promoter, and the hly 5′ UTR sequence, which was shown to enhance expression of cis-associated genes, possibly through a posttranscriptional mechanism (26). However, we replaced the gfp gene in pH-hly gfp-PL3 (5) with the gene encoding GFPmut2, which has greater fluorescence intensity, or with the genes encoding CFP, YFP, and mCherry. In summary, we constructed four plasmids, pAD1-cGFP, pAD1-cCFP, pAD1-cYFP, and pAD1-cCherry, which constitutively express GFP, CFP, YFP, and mCherry, respectively (Fig. 1 and Table 1), all of which (except the mCherry constructs) were used in assays described below.
In addition, we constructed four multicopy plasmids, pAT18-cGFP, pAT18-cYFP, pAT18-cCFP, and pAT18-cCherry, in which the markers are also under control of the Phyper promoter and downstream of the 5′UTR of hly, which allowed constitutive green, yellow, and blue labeling of Listeria (Table 1). Due to the multicopy nature of these plasmids, they induce stronger fluorescent labeling of bacteria (data not shown). However, they require antibiotic pressure for maintenance and therefore can be used only for short-term experiments.
Generation of transcriptional fusions.
Transcriptional reporter vectors with β-galactosidase or cat (chloramphenicol acetyltransferase) genes are extensively used to assess gene expression. However, these reporters cannot be used to assess regulatory processes at the single-bacterium level because they rely on assays carried out with a bacterial population. Thus, fluorescent reporter systems integrated into the bacterial chromosome are necessary to study gene expression at the single-bacterium level.
We constructed five plasmids harboring transcriptional fusions with the PactA and PinlC promoters. These promoters are regulated by PrfA and are induced at different levels when bacteria enter cells (3, 29). We constructed three transcriptional fusions with the PactA promoter (fused to GFP, CFP, or YFP) and two transcriptional fusions with the PinlC promoter (fused to GFP and YFP) (Fig. 1 and Table 1), which were used in assays described below.
The plasmids that we constructed allow easy insertion of any other promoter of interest and are therefore very versatile. Promoters of interest can be inserted in place of PactA at the EagI or BamHI/SalI sites of plasmids pAD3-PactA-GFP, pAD3-PactA-CFP, and pAD3-PactA-YFP, which allows fusion with each of the markers present in the plasmid (Fig. 1 and Table 1).
Tools for generation of translational fusions.
In order to analyze the expression and the fate of a protein encoded by a gene of interest, we also constructed plasmids to generate translational fusions. We constructed four vectors harboring the fluorescent protein-encoding genes (pTL-GFP, pTL-CFP, pTL-YFP, and pTL-Cherry) without the hly UTR and their start codon (Table 1). Each of these plasmids can be used to construct translational fusions and cloned in the pAD plasmids described above.
Briefly, the fluorescence genes are first liberated from the pTL cloning vector using PstI and SalI. The gene of interest is amplified with 5′ and 3′ primers bearing an XmaI site and a PstI site, respectively, at the 5′ end. Three-fragment ligation with the XmaI/PstI-digested PCR product, the PstI/SalI fluorescence-encoding gene, and the XmaI/SalI-digested pAD-based plasmid (pAD3-PactA-GFP, for instance) is then performed to generate the translational fusion. The expression and localization of the encoded protein in live or fixed L. monocytogenes can then be determined.
Detection of constitutive and inducible GFP-, YFP-, and CFP-tagged L. monocytogenes.
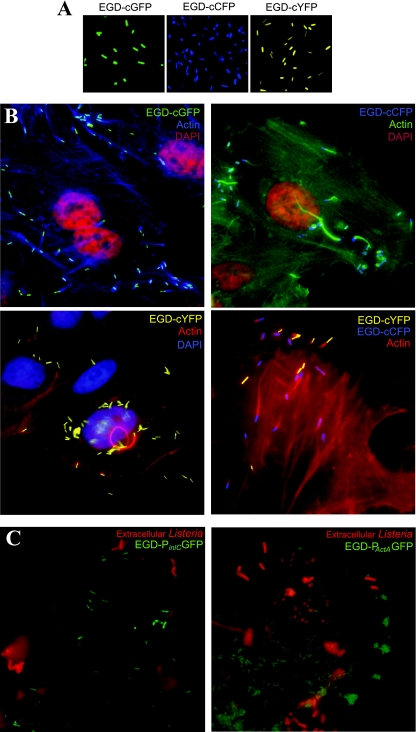
To determine whether a single copy of the fluorescent marker gene expressed under control of the Phyper promoter was sufficient for detection by epifluorescence microscopy, we transformed L. monocytogenes (strain EGD-e or EGD) with four integrative plasmids (pAD1-cGFP, pAD1-cCFP, pAD1-cYFP, and pAD1-cCherry), each expressing a different fluorescence gene under control of a constitutive promoter. Figure 2 shows that L. monocytogenes strains EGD-cGFP, EGD-cCFP, and EGD-cYFP were fluorescent and were detected by immunofluorescence microscopy. Furthermore, the colors are spectrally different; therefore, the bacteria in a mixture containing different types of bacteria, each expressing a different color, can be distinguished from one another and used in combinations (Fig. 2). As described below, the L. monocytogenes EGD-cGFP strain was also used to perform FACS analyses.
FIG. 2.
Fluorescence microscopy of L. monocytogenes. (A) Exponentially growing L. monocytogenes EGD-cGFP, EGD-cCFP, and EGD-cYFP were harvested at an OD600 of 1 and observed by using fluorescence microscopy. (B) Jeg-3 cells were infected with EGD-cGFP, EGD-cCFP, or EGD-cYFP or a mixture of EGD-cYFP and EGD-cCFP for 3 h. Cells were fixed, permeabilized, and marked with 4′,6′-diamidino-2-phenylindole (DAPI) and phalloidin (Alexa 647, 546, or 488). (C) Jeg-3 cells were infected with EGD-PinlC or EGD-PactA for 3 h. Extracellular bacteria were distinguished from intracellular bacteria by staining with an anti-Listeria antibody (red) prior to permeabilization.
We also infected Jeg-3 cells with L. monocytogenes strains EGD-cGFP, EGD-cCFP, and EGD-cYFP. Figure 2 shows intracellular bacteria as they polymerize actin tails. These intracellular bacteria could also be detected by their different colors, and therefore they expressed sufficient fluorescence that they could be detected when they were located intracellularly. Additionally, we could coinfect cells with L. monocytogenes EGD-cCFP and EGD-cYFP and detect the two distinct populations in the cytoplasm. This is a powerful tool for comparing the invasiveness and infection potentials of different strains.
We also compared the expression of two genes, actA and inlC, both of which are, as indicated above, regulated by the major virulence transcription factor PrfA (3, 29). To do this, we transformed the pAD2-PinlC-GFP and pAD3-PactA-GFP plasmids into wild-type L. monocytogenes strain EGD and infected HeLa cells. We examined the fluorescence of the strains during infection in order to determine the location of gene expression. Figure 2 shows that the strain expressing PactA-GFP was fluorescent both before and after invasion, which is consistent with results obtained previously (21). In contrast, the strain expressing PinlC-GFP was fluorescent only after the bacteria entered the host cell, in agreement with the results of previous transcriptional studies (9). Thus, although the two genes are regulated by the same transcription factor, inlC expression is restricted to the cytosol of the host cell, at least in the EGD strain used in this study. In addition, we showed that the inlC promoter fused to the CFP marker is also readily detectable inside cells. Therefore, a single copy of a fluorescent reporter gene fused to either PactA or PinlC is easily detected during infection and can be used to detect the precise cellular compartment where gene expression occurs.
Measurement of GFP-tagged L. monocytogenes internalization using flow cytometry.
We took advantage of our chromosomally tagged L. monocytogenes strain constitutively expressing GFP to develop a rapid method to determine the proportion of infected host cells. Indeed the time-consuming method of counting the number of L. monocytogenes CFU in the gentamicin survival assay determines only the percentage of bacteria that have invaded the host cell and gives no indication of the proportion of infected cells. To measure the number of cells harboring intracellular bacteria, we infected Caco-2 cells with L. monocytogenes EGD-cGFP, trypsinized and fixed them at various time points after infection, and used FACS analysis to detect fluorescent infected cells.
In a representative experiment in which Caco-2 cells were infected using a multiplicity of infection of 50 bacteria per cell, 21% of the cells were fluorescent as early as 1.5 h after infection. Thus, L. monocytogenes invaded 21% of the Caco-2 cells. To demonstrate that the fluorescence detected at early times during infection was really due to invasion, we treated cells with cytochalasin D, a drug that inhibits actin polymerization and prevents entry of bacteria. Cytochalasin D pretreatment decreased the number of infected cells to the background level, showing that the fluorescence detected in this assay was due to intracellular bacteria. Therefore, at early time points after infection, this method allows measurement of the proportion of infected cells and the efficiency of bacterial invasion.
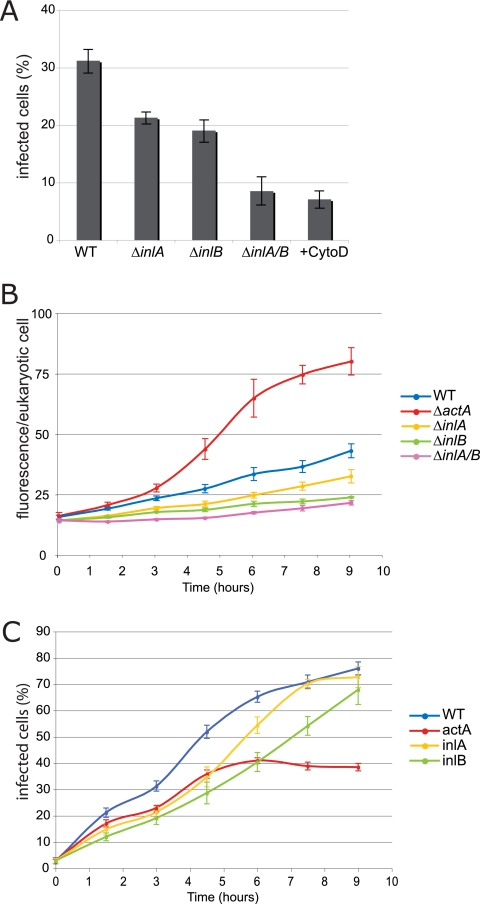
To determine the sensitivity of this method, we tested three mutants known to have an invasion defect. InlA and InlB are two proteins that are important for invasion and that bind E-cadherin and Met receptors, respectively (2, 15, 34). We tested inlA or inlB single mutants and a inlAB double mutant to determine their capacities to invade Caco-2 cells. Figure 3A shows that as early as 1.5 h postinfection we detected a partial entry defect for the inlA and inlB mutants, which infected 15% and 11% of the cells, respectively (compared to 21% for a wild-type strain). A greater entry defect was detected for the inlAB double mutant, as only 7% of the cells were infected. Therefore, infection with GFP-expressing L. monocytogenes can be quantified by flow cytometry, a powerful tool for measuring internalization defects.
FIG. 3.
Flow cytometry analysis of infection. (A) Bacterial cell invasion at 3 h postinfection. Semiconfluent Caco-2 cells were infected with chromosomally GFP-tagged L. monocytogenes wild-type strain EGD and ΔinlA, ΔinlB, and ΔinlAB mutants (MOI, 50). Caco-2 cells treated with cytochalasin D (CytoD) were infected with strain EGD. At 3 h postinfection, Caco-2 cells were treated with trypsin and resuspended in PBS. Caco-2 cells that were infected with GFP-tagged strains were analyzed by FACS. The percentages of infected cells in three independent experiments are shown. The error bars indicate standard errors of the means. (B) Kinetics of intracellular replication of L. monocytogenes wild-type strain EGD and ΔactA, ΔinlA, ΔinlB, and ΔinlAB mutants during infection. The y axis indicates the geometric means of GFP-positive cells as determined by FlowJo software analysis. Each symbol indicates the average fluorescence of cells collected in at least three independent experiments. The error bars indicate standard errors of the means. (C) Kinetics of cell invasion by L. monocytogenes wild-type strain EGD and ΔinlA, ΔinlB, and ΔinlAB mutants during infection. The y axis indicates the percentages of infected cells as determined by FlowJo software analysis. Each symbol indicates the average for 10,000 cells collected in at least three independent experiments. The error bars indicate standard errors of the means. WT, wild type.
Analysis of intracellular replication and cell-to-cell spread by flow cytometry.
The same protocol of infection of Caco-2 cells with L. monocytogenes was used to study other phenotypes, such as intracellular replication and cell-to-cell spread. In our assay, to measure intracellular replication, gentamicin was added to the culture at 1 h postinfection to stop internalization. Therefore, any increase in the number of intracellular bacteria was due solely to replication and spreading to neighboring cells. We performed a FACS analysis like that described above and evaluated intracellular replication by quantifying the intensity of fluorescence in infected cells (geometric mean of the fluorescence values for GFP-positive cells). The results of a representative experiment are shown in Fig. 3B, which shows the average fluorescence intensity for infected cells over the course of infection. Cells infected with wild-type L. monocytogenes and with inlA, inlB, and inlAB mutants exhibited similar fluorescence intensities, which did not increase over the course of infection. In contrast, cells infected with an actA mutant, which could not spread from cell to cell, exhibited a >2-fold increase in fluorescence, reflecting the well-described actA bacterial microcolonies that accumulate in infected cells.
Cell-to-cell spreading can also be monitored using this method, although a more thorough method is described below. Indeed, in the FACS assay described above, the number of infected cells increased steadily over time. Since extracellular bacteria were cleared by the gentamicin added 1 h after the start of infection, the increase in the percentage of infected cells was due mostly to cell-to-cell spreading. Figure 3C shows that over time there was an increase in the number of cells that were infected with wild-type L. monocytogenes, and up to 75% of the cells were infected by 9 h after infection. Similar results were obtained for the inlA and inlB mutants, which did not have a cell-to-cell spread defect, and by 9 h after infection these strains had infected 70% of the cells. In contrast, an actA mutant, which was defective in actin polymerization, infected a significantly lower number of cells than the wild-type strain. Therefore, FACS analysis of cells infected with L. monocytogenes expressing GFP can also be used to monitor cell-to-cell spread.
Measurement of cell-to-cell spread using L. monocytogenes EGD-cGFP in a “two-cell” infection assay.
Although cell-to-cell spread can be evaluated as described above (Fig. 3), we developed a more thorough assay which allowed us to circumvent potential initial entry defects. This assay is based on a previously described protocol (8), in which we used our fluorescent L. monocytogenes EGD-cGFP strain and flow cytometry. In this assay, in order to bypass entry defects, J774A.1 macrophages were infected with fluorescent bacteria (Fig. 4A). The infected macrophages were then placed on a monolayer of fluorescently labeled epithelial cells (Cy3) that had been transfected with a Cy3-labeled control siRNA. Cell-to-cell spread from the macrophages to the epithelial cells could then occur. At the end of the assay, cells were collected and analyzed by FACS to determine the number of infected epithelial cells, which were the cells labeled with both GFP and Cy3. The results obtained in a representative experiment using this technique are shown in Fig. 4B and C. Macrophages were infected with the wild-type strain or the ΔactA or ΔinlAB strain, all constitutively expressing GFP. As shown in Fig. 4B, at 3 h postinfection, all macrophages exhibited the same fluorescence, which shows that all of the strains infected the macrophages similarly. Subsequently, the infected macrophages were added to confluent HeLa cells that were previously transfected with the Cy3-labeled “control” siRNA. As shown in Fig. 4C, 97% of the HeLa cells were Cy3 positive, demonstrating that the transfection efficiency was high. After 18 h of contact between wild-type EGD-infected macrophages and Cy3-labeled HeLa cells, FACS analysis was performed; 40% of the HeLa cells were fluorescent (green), indicating that 40% of the HeLa cells were infected through cell-to-cell spreading. Similar results were obtained with a mutant defective for both inlA and inlB, which had an entry defect but no cell-to-cell spread defect. In contrast, when the actA mutant, which was defective in cell-to-cell spread, was used, only a small percentage of HeLa cells were infected (5%, compared with 40% for the wild-type strain).
FIG. 4.
Macrophage overlay method and measurement of L. monocytogenes cell-to-cell spread by flow cytometry. (A) Schematic diagram of the experimental procedure. (B) Flow cytometry analysis of macrophages infected with chromosomally GFP-tagged L. monocytogenes wild-type strain EGD and ΔactA and ΔinlAB mutants. Three hours after infection, macrophage infection was analyzed by FACS; 10,000 cells were collected for each sample. (C) Flow cytometry analysis of HeLa cells infected by bacterial spreading from macrophages. Dot plots representing fluorescence from 10,000 cells were generated using FlowJo software. The data are representative of the results of at least three independent experiments.
Together, these results demonstrate that this technique is a sensitive method for analyzing the abilities of different L. monocytogenes strains to spread, independently of their entry phenotype. It could also be useful for high-throughput screening after mutagenesis or RNA interference in order to identify bacterial and host factors required for cell-to-cell spreading (which is currently being studied).
In vivo detection of L. monocytogenes in organs.
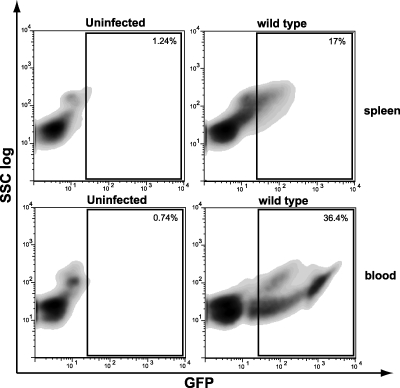
The considerable advantage of the integrative and constitutively fluorescent L. monocytogenes strains and plasmids constructed in this study is that antibiotic pressure is not required, which allows long-term in vivo experiments to be performed. To determine whether L. monocytogenes with GFP could be detected in the organs of infected animals, we infected BALB/c mice for 3 days. We injected a sublethal dose (105 bacteria) intravenously, and 72 h later spleens were collected, homogenized, and analyzed by flow cytometry. Figure 5 shows that a significant proportion of the cells (17%) in infected mice displayed green fluorescence, which was indicative of infection. Therefore, 72 h after inoculation, cells infected by fluorescent L. monocytogenes in vivo can be unambiguously detected.
FIG. 5.
In vivo and ex vivo infection by L. monocytogenes EGD-cGFP: flow cytometry data for uninfected and infected spleens (in vivo) and blood (ex vivo). BALB/c mice were infected with 105 CFU of L. monocytogenes wild-type strain EGD intravenously for 3 days. Blood was collected from BALB/c mice and infected with 109 wild-type L. monocytogenes CFU/ml of blood. Spleen and blood cells were collected and analyzed by FACS. At least 20,000 cells were analyzed for each sample, and the data are representative of the results of three experiments. Dot plots and gates were obtained using FlowJo software.
Further experiments were done to determine whether after incubation of bacteria ex vivo in blood, blood cells infected with L. monocytogenes containing GFP could be detected. Figure 5 shows that 34% of the cells were fluorescent after infection with L. monocytogenes, demonstrating that blood cells were heavily infected.
DISCUSSION
In this study we developed novel techniques based on a combination of FACS analysis and chromosomally tagged fluorescent bacteria to study the different steps of the Listeria infectious process. We constructed 13 fluorescence plasmids, which were subsequently transformed into L. monocytogenes and used for detection during infection. Specifically, we constructed nine integrative plasmids in which a fluorescence-encoding gene was placed under control of a constitutive promoter (Phyper) (23) or under control of a specific listerial promoter. The resulting strains were examined either by using fluorescence microscopy techniques or by performing flow cytometry analyses. The results obtained demonstrated the utility, versatility, and efficacy of the constructed plasmids.
Transcriptional fusions.
In this study we constructed plasmids for transcriptional fusion in order to study gene expression at the single-cell level. We used the PactA and PinlC promoters as examples and showed by using immunofluorescence that the inlC gene is tightly regulated and expressed only after invasion of host cells, whereas the actA gene, although also induced inside cells, is expressed more ubiquitously. Therefore, the PinlC promoter can be used as an intracellular reporter, marking intracytoplasmic bacteria. The ability to distinguish intracellular and extracellular bacteria based only on expression of a fluorescence marker is very useful and can be used to screen bacterial strains defective for invasion or host cells defective for components important for bacterial invasion.
It is important to note that the two transcriptional fusion plasmids were designed for versatile use with any other promoter. Indeed, release of PactA or PinlC and insertion of another promoter can be accomplished by simple restriction digestion with SmaI and either BamHI or EagI. Furthermore, although we studied expression of the actA and inlC genes only in vitro, these constructs should also be functional in vivo for studying expression in infected organs. In a similar manner, translational fusions can be constructed using the tools described here to determine the expression or fate of a specific protein in vitro or in vivo.
Flow cytometry methods.
Here we describe methods to analyze L. monocytogenes internalization and further steps in the infectious process by flow cytometry and show that this technique is sensitive and can detect different degrees of internalization defects. Indeed, an inlA or inlB mutant has a small entry defect, whereas the inlAB double mutant has a much larger defect, as previously described (7, 11). We also analyzed cell-to-cell spreading and intracellular replication. We used this new technique to confirm that inlA and inlB mutants did not have cell-to-cell spreading defects and ultimately infected the same number of cells as the wild-type strain, whereas much higher numbers of cells of an actA mutant, which cannot spread, accumulated inside host cells. A closer analysis of this technique suggested that it is not as effective for detecting a cell-to-cell spreading defect as the “two-cell infection assay” also described here. Indeed, Fig. 3C shows that the number of cells infected with an actA mutant increased until 6 h after the start of infection, even though gentamicin was added after 1 h of infection. There are two possible explanations for this observation: (i) the gentamicin concentration used in our study was not high enough to completely kill extracellular L. monocytogenes, and reinfection occurred over time; or (ii) at 1.5 h after infection we detected only a fraction of infected cells, the cells containing a significant number of bacteria per cell, and cells infected with only a few bacteria were detectable only later in infection, when the number of bacteria per cell reached the detection threshold. However, despite these limitations this assay still allowed detection of a cell-to-cell spreading defect, which could be more precisely analyzed using the “two-cell infection assay” described here.
The flow cytometry methods that we describe in this paper, in combination with the new plasmids constructed, are powerful tools for single-cell high-throughput studies. To analyze infected cells by flow cytometry, the cells must be fluorescently marked so that they can be distinguished from noninfected cells. The integrative plasmids described here avoid the lengthy immunofluorescence staining techniques used for these types of analyses. Furthermore, only one copy of each of the plasmids is present on the chromosome; therefore, the fluorescence measured is a direct measure of the number of bacteria. The main advantage of using flow cytometry is that a high number of cells can be analyzed in a short time. Indeed, a standard FACS analysis is able to analyze approximately 1,000 cells per second, which allows rapid acquisition of data for 10,000 or 20,000 cells. With such a large sample size, statistical analysis is very powerful. Furthermore, as demonstrated here, in a single experiment carried out over a time course of infection, we can study all of the cell cycle-associated phenotypes (i.e., entry, intracellular replication, and cell-to-cell spreading).
Finally, by using a cell sorter, it is possible to separate the labeled infected cells from the noninfected cells. Thus, with this method one can generate a pure culture of infected cells at any time point during infection (data not shown) in order to perform subsequent experiments, such as biochemical analyses. These fast and sensitive techniques are a considerable improvement compared with the tools currently at our disposal. We believe that these techniques could be applied to other intracellular pathogens, like Shigella and Salmonella, and should be important for high-throughput screening.
Two-cell infection assay.
The plaque assay is typically used to evaluate L. monocytogenes cell-to-cell spreading efficacy in vitro. The size and appearance of the plaques compared to those formed by the wild-type strain indicate a defect in cell-to-cell spreading. This technique, although reliable, has been used mainly with fibroblast cell lines, and weak spreading defects are difficult to detect. Taking advantage of our fluorescence-tagged bacteria, we adapted a previously described technique (8) to measure cell-to-cell spreading. This assay can be performed with any cell type and bypasses potential bacterial entry defects. The first step in this assay is to infect macrophages with L. monocytogenes. In this cell type, L. monocytogenes is phagocytosed and it does not use active entry mechanisms. This initial step, therefore, bypasses active invasion of cells and allows exclusive study of cell-to-cell spreading. This assay could, for example, be important for measuring the role in cell-to-cell spreading of proteins that otherwise are important for entry of L. monocytogenes. The second step in our assay is to overlay the infected macrophages on a layer of epithelial cells, such as HeLa cells. To be able to differentiate the infected HeLa cells from the macrophages, HeLa cells are labeled by transfection with a Cy3-labeled siRNA, and cell-to-cell spreading is measured by enumerating the number of cells labeled with both Cy3 and GFP. Our results show that with this novel method, cell-to-cell spreading can be precisely quantified and bacterial or host factors required for this process can be determined. Indeed, because a large number of cells can be observed in a short time, this method can reveal subtle phenotypes that are difficult to detect using plaque assays. This method should also be convenient for RNA interference-based large-scale screens for host cell factors involved in cell-to-cell spreading.
Red fluorescent L. monocytogenes.
We constructed L. monocytogenes strains expressing GFP, CFP, and YFP. However, in contrast to the results of Andersen et al. (1), we were not able to generate an L. monocytogenes strain expressing red fluorescence. Indeed, plasmids harboring the tetrameric DsRed-, dimeric HcRed-, monomeric mRFP1-, or mCherry-encoding genes were constructed and tested in L. monocytogenes (both the EGD and EGD-e strains), but no fluorescence was detected, whereas fluorescence was easily detected in E. coli transformed with the same plasmids. We hypothesized that the amount of red protein produced from a single chromosomal copy was not sufficient to label Listeria. To test this possibility, we cloned the same constructs into the multicopy plasmid pAT18 (30). Similarly, although the E. coli strain harboring pAT18-Cherry was fluorescent, the corresponding L. monocytogenes strain was not fluorescent (data not shown). The Phyper promoter, in contrast to the dlt promoter used by Andersen et al. (1), is probably not strong enough to label Listeria red even when a multicopy plasmid is used.
In vivo studies.
Fluorescent Listeria strains have been used previously to study specific infected host cell populations in the central nervous system of infected mice (17). However, this study was limited to early time points due to the use of multicopy plasmids. Our results show that we could detect fluorescent L. monocytogenes in spleens after 72 h of infection. We are currently investigating the specific infected spleen cell populations by labeling cells with cell-specific markers.
In summary, our integrative plasmids encoding fluorescent reporters are new, convenient, and powerful tools for analyzing the different steps of the L. monocytogenes infectious process by fluorescence microscopy or flow cytometry and also gene expression or protein localization at the single-bacterial-cell level.
Acknowledgments
We are grateful to Darren Higgins for providing pH-hly gfp-PL3, to Jens B. Andersen and Tine R. Licht for providing pJEBAM2 and -3, and to Jean-Marc Ghigo for providing E. coli TG1 λ-att cat-gfp.
This work was supported by Institut Pasteur, INSERM, INRA, and ERC (advanced grant 233348). A.T.-A. was an EMBO long-term fellow. D.B. was supported by INRA. L.D. was supported by Fondation pour la Recherche Médicale (FRM). P.C. is an international research scholar of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Andersen, J. B., B. B. Roldgaard, A. B. Lindner, B. B. Christensen, and T. R. Licht. 2006. Construction of a multiple fluorescence labelling system for use in co-invasion studies of Listeria monocytogenes. BMC Microbiol. 6:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonazzi, M., M. Lecuit, and P. Cossart. 2010. Listeria monocytogenes internalin and E-cadherin: from structure to pathogenesis. Cell. Microbiol. 11:693-702. [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chudakov, D. M., S. Lukyanov, and K. A. Lukyanov. 2005. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 23:605-613. [DOI] [PubMed] [Google Scholar]
- 5.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 6.Cossart, P., and A. Toledo-Arana. 2008. Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect. 10:1041-1050. [DOI] [PubMed] [Google Scholar]
- 7.Dramsi, S., I. Biswas, E. Maguin, L. Braun, P. Mastroeni, and P. Cossart. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Mol. Microbiol. 16:251-261. [DOI] [PubMed] [Google Scholar]
- 8.Dramsi, S., S. Levi, A. Triller, and P. Cossart. 1998. Entry of Listeria monocytogenes into neurons occurs by cell-to-cell spread: an in vitro study. Infect. Immun. 66:4461-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelbrecht, F., S. K. Chun, C. Ochs, J. Hess, F. Lottspeich, W. Goebel, and Z. Sokolovic. 1996. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 21:823-837. [DOI] [PubMed] [Google Scholar]
- 10.Fortineau, N., P. Trieu-Cuot, O. Gaillot, E. Pellegrini, P. Berche, and J. L. Gaillard. 2000. Optimization of green fluorescent protein expression vectors for in vitro and in vivo detection of Listeria monocytogenes. Res. Microbiol. 151:353-360. [DOI] [PubMed] [Google Scholar]
- 11.Gaillard, J. L., and B. B. Finlay. 1996. Effect of cell polarization and differentiation on entry of Listeria monocytogenes into the enterocyte-like Caco-2 cell line. Infect. Immun. 64:1299-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 13.Gouin, E., J. Mengaud, and P. Cossart. 1994. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect. Immun. 62:3550-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundling, A., L. S. Burrack, H. G. Bouwer, and D. E. Higgins. 2004. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc. Natl. Acad. Sci. U. S. A. 101:12318-12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamon, M., H. Bierne, and P. Cossart. 2006. Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 4:423-434. [DOI] [PubMed] [Google Scholar]
- 16.Hastings, J. W., and J. G. Morin. 1969. Calcium-triggered light emission in Renilla. A unitary biochemical scheme for coelenterate bioluminescence. Biochem. Biophys. Res. Commun. 37:493-498. [DOI] [PubMed] [Google Scholar]
- 17.Join-Lambert, O. F., S. Ezine, A. Le Monnier, F. Jaubert, M. Okabe, P. Berche, and S. Kayal. 2005. Listeria monocytogenes-infected bone marrow myeloid cells promote bacterial invasion of the central nervous system. Cell. Microbiol. 7:167-180. [DOI] [PubMed] [Google Scholar]
- 18.Lauer, P., M. Y. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levraud, J. P., O. Disson, K. Kissa, I. Bonne, P. Cossart, P. Herbomel, and M. Lecuit. 2009. Real-time observation of Listeria monocytogenes-phagocyte interactions in living zebrafish larvae. Infect. Immun. 77:3651-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lun, S., and P. J. Willson. 2004. Expression of green fluorescent protein and its application in pathogenesis studies of serotype 2 Streptococcus suis. J. Microbiol. Methods 56:401-412. [DOI] [PubMed] [Google Scholar]
- 21.Moors, M. A., B. Levitt, P. Youngman, and D. A. Portnoy. 1999. Expression of listeriolysin O and ActA by intracellular and extracellular Listeria monocytogenes. Infect. Immun. 67:131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pentecost, M., G. Otto, J. A. Theriot, and M. R. Amieva. 2006. Listeria monocytogenes invades the epithelial junctions at sites of cell extrusion. PLoS Pathog. 2:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quisel, J. D., W. F. Burkholder, and A. D. Grossman. 2001. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J. Bacteriol. 183:6573-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Shaner, N. C., M. Z. Lin, M. R. McKeown, P. A. Steinbach, K. L. Hazelwood, M. W. Davidson, and R. Y. Tsien. 2008. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen, A., and D. E. Higgins. 2005. The 5′ untranslated region-mediated enhancement of intracellular listeriolysin O production is required for Listeria monocytogenes pathogenicity. Mol. Microbiol. 57:1460-1473. [DOI] [PubMed] [Google Scholar]
- 27.Shimomura, O., F. H. Johnson, and Y. Saiga. 1962. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 59:223-239. [DOI] [PubMed] [Google Scholar]
- 28.Thone, F., B. Schwanhausser, D. Becker, M. Ballmaier, and D. Bumann. 2007. FACS-isolation of Salmonella-infected cells with defined bacterial load from mouse spleen. J. Microbiol. Methods 71:220-224. [DOI] [PubMed] [Google Scholar]
- 29.Toledo-Arana, A., O. Dussurget, G. Nikitas, N. Sesto, H. Guet-Revillet, D. Balestrino, E. Loh, J. Gripenland, T. Tiensuu, K. Vaitkevicius, M. Barthelemy, M. Vergassola, M. A. Nahori, G. Soubigou, B. Regnault, J. Y. Coppee, M. Lecuit, J. Johansson, and P. Cossart. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950-956. [DOI] [PubMed] [Google Scholar]
- 30.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. An integrative vector exploiting the transposition properties of Tn1545 for insertional mutagenesis and cloning of genes from gram-positive bacteria. Gene 106:21-27. [DOI] [PubMed] [Google Scholar]
- 31.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 102:99-104. [DOI] [PubMed] [Google Scholar]
- 32.Valdivia, R. H., A. E. Hromockyj, D. Monack, L. Ramakrishnan, and S. Falkow. 1996. Applications for green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene 173:47-52. [DOI] [PubMed] [Google Scholar]
- 33.Vazquez-Torres, A., J. Jones-Carson, A. J. Baumler, S. Falkow, R. Valdivia, W. Brown, M. Le, R. Berggren, W. T. Parks, and F. C. Fang. 1999. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401:804-808. [DOI] [PubMed] [Google Scholar]
- 34.Veiga, E., J. A. Guttman, M. Bonazzi, E. Boucrot, A. Toledo-Arana, A. E. Lin, J. Enninga, J. Pizarro-Cerda, B. B. Finlay, T. Kirchhausen, and P. Cossart. 2007. Invasive and adherent bacterial pathogens co-opt host clathrin for infection. Cell Host Microbe 2:340-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warrens, A. N., M. D. Jones, and R. I. Lechler. 1997. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186:29-35. [DOI] [PubMed] [Google Scholar]
- 36.Wilson, R. L., A. R. Tvinnereim, B. D. Jones, and J. T. Harty. 2001. Identification of Listeria monocytogenes in vivo-induced genes by fluorescence-activated cell sorting. Infect. Immun. 69:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]