Abstract
The assembly of 20,000 sequencing reads obtained from shotgun and chromosome-specific libraries of the Spiroplasma citri genome yielded 77 chromosomal contigs totaling 1,674 kbp (92%) of the 1,820-kbp chromosome. The largest chromosomal contigs were positioned on the physical and genetic maps constructed from pulsed-field gel electrophoresis and Southern blot hybridizations. Thirty-eight contigs were annotated, resulting in 1,908 predicted coding sequences (CDS) representing an overall coding density of only 74%. Cellular processes, cell metabolism, and structural-element CDS account for 29% of the coding capacity, CDS of external origin such as viruses and mobile elements account for 24% of the coding capacity, and CDS of unknown function account for 47% of the coding capacity. Among these, 21% of the CDS group into 63 paralog families. The organization of these paralogs into conserved blocks suggests that they represent potential mobile units. Phage-related sequences were particularly abundant and include plectrovirus SpV1 and SVGII3 and lambda-like SpV2 sequences. Sixty-nine copies of transposases belonging to four insertion sequence (IS) families (IS30, IS481, IS3, and ISNCY) were detected. Similarity analyses showed that 21% of chromosomal CDS were truncated compared to their bacterial orthologs. Transmembrane domains, including signal peptides, were predicted for 599 CDS, of which 58 were putative lipoproteins. S. citri has a Sec-dependent protein export pathway. Eighty-four CDS were assigned to transport, such as phosphoenolpyruvate phosphotransferase systems (PTS), the ATP binding cassette (ABC), and other transporters. Besides glycolytic and ATP synthesis pathways, it is noteworthy that S. citri possesses a nearly complete pathway for the biosynthesis of a terpenoid.
Spiroplasmas are arthropod-associated bacteria belonging to the class Mollicutes, a group of wall-less microorganisms phylogenetically related to low-G+C-content, Gram-positive bacteria (51). Spiroplasma citri is a helical plant-pathogenic mollicute responsible for the “stubborn” disease of citrus (39). It inhabits the phloem sap of infected plants to which it is transmitted by sap-sucking hemipteran insect in a circulative and propagative manner (31, 32). S. citri can infect a wide range of plant species, including crop and wild plants, as it is transmitted by polyphagous leafhoppers (13). Spiroplasmas are available in pure culture, and their study has therefore benefited from the use of molecular genetics. In particular, the relationships of spiroplasmas with their two hosts, the plant and the leafhopper vector, have been extensively studied (11, 22). In S. citri, the inactivation of genes and functional complementation of mutants have shown that (i) fructose consumption by the spiroplasma is a major cause for symptom production in plants, (ii) the solute binding protein of a putative ABC-type transporter is involved in the insect transmission process, and (iii) spiralin, the major membrane protein, is not essential for helicity, motility, and pathogenicity but is required for efficient transmission by the leafhopper vector (10, 19, 23, 24, 28). To characterize other spiroplasma genes potentially involved in insect transmission and pathogenicity, the genome of S. citri strain GII3-3X is currently being deciphered.
The S. citri genome is characterized by an abundance of extrachromosomal elements, including seven plasmids, pSciA and pSci1 to pSci6, present as 10 to 14 copies per cell. These plasmids are vertically inherited, but some of them could also be horizontally transferred, as they encode proteins involved in partitioning and the cell-to-cell transfer of DNA molecules (12, 40). Plasmids pSci1 to pSci5 encode surface proteins of the S. citri adhesion-related protein (ScARP) family, and pSci6 was previously shown to confer insect transmissibility (9). Therefore, it is likely that the abundance and diversity of plasmids could provide S. citri strain GII3-3X with the ability to quickly adapt to various vector insects and, hence, to be transmitted to diverse host plants. However, chromosome-encoded determinants are also expected to play a role in spiroplasma biology. In S. citri, the chromosome sizes vary from 1.6 to 1.9 Mbp among strains (53, 54), and part of the size variation is thought to result from different amounts of prophage sequences (35). Many S. citri strains are infected by single-stranded DNA-containing filamentous phages (Plectrovirus), whose sequences also occur as partial or full-length prophages integrated into the spiroplasma chromosome (7, 35, 38). Here we report the partial chromosome sequence of S. citri strain GII3-3X and the functional assignment of the predicted coding sequences.
MATERIALS AND METHODS
Spiroplasma strain and cultivation.
S. citri strain GII3 was originally isolated from the leafhopper Circulifer haematoceps, collected in Morocco in 1980 (49). A triply cloned strain was further produced by plating onto SP4 medium, and one of the clones was further propagated as GII3-3X. Spiroplasmas were grown at 32°C in SP4 medium (48).
Chromosome mapping.
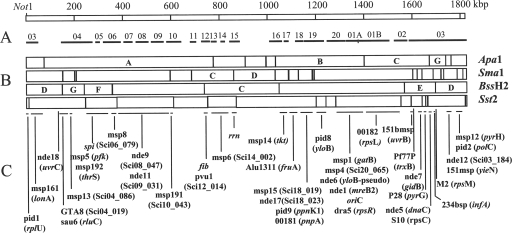
The S. citri chromosome was digested with NotI, ApaI, SmaI, BssHII, and SstII, and the restriction fragments were separated by pulsed-field gel electrophoresis (PFGE) with a Bio-Rad (Marnes-la-Coquette, France) CHEF DRIII apparatus. The results indicated that the chromosome contained one single site for the NotI endonuclease and 9, 19, 7, and 15 sites for ApaI, SmaI, BssHII, and SstII, respectively. The full-size circular chromosome was estimated to be 1,820 kbp. Southern blot hybridizations of the DNA fragments were performed with 43 different probes (see Fig. S1 in the supplemental material). From these data, a physical and genetic map comprising 51 restriction sites and 43 genetic markers was constructed (Fig. 1B and C). The previously reported replication origin oriC (55) was located at 1,380 kbp on the ApaB fragment.
FIG. 1.
Linear representation of the restriction and genetic map of the S. citri circular chromosome. The origin of the map has been arbitrarily located at the NotI site. (A) Distribution of contigs along the chromosome. (B) Restriction map. Fragments with letters have been used for map-based plasmid libraries. (C) Positions of genetic markers.
Sequence assembly.
Sequencing data were produced following a chromosome map-based approach and a classical shotgun strategy completed by the end sequencing of inserts from a mini bacterial artificial chromosome (miniBAC) library. A first pSMART library of 4,000 clones with 3- to 4-kbp inserts (prepared by Amplicon Express, Pullman, WA) and a second pBluescript library of 2,400 clones with 1- to 3-kbp inserts were produced and end sequenced. Additional data were produced by the end sequencing of a miniBAC library of 900 clones in pECBAC1 with inserts of 15 to 25 kbp obtained by limited Sau3A digestion. In addition, 12 Sau3A plasmid libraries were constructed from restriction fragments (indicated by letters in Fig. 1B) eluted from PFGE gels. After the primary assembly of the data, gap filling was performed by primer walking, PCR, and genome walking. The assembly and editing of 20,000 sequencing reads were performed with the phred-phrap-consed package (20, 21, 26). Incorrect assemblies of repeated sequences were detected by phrap on the basis of abnormally long distances between insert extremities, for instance, those exceeding 4 kbp for plasmid inserts. To overcome these difficulties, the misassembled DNA regions were assembled separately and completed by primer walking. The resulting consensus sequences were reintroduced into the general assembling process to resolve repeated regions and restore a normal scaffold. The phrap assembly yielded 77 chromosomal contigs totaling 1,674 kbp of the 1,820-kbp chromosome and 8 circular contigs, which proved to be an SVTS2-like viral DNA (SVGII3) and seven plasmids, pSciA and pSci1 to pSci6, with sizes ranging from 7.8 to 35.3 kbp (40). Based on the relative positions of the restriction sites and genetic markers, the 20 largest contigs were organized on the chromosome map (Fig. 1A). Eighteen additional contigs, NP01 to NP18, totaling 165 kbp, could not be located on the physical map.
Functional annotation and comparative analyses.
Sequence analyses and annotation were managed with the iANT (Integrated Annotation Tool) Web-based annotation environment developed for Ralstonia solanacearum genome annotation (41). Coding sequences (CDS) were predicted by using the FrameD program (42) trained with known S. citri genes. For the structural and functional assignment of the CDS, they were compared to data in the SP-TrEMBL database using the BLASTP program with a cutoff E value of e−10 (1), submitted to TMPRED for membrane-spanning domains, and aligned to data in the ProDom and Prosite databases for conserved protein domains (5, 16). Signal peptides were predicted by using SignalP 3.0 (8), and transmembrane topology was predicted by using TMHMM (29). The prediction of tRNA was conducted with tRNA-ScanSE (33). The genome of Bacillus subtilis was taken as a reference for the gene product and gene name. The gene product and gene name were propagated when at least 50% of each protein was involved in the alignment with at least 20% identity at the amino acid level. Levels of confidence were “blank,” probable, putative, and hypothetical when identity levels were above 80%, 60%, 40%, and 20%, respectively, for a reciprocal coverage of 80%. For reciprocal coverage in the range of 50 to 80%, levels of confidence were probable, putative, and hypothetical when identity levels were above 80%, 60%, and 40%, respectively. Annotated sequence data can be accessed at http://iant.toulouse.inra.fr/bacteria/annotation/cgi/spici.cgi?. Comparisons of proteomes were performed with Molligen 3.0 (http://cbi.labri.fr/outils/molligen/). To determine the number of orthologous CDS between two genomes, the bidirectional best hit was computed from Smith and Waterman SSEARCH local alignments. Best reciprocal hits below a cutoff E value of e−4 were considered significant when more than 50% of the longest protein was involved in the alignment.
Nucleotide sequence accession numbers.
The 38 largest contigs were annotated and deposited in the EMBL nucleotide sequence database under the accession numbers reported in Table 1.
TABLE 1.
S. citri chromosomal contigs and accession numbers
| Contig | GenBank accession no. | Size (bp) |
|---|---|---|
| Located on the S. citri chromosome genetic mapa | ||
| 01A | AM285301 | 52,177 |
| 01B | AM285302 | 127,324 |
| 02 | AM285303 | 55,702 |
| 03 | AM285304 | 290,001 |
| 04 | AM285305 | 130,001 |
| 05 | AM285306 | 19,000 |
| 06 | AM285307 | 74,020 |
| 07 | AM285308 | 34,700 |
| 08 | AM285309 | 53,900 |
| 09 | AM285310 | 53,400 |
| 10 | AM285311 | 61,960 |
| 11 | AM285312 | 25,500 |
| 12 | AM285313 | 35,850 |
| 13 | AM285314 | 24,400 |
| 14 | AM285315 | 11,100 |
| 15 | AM285316 | 43,700 |
| 16 | AM285317 | 52,860 |
| 17 | AM285318 | 20,400 |
| 18 | AM285319 | 34,200 |
| 19 | AM285320 | 83,681 |
| 20 | AM285321 | 76,301 |
| Could not be located on the S. citri chromosome genetic map | ||
| NP01 | AM285322 | 19,320 |
| NP02 | AM285323 | 17,551 |
| NP03 | AM285324 | 10,404 |
| NP04 | AM285325 | 8,700 |
| NP05 | AM285326 | 10,144 |
| NP06 | AM285327 | 9,000 |
| NP07 | AM285328 | 6,800 |
| NP08 | AM285329 | 4,593 |
| NP09 | AM285330 | 5,916 |
| NP10 | AM285331 | 4,076 |
| NP11 | AM285332 | 9,000 |
| NP12 | AM285333 | 15,184 |
| NP13 | AM285334 | 9,597 |
| NP14 | AM285335 | 9,500 |
| NP15 | AM285336 | 5,800 |
| NP16 | AM285337 | 6,000 |
| NP17 | AM285338 | 7,000 |
| NP18 | AM285339 | 6,994 |
See Fig. 1.
RESULTS AND DISCUSSION
General features of the S. citri GII3-3X chromosome: repeated sequences and gene decay.
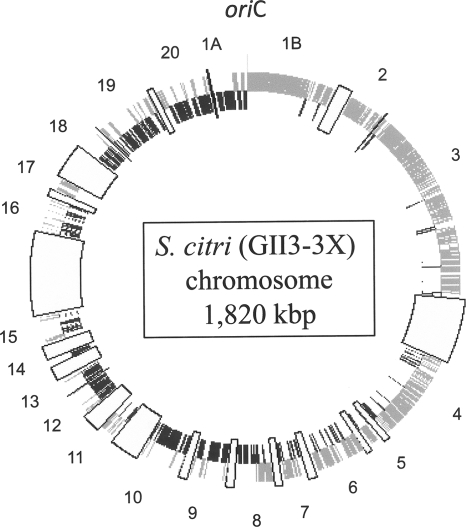
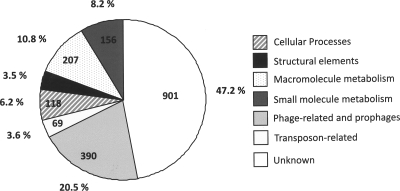
The S. citri GII3-3X chromosome has a G+C content of 26.1%. It encodes one single 16S-23S-5S rRNA operon and 32 tRNAs, including the two tryptophan tRNAs, tRNATrp(CCA) and tRNATrp(UCA) (14). The tRNA genes are organized into eight clusters. The 38 contigs comprise 1,908 CDS representing an overall coding density of 74%. CDS are symmetrically distributed on both sides of oriC, with a strong bias for the leading strands of chromosome replication (Fig. 2). Every predicted CDS was assigned to a functional category. CDS encoding components of cellular processes, cell metabolism, and structural elements account for 6.2%, 20%, and 3.5% of the CDS, respectively (Fig. 3). CDS of external origin such as viruses and mobile elements represent 20.5% and 3.6% of the coding capacity, respectively, and CDS of unknown function represent 47% of the coding capacity. Among these CDS of unknown function, 21% group into 63 paralog families that cluster mainly in a large region of the chromosome at the opposite side of oriC. The largest family comprises 23 paralogs with an N-terminal A(D/N)LXX pentapeptide repeat motif (PPA) of unknown function. Paralogous CDS group into repeated blocks of 7 to 12 ordered units each (see Fig. S2 in the supplemental material). These blocks are arranged into four distinct types depending on whether they contain the P70, P25, P31, or P29 paralog. The most conserved CDS are found in the PPA and P16 families. The organization of these paralogs into conserved blocks of various sizes suggests that they may represent remnants of former mobile units having undergone many insertion/deletion/recombination events (Fig. S2). No integrase/transposase-encoding CDS were identified in these blocks. Phage-related sequences were homologous to plectrovirus SpV1 (38) and SVTS2 (43) as well as those presumed to be from SpV2, a lambda-like phage found in S. citri cultures (15). These viral sequences are dispersed into more than 50 sites all along the chromosome. It is noteworthy that most (13/14) of the regions containing plectrovirus sequences larger than 7 kbp (i.e., that might represent full-length prophages) contain truncated CDS. Only one seems to correspond to a linearized, entire SpV1 genome. The integration of a plectrovirus genome into the S. citri chromosome has been shown to occur upon virus infection and to be associated with resistance to subsequent infection (44).
FIG. 2.
Distribution of CDS along the circular chromosome. Gray and dark gray boxes indicate CDS in sense and antisense orientations, respectively. Large boxes indicate sequencing gaps. Contig numbers are clockwise.
FIG. 3.
Distribution of the S. citri chromosomal CDS in various functional categories.
Sixty-nine CDS encoding truncated and full-length transposases were detected. Based on sequence similarities with transposases in the ISFinder database (45), these transposases belong to four distinct insertion sequence (IS) families (IS30, IS431, IS3, and ISNCY). Whereas those of the IS30 and IS431 (formerly classified as IS3) families are all associated with SpV1 sequences, those of the IS3 (one copy) and ISNCY (seven copies of IS1202) families are not.
Similarity analyses showed that 21% of the S. citri chromosomal CDS were truncated compared to their bacterial orthologs, revealing important gene decay. In addition to many transposases and viral CDS, which appeared C-terminally or N-terminally truncated, housekeeping genes were also found to be incomplete. Among these are recA, involved in homologous recombination and shown to be truncated in other strains of S. citri (34); dinB, encoding DNA polymerase IV; and pyrP, encoding uracil permease.
Membrane proteins, transport, and metabolism.
Transmembrane domains, including signal peptides, were predicted for 599 CDS, of which 58 were putative lipoproteins with a cysteine residue at positions 22 to 26. S. citri has a Sec-dependent protein export pathway, as indicated by the presence of the secY, secA, ftsY, and ffh (SRP54) genes and homologs to secE (SPICI03_054) and oxaA or yidC (SPICI01A_062). Eighty-four CDS were involved in the acquisition of various metabolites from the host/environment. They encode components of phosphoenolpyruvate phosphotransferase systems (PTS), ATP binding cassette (ABC) transporters, and various other transporters. In addition to the general enzymes EI and HPr and the glucose-, fructose-, and trehalose-specific PTS permeases that were previously shown to be functional (2, 3, 18, 25), the S. citri genome also encodes several other hypothetical PTS permeases of unknown substrates, most of which are truncated. The occurrence of PTS in spiroplasma genomes is a striking difference from the genomes of other phloem-limited bacteria such as phytoplasmas (30, 36, 47) and the phloem-restricted proteobacterium “Candidatus Liberibacter asiaticus” (17), which do not encode PTS. In the S. citri GII3-3X genome, 32 CDS were predicted to encode components of ABC transporters (27), including 12 permeases and 15 ATP binding and 5 solute binding components, 2 of which are specific for phosphate and oligopeptide, respectively. Other transporters include an Na+-dependent transporter, three cation transporters, one arginine/ornithine antiporter, two amino acid and polyamine permeases, one formate/nitrite transporter, ferritin, a malate permease, an Na+/dicarboxylate symporter, and a NAD transporter. Like the other mollicutes, S. citri has limited synthesis capabilities. In particular, except for the glycine hydroxymethyltransferase, which converts glycine into serine, it lacks most of the enzymes involved in amino acid metabolism. In contrast, many genes of the purine and pyrimidine metabolism pathways are present.
Besides the PTS to import sugars, S. citri GII3-3X also possesses a complete glycolytic pathway and lactate dehydrogenase to promote fermentation. Regarding the pentose-phosphate pathway, S. citri GII3-3X lacks the two genes encoding the enzymes that convert glucose-6P into ribulose-5P. Therefore, this pathway appeared deficient for the oxidative phase. In turn, the S. citri GII3-3X genome encodes four enzymes of the nonoxidative phase of this pathway, namely, ribulose-phosphate 3-epimerase (rpe; EC 5.1.3.1), transketolase (tkt; EC 2.2.1.1), ribose-phosphate pyrophosphokinase (prs; EC 2.7.6.1), and, possibly, ribose-5-phosphate isomerase B (rpiB; EC 5.3.1.6). These four genes are present in mycoplasmas but are totally absent in phytoplasmas.
The S. citri genome contains CDS homologous to Clostridium perfringens arcA, arcB, arcC, and arcD, which encode proteins homologous to arginine deiminase (ADI), ornithine carbamoyltransferase (OTC), carbamate kinase (CK), and the arginine/ornithine antiporter (ArcD), respectively. The presence of the ADI operon is consistent with our previous data showing the ability of S. citri GII3-3X to metabolize arginine (18), a feature shared by other mollicutes, including Mycoplasma penetrans and Mycoplasma hominis (37).
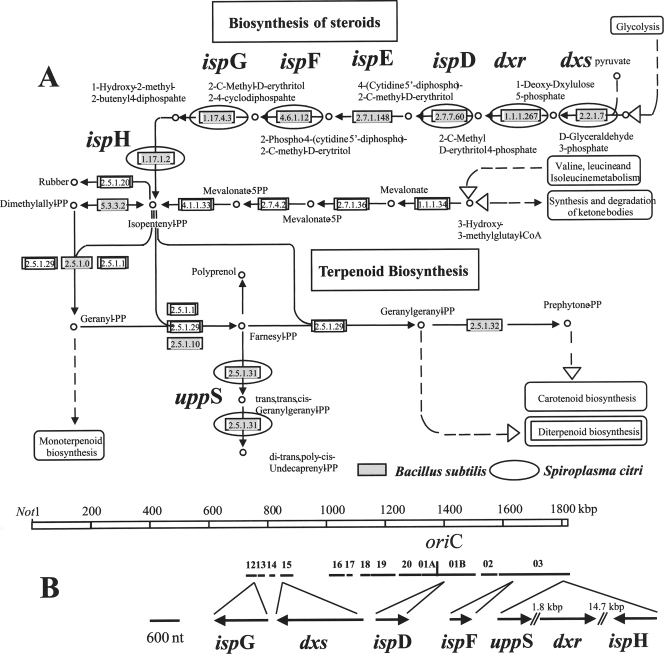
A large operon made of eight CDS was predicted to represent the operon of atpB, atpE, atpF, atpH, atpA, atpG, atpD, and atpC, encoding the eight subunits of the classical FoF1-ATP synthase, which is thought to be essential for ATP production. Surprisingly, this ATP synthase is absent in sequenced phytoplasma genomes (30, 36, 47). The S. citri GII3-3X chromosome also encodes a nearly complete pathway for the biosynthesis of a C55 terpenoid (Fig. 4A). Six genes (dxs, dxr, ispD, ispF, ispG, and ispH) dedicated to the synthesis of isopentenyl pyrophosphate through the 2-C-methyl-d-erythritol 4-phosphate/1-deoxy-d-xylulose 5-phosphate (MEP/DOXP) pathway were identified. Only ispE is missing for the pathway to be complete, as is the case for Bacillus subtilis (Fig. 4A). A comparison with B. subtilis also reveals that both bacteria lack the mevalonate pathway for isopentenyl-pyrophosphate (PP) synthesis (Fig. 4A). Other bacteria, such as Xylella fastidiosa and Wolbachia pipiensis, which are also associated with plants or arthropods, have a complete MEP/DOXP pathway, and in the class Mollicutes it is also the case for M. penetrans and Mycoplasma gallisepticum. On the contrary, other phloem-restricted bacteria like phytoplasmas and “Ca. Liberibacter asiaticus” lack genes for the MEP/DOXP pathway. Note that in S. citri, the genes involved in the MEP/DOXP pathway are not conserved due to their clustering in an operon, but instead, they are scattered over 1 Mbp of the S. citri chromosome (Fig. 4B). A possible use of isopentenyl-PP in spiroplasma cells is the modification of tRNA. Indeed, S. citri has an miaA gene, which encodes the tRNA isopentenyl transferase, which catalyzes the first step of the modification of an adenosine near the tRNA anticodon. Even though S. citri has the uppS gene, which transforms farnesyl-PP into undecaprenyl-PP, it lacks the gene encoding the enzyme that transforms isopentenyl-PP into farnesyl-PP. Therefore, the capability of S. citri to synthesize the C55 terpenoid undecaprenyl pyrophosphate remains to be established.
FIG. 4.
(A) Steroid and terpenoid synthesis pathways according to the KEGG encyclopedia with predicted S. citri pathways. Predicted S. citri genes are indicated by black circles, while enzymes present in Bacillus subtilis are indicated by gray boxes. (B) Chromosomal distribution of genes involved in steroid and terpenoid synthesis in S. citri. nt, nucleotides.
Comparison with other mollicute genomes.
In summary, S. citri, like other members of the Mollicutes (46), has undergone a reductive genome evolution resulting in the lack of many biosynthetic pathways. Unlike phytoplasmas (30, 36, 47) and like mycoplasmas, S. citri has retained the ability to import sugars via the PTS and to synthesize ATP using an FoF1-ATP synthase. The S. citri chromosome is roughly 1 Mbp larger than most phytoplasma genomes. It contains many CDS of plectrovirus prophages and conserved blocks of unknown function that could also be remnants of viral sequence insertions, a feature that is common with phytoplasma genomes, as they contain repeated clusters of genes that could be mobile genetic elements or remnants of ancient phage attacks (4, 50). In S. citri GII3-3X, the finding of hundreds of truncated CDS reveals an important gene decay. A similar gene decay was also reported for a published 85-kbp region of the Spiroplasma kunkelii chromosome (56). It may be possible that the reductive evolution of the spiroplasma genome is still ongoing on the way to smaller genomes with the loss of genes that are not necessary. The resulting genomes would have a size closer to that of related mollicutes, around 1 Mbp. By comparison of the annotated region of the S. kunkelii chromosome with the homologous region in the S. citri GII3-3X chromosome, over 100 predicted CDS, 66 CDS, and a cluster of two tRNAs were found conserved and organized in the same order. Despite this overall synteny, S. kunkelii has 18 additional small CDS, most of which encode hypothetical proteins of unknown function, while S. citri possesses 8 additional CDS. Both spiroplasmas have a disrupted uraA or pyrP gene for uracil permease, but S. kunkelii presented three truncated CDS for RNase HII (rnhB), whereas S. citri had the complete rnhB gene. Apart from the important gene decay, the S. citri chromosome is also characterized by an important proportion of species-specific CDS of unknown function (47%). Comparison of the predicted proteome of S. citri GII3-3X with those of the phylogenetically related Mycoplasma mycoides SC PG1 and Mesoplasma florum showed that they have 425 and 444 orthologous CDS in common, respectively. For comparisons to mollicutes of other phylogenetic branches, the figures vary, from 290 (Mycoplasma genitalium) to 322 (Mycoplasma pulmonis) and 359 (Acholeplasma laidlawii) orthologous CDS. Among the plant-pathogenic mollicutes, which are transmitted by hemipteran insects, S. citri shares only 223, 240, and 242 orthologous CDS with “Candidatus Phytoplasma mali,” “Ca. Phytoplasma asteris,” and “Ca. Phytoplasma australiense,” respectively. These data indicate that in the S. citri genome, the overall gene conservation is more related to the phylogenetic origin than to the ecological niche. Therefore, spiroplasmas and phytoplasmas, despite sharing a similar biological cycle between plants and insects, have rather divergent genome information.
Due to the extremely large number of repeated sequences, the S. citri chromosome sequence could not be completed. However, it is likely that most of the missing data correspond to repeated sequences of viral origin. Despite its abundant direct and inverted repeats, the genome of S. citri is quite stable, certainly due to its being recA deficient. A comparison of the S. citri and Spiroplasma melliferum chromosome maps showed that very few large-scale rearrangements took place during their evolution (52). The sequence data described here sustain the current efforts to establish the S. citri proteome and produce new targeted mutants for a better understanding of the interactions of S. citri with its leafhopper vector. It will also contribute to comparative genomics in the class Mollicutes (6, 46) and may help in an understanding of genome evolution in this class of small bacteria with reduced genomes and provide clues for the identification of the function of the numerous cryptic genes that have been described in the past 15 years.
Supplementary Material
Acknowledgments
This work was funded by INRA, Fundecitrus, and the Conseil Régional d'Aquitaine.
We thank Audrey Henri and Marie Weber for excellent technical assistance and Pascal Sirand-Pugnet for helpful discussions.
Footnotes
Published ahead of print on 2 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.André, A., W. Maccheroni, F. Doignon, M. Garnier, and J. Renaudin. 2003. Glucose and trehalose PTS permeases of Spiroplasma citri probably share a single IIA domain, enabling the spiroplasma to adapt quickly to carbohydrate changes in its environment. Microbiology 149:2687-2696. [DOI] [PubMed] [Google Scholar]
- 3.André, A., M. Maucourt, A. Moing, D. Rolin, and J. Renaudin. 2005. Sugar import and phytopathogenicity of Spiroplasma citri: glucose and fructose play distinct roles. Mol. Plant Microbe Interact. 18:32-42. [DOI] [PubMed] [Google Scholar]
- 4.Bai, X. D., J. H. Zhang, A. Ewing, S. A. Miller, A. J. Radek, D. V. Shevchenko, K. Tsukerman, T. Walunas, A. Lapidus, J. W. Campbell, and S. A. Hogenhout. 2006. Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 188:3682-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bairoch, A. 1992. Prosite—a dictionary of sites and patterns in proteins. Nucleic Acids Res. 20:2013-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barré, A., A. de Daruvar, and A. Blanchard. 2004. MolliGen, a database dedicated to the comparative genomics of Mollicutes. Nucleic Acids Res. 32:D307-D310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bébéar, C. M., P. Aullo, J. M. Bové, and J. Renaudin. 1996. Spiroplasma citri virus SpV1: characterization of viral sequences present in the spiroplasma host chromosome. Curr. Microbiol. 32:134-140. [Google Scholar]
- 8.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 9.Berho, N., S. Duret, J. L. Danet, and J. Renaudin. 2006. Plasmid pSci6 from Spiroplasma citri GII-3 confers insect transmissibility to the non-transmissible strain S. citri 44. Microbiology 152:2703-2716. [DOI] [PubMed] [Google Scholar]
- 10.Boutareaud, A., J. L. Danet, M. Garnier, and C. Saillard. 2004. Disruption of a gene predicted to encode a solute binding protein of an ABC transporter reduces transmission of Spiroplasma citri by the leafhopper Circulifer haematoceps. Appl. Environ. Microbiol. 70:3960-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bové, J. M., J. Renaudin, C. Saillard, X. Foissac, and M. Garnier. 2003. Spiroplasma citri, a plant pathogenic mollicute: relationships with its two hosts, the plant and the leafhopper vector. Annu. Rev. Phytopathol. 41:483-500. [DOI] [PubMed] [Google Scholar]
- 12.Breton, M., S. Duret, N. Arricau-Bouvery, L. Beven, and J. Renaudin. 2008. Characterizing the replication and stability regions of Spiroplasma citri plasmids identifies a novel replication protein and expands the genetic toolbox for plant-pathogenic spiroplasmas. Microbiology 154:3232-3244. [DOI] [PubMed] [Google Scholar]
- 13.Calavan, E. C., and J. M. Bové. 1989. Ecology of Spiroplasma citri, p. 425-487. In R. F. Whitcomb and J. G. Tully (ed.), The mycoplasmas, vol. 5. Academic Press, New York, NY. [Google Scholar]
- 14.Citti, C., L. Marechal-Drouard, C. Saillard, J. H. Weil, and J. M. Bové. 1992. Spiroplasma citri UGG and UGA tryptophan codons: sequence of the two tryptophanyl-tRNAs and organization of the corresponding genes. J. Bacteriol. 174:6471-6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole, R. M., J. G. Tully, T. J. Popkin, and J. M. Bové. 1973. Morphology, ultrastructure and bacteriophage infection of the helical mycoplasma-like organism (Spiroplasma citri gen. nov., sp. nov.) cultured from “stubborn” disease of citrus. J. Bacteriol. 115:367-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corpet, F., J. Gouzy, and D. Kahn. 1998. The ProDom database of protein domain families. Nucleic Acids Res. 26:323-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan, Y. P., L. J. Zhou, D. G. Hall, W. B. Li, H. Doddapaneni, H. Lin, L. Liu, C. M. Vahling, D. W. Gabriel, K. P. Williams, A. Dickerman, Y. J. Sun, and T. Gottwald. 2009. Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plant Microbe Interact. 22:1011-1020. [DOI] [PubMed] [Google Scholar]
- 18.Duret, S., A. André, and J. Renaudin. 2005. Specific gene targeting in Spiroplasma citri: improved vectors and production of unmarked mutations using site-specific recombination. Microbiology 151:2793-2803. [DOI] [PubMed] [Google Scholar]
- 19.Duret, S., N. Berho, J. L. Danet, M. Garnier, and J. Renaudin. 2003. Spiralin is not essential for helicity, motility, or pathogenicity but is required for efficient transmission of Spiroplasma citri by its leafhopper vector Circulifer haematoceps. Appl. Environ. Microbiol. 69:6225-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 21.Ewing, B., L. Hillier, M. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 22.Fletcher, J., A. Wayadande, U. Melcher, and F. Ye. 1998. The phytopathogenic mollicute-insect vector interface: a closer look. Phytopathology 88:1351-1358. [DOI] [PubMed] [Google Scholar]
- 23.Foissac, X., J. L. Danet, C. Saillard, P. Gaurivaud, F. Laigret, C. Pare, and J. M. Bové. 1997. Mutagenesis by insertion of Tn4001 into the genome of Spiroplasma citri: characterization of mutants affected in plant pathogenicity and transmission to the plant by the leafhopper vector Circulifer haematoceps. Mol. Plant Microbe Interact. 10:454-461. [Google Scholar]
- 24.Gaurivaud, P., J. L. Danet, F. Laigret, M. Garnier, and J. M. Bové. 2000. Fructose utilization and phytopathogenicity of Spiroplasma citri. Mol. Plant Microbe Interact. 13:1145-1155. [DOI] [PubMed] [Google Scholar]
- 25.Gaurivaud, P., F. Laigret, M. Garnier, and J. M. Bové. 2000. Fructose utilization and pathogenicity of Spiroplasma citri: characterization of the fructose operon. Gene 252:61-69. [DOI] [PubMed] [Google Scholar]
- 26.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 27.Higgins, C. F., S. C. Hyde, M. M. Mimmack, U. Gileadi, D. R. Gill, and M. P. Gallagher. 1990. Binding protein-dependent transport systems. J. Bioenerg. Biomembr. 22:571-592. [DOI] [PubMed] [Google Scholar]
- 28.Killiny, N., M. Castroviejo, and C. Saillard. 2005. Spiroplasma citri spiralin acts in vitro as a lectin binding to glycoproteins from its insect vector Circulifer haematoceps. Phytopathology 95:541-548. [DOI] [PubMed] [Google Scholar]
- 29.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 30.Kube, M., B. Schneider, H. Kuhl, T. Dandekar, K. Heitmann, A. M. Migdoll, R. Reinhardt, and E. Seemuller. 2008. The linear chromosome of the plant-pathogenic mycoplasma ‘Candidatus Phytoplasma mali.’ BMC Genomics 9:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, H. Y., D. J. Gumpf, G. N. Oldfield, and E. C. Calavan. 1983. The relationship of Spiroplasma citri and Circulifer tenellus. Phytopathology 73:585-590. [Google Scholar]
- 32.Liu, H. Y., D. J. Gumpf, G. N. Oldfield, and E. C. Calavan. 1983. Transmission of Spiroplasma citri by Circulifer tenellus. Phytopathology 73:582-585. [Google Scholar]
- 33.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marais, A., J. M. Bové, and J. Renaudin. 1996. Characterization of the recA gene regions of Spiroplasma citri and Spiroplasma melliferum. J. Bacteriol. 178:7003-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melcher, U., Y. Sha, F. Ye, and J. Fletcher. 1999. Mechanisms of spiroplasma genome variation associated with SpV1-like viral DNA inferred from sequence comparisons. Microb. Comp. Genomics 4:29-46. [DOI] [PubMed] [Google Scholar]
- 36.Oshima, K., S. Kakizawa, H. Nishigawa, H. Y. Jung, W. Wei, S. Suzuki, R. Arashida, D. Nakata, S. Miyata, M. Ugaki, and S. Namba. 2004. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat. Genet. 36:27-29. [DOI] [PubMed] [Google Scholar]
- 37.Pereyre, S., P. Sirand-Pugnet, L. Beven, A. Charron, H. Renaudin, A. Barre, P. Avenaud, D. Jacob, A. Couloux, V. Barbe, A. de Daruvar, A. Blanchard, and C. Bebear. 2009. Life on arginine for Mycoplasma hominis: clues from its minimal genome and comparison with other human urogenital mycoplasmas. PLoS Genet. 5:e1000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renaudin, J., and J. M. Bové. 1994. Spv1 and Spv4, spiroplasma viruses with circular, single-stranded-DNA genomes, and their contribution to the molecular biology of spiroplasmas. Adv. Virus Res. 44:429-463. [DOI] [PubMed] [Google Scholar]
- 39.Saglio, P., M. L'Hospital, D. Laflèche, G. Dupont, J. M. Bové, J. G. Tully, and E. A. Freundt. 1973. Spiroplasma citri gen. and sp. nov.: a mycoplasmalike organism associated with “stubborn” disease of citrus. Int. J. Syst. Bacteriol. 23:191-204. [Google Scholar]
- 40.Saillard, C., P. Carle, S. Duret-Nurbel, R. Henri, N. Killiny, S. Carrère, J. Gouzy, J. M. Bové, J. Renaudin, and X. Foissac. 2008. The abundant extrachromosomal DNA content of the Spiroplasma citri GII3-3X genome. BMC Genomics 9:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 42.Schiex, T., J. Gouzy, A. Moisan, and Y. de Oliveira. 2003. FrameD: a flexible program for quality check and gene prediction in prokaryotic genomes and noisy matured eukaryotic sequences. Nucleic Acids Res. 31:3738-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sha, Y., U. Melcher, R. E. Davis, and J. Fletcher. 2000. Common elements of spiroplasma plectroviruses revealed by nucleotide sequence of SVTS2. Virus Genes 20:47-56. [DOI] [PubMed] [Google Scholar]
- 44.Sha, Y. H., U. Melcher, R. E. Davis, and J. Fletcher. 1995. Resistance of Spiroplasma citri lines to the virus SVTS2 is associated with integration of viral DNA sequences into host chromosomal and extrachromosomal DNA. Appl. Environ. Microbiol. 61:3950-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siguier, P., J. Perochon, L. Lestrade, J. Mahillon, and M. Chandler. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34:D32-D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sirand-Pugnet, P., C. Citti, A. Barre, and A. Blanchard. 2007. Evolution of mollicutes: down a bumpy road with twists and turns. Res. Microbiol. 158:754-766. [DOI] [PubMed] [Google Scholar]
- 47.Tran-Nguyen, L. T. T., M. Kube, B. Schneider, R. Reinhardt, and K. S. Gibb. 2008. Comparative genome analysis of “Candidatus Phytoplasma australiense” (subgroup tuf-Australia I; rp-A) and “Ca. Phytoplasma asteris” strains OY-M and AY-WB. J. Bacteriol. 190:3979-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tully, J. G., R. F. Whitcomb, H. F. Clark, and D. L. Williamson. 1977. Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science 195:892-894. [DOI] [PubMed] [Google Scholar]
- 49.Vignault, J. C., J. M. Bové, C. Saillard, R. Vogel, A. Faro, L. Venegas, W. Stemmer, S. Aoki, R. E. McCoy, A. S. Albeldawi, M. Larue, M. Tuzcu, M. Ozsan, A. Nhami, M. Abassi, J. Bonfils, G. Moutous, A. Fos, F. Poutiers, and G. Viennot-Bourgin. 1980. Mise en culture de spiroplasmes à partir de matériel végétal et d'insectes provenant des pays circum-Méditerranéens et du Proche-Orient. C. R. Acad. Sci. D 290:775-778. [Google Scholar]
- 50.Wei, W., R. E. Davis, R. Jomantiene, and Y. Zhao. 2008. Ancient, recurrent phage attacks and recombination shaped dynamic sequence-variable mosaics at the root of phytoplasma genome evolution. Proc. Natl. Acad. Sci. U. S. A. 106:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weisburg, W. G., J. G. Tully, D. L. Rose, J. P. Petzel, H. Oyaizu, D. Yang, L. Mandelco, J. Sechrest, T. G. Lawrence, J. V. Etten, J. Maniloff, and C. R. Woese. 1989. A phylogenetic analysis of the mycoplasmas: basis for their classification. J. Bacteriol. 171:6455-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye, F. C., F. Laigret, and J. M. Bové. 1994. A physical and genomic map of the prokaryote Spiroplasma melliferum and its comparison with the Spiroplasma citri map. C. R. Acad. Sci. III 317:392-398. [Google Scholar]
- 53.Ye, F. C., F. Laigret, P. Carle, and J. M. Bové. 1995. Chromosomal heterogeneity among various strains of Spiroplasma citri. Int. J. Syst. Bacteriol. 45:729-734. [DOI] [PubMed] [Google Scholar]
- 54.Ye, F. C., F. Laigret, J. C. Whitley, C. Citti, L. R. Finch, P. Carle, J. Renaudin, and J. M. Bové. 1992. A physical and genetic map of the Spiroplasma citri genome. Nucleic Acids Res. 20:1559-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye, F. C., J. Renaudin, J. M. Bové, and F. Laigret. 1994. Cloning and sequencing of the replication origin (Oric) of the Spiroplasma citri chromosome and construction of autonomously replicating artificial plasmids. Curr. Microbiol. 29:23-29. [DOI] [PubMed] [Google Scholar]
- 56.Zhao, Y., R. W. Hammond, R. Jomantiene, E. L. Dally, I. M. Lee, H. Jia, H. Wu, S. Lin, P. Zhang, S. Kenton, F. Z. Najar, A. Hua, B. A. Roe, J. Fletcher, and R. E. Davis. 2003. Gene content and organization of an 85-kb DNA segment from the genome of the phytopathogenic mollicute Spiroplasma kunkelii. Mol. Gen. Genomics 269:592-602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.