Abstract
Biofilm formation results in medical threats or economic losses and is therefore a major concern in a variety of domains. In two-species biofilms of marine bacteria grown under dynamic conditions, Pseudoalteromonas sp. strain 3J6 formed mixed biofilms with Bacillus sp. strain 4J6 but was largely predominant over Paracoccus sp. strain 4M6 and Vibrio sp. strain D01. The supernatant of Pseudoalteromonas sp. 3J6 liquid culture (SN3J6) was devoid of antibacterial activity against free-living Paracoccus sp. 4M6 and Vibrio sp. D01 cells, but it impaired their ability to grow as single-species biofilms and led to higher percentages of nonviable cells in 48-h biofilms. Antibiofilm molecules of SN3J6 were able to coat the glass surfaces used to grow biofilms and reduced bacterial attachment about 2-fold, which might partly explain the biofilm formation defect but not the loss of cell viability. SN3J6 had a wide spectrum of activity since it affected all Gram-negative marine strains tested except other Pseudoalteromonas strains. Biofilm biovolumes of the sensitive strains were reduced 3- to 530-fold, and the percentages of nonviable cells were increased 3- to 225-fold. Interestingly, SN3J6 also impaired biofilm formation by three strains belonging to the human-pathogenic species Pseudomonas aeruginosa, Salmonella enterica, and Escherichia coli. Such an antibiofilm activity is original and opens up a variety of applications for Pseudoalteromonas sp. 3J6 and/or its active exoproducts in biofilm prevention strategies.
Biofilms are defined as microbial communities of cells that are irreversibly attached to a substratum, to an interface, or to each other and are embedded into a matrix of extracellular polymeric substances that they have produced (8). It is now considered that most (if not all) bacteria are capable of forming biofilms and that this is their predominant bacterial life-style. Biofilm formation is a complex biological phenomenon and has been generally described as a temporal process involving a succession of distinct stages: a reversible and then irreversible attachment of planktonic bacteria onto a surface, the formation of microcolonies either by the clonal growth of attached cells or by the active translocation of cells across the surface, the coalescence of growing microcolonies to form a macrocolony, and cell dispersal. It should, however, be noted that this developmental model still requires further experimental validation, especially concerning the possibility of a hierarchical order of genetic pathways (26). Furthermore, Karatan and Watnick (17) pointed out that there are as many different types of biofilms as there are bacteria and that a single bacterium may even make several different types of biofilms under different environmental conditions. Biofilm formation is associated with the virulence of pathogenic bacteria, and cells included within a biofilm are generally more resistant (up to 1,000-fold) to antibiotics and disinfectants than free-living bacteria (8, 26). Biofilms are therefore a major concern in medicine and in medical environments but also in all domains where their growth constitutes a source of contamination for humans or animals (food industry, cooling towers, and water pipes, etc.) or leads to economical losses (biofouling of boats and immersed structures and material biocorrosion, etc.). The development of antibiofilm strategies is therefore of major interest and currently constitutes an important field of investigation in which environmentally friendly antibiofilm molecules or organisms are highly valuable (5, 7, 9).
Marine bacteria belonging to the genus Pseudoalteromonas of the class Gammaproteobacteria are often found in association with marine eukaryotes, and their ability to produce a variety of biological activities has attracted particular attention (2, 11, 13, 15, 28). We previously isolated marine bacteria attached to solid surfaces (glass in most cases) immersed for 3 or 6 h in the Morbihan Gulf or in the Bay of Brest, France (10, 20, 21, 27). Out of the three Pseudoalteromonas strains isolated, we were able to tag strain 3J6 with a green fluorescent protein (GFP)-encoding plasmid. This allowed us to investigate whether Pseudoalteromonas sp. strain 3J6 affected the biofilm growth of other marine bacterial isolates. Here, we report that strain 3J6 predominated in two-species biofilms over Paracoccus sp. strain 4M6 and Vibrio sp. strain D01. Although devoid of antibacterial activity against planktonic cells, Pseudoalteromonas sp. 3J6 exoproducts impaired biofilm formation by Paracoccus sp. 4M6 and Vibrio sp. D01. We characterized the effects of these exoproducts on the latter strains and on other bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this work are listed in Table 1. Strain 3J6 was affiliated with the genus Pseudoalteromonas (10). Its 16S rRNA gene sequence (GenBank accession number FJ966949) is most closely related (95.5% identity) to that of Pseudoalteromonas sp. strain SM9913, which belongs to a newly discovered species (28). Pseudoalteromonas sp. 3J6 and most of the strains from marine origin that we used were isolated from the same location in the Morbihan Gulf within a 2-month period (10) and therefore shared the same natural habitat. Marine bacteria were grown in Vaatanen nine-salt solution (VNSS) (24) at 20°C with shaking (120 rpm). Pseudoalteromonas sp. 3J6 was selected, when required, with 200 μg ml−1 streptomycin, and Pseudoalteromonas sp. 3J6(pCJS10) was grown with 125 μg ml−1 chloramphenicol. Escherichia coli DH5α cells containing pCJS10 or pRK2013 were grown at 37°C with shaking (180 rpm) in Luria-Bertani (LB) broth containing 50 μg ml−1 chloramphenicol or kanamycin, respectively. Pseudomonas aeruginosa PAO1YFP, E. coli CC118DsRed, and Salmonella enterica MB1409HcRed were grown with shaking (180 rpm) at 37°C in LB medium (34) supplemented with 50 μg ml−1 gentamicin, 15 μg ml−1 gentamicin, and a mix of 50 μg ml−1 kanamycin and 10 μg ml−1 chloramphenicol, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Origin or construction | Characteristic(s)d | Reference(s) and/or source |
|---|---|---|---|
| Strains | |||
| Pseudoalteromonas sp. 3J6 | Morbihan Gulf, Francea | Wild-type strain; Smr | 10 |
| Paracoccus sp. 4M6 | Morbihan Gulfa | Wild-type strain | 10 |
| Bacillus sp. 4J6 | Morbihan gulfa | Wild-type strain | 10 |
| Vibrio sp. D01 | Bay of Brest, Franceb | Wild-type strain | 27 |
| Algibacter sp. 1M6 | Morbihan Gulfa | Wild-type strain; initially affiliated with the genus Cytophaga | 10 |
| Colwellia sp. 4J3 | Morbihan Gulfa | Wild-type strain | 10 |
| Sulfitobacter sp. 8J6 | Morbihan Gulfa | Wild-type strain | 10 |
| Vibrio sp. D66 | Bay of Brestb | Wild-type strain | IFREMER collection |
| Alteromonas sp. 1J3 | Morbihan Gulfa | Wild-type strain | LBCMe collection |
| Pseudoalteromonas sp. 3J3 | Morbihan Gulfa | Wild-type strain | 10 |
| Pseudoalteromonas sp. D41 | Bay of Brestc | Wild-type strain | 20, 21, 27 |
| Escherichia coli DH5α | supE44 ΔlacU169(φlacZΔM15) recA1 endA1 hsdR17 thi-1 gyrA96 relA1 | Biomedal S.L., Spain | |
| Pseudomonas aeruginosa PAO1YFP | Wild-type PAO1 strain tagged with eYFP in a mini-Tn7 construct | Mini-Tn7(Gm)PA1/04/03-eyfp-a; Gmr | 18 |
| Escherichia coli CC118DsRed | CC118(λpir) strain containing pUT-miniTn5(Gm)PrrnB P1 DsRed | DsRed; Gmr | T. Tolker-Nielsen |
| Salmonella enterica MB1409HcRed | Wild-type MB1409 strain containing pBK-miniTn7(Km Sm)PA1/04/03-HcRed-a | HcRed; Kanr Cmr | 19, 25; G. Legendre |
| Plasmids | |||
| pCJS10 | gfpmut3 gene cloned into plasmid RSF1010 | GFP; Cmr | 30 |
| pRK2013 | Helper plasmid for pCJS10 transfer | oriColE1 RK2-Mob+ RK2-Tra+; Kanr | Biomedal S.L., Spain |
Isolated from a glass slide immersed for 3 or 6 h in the Morbihan Gulf, France.
Isolated from a glass slide immersed for 6 h in the Bay of Brest, France.
Isolated from a Teflon coupon immersed for 24 h in the Bay of Brest, France.
Cmr, chloramphenicol resistance; Kanr, kanamycin resistance; Smr, streptomycin resistance; Gmr, gentamicin resistance.
LBCM, Laboratoire de Biotechnologie et Chimie Marines, Université de Bretagne-Sud, France.
Pseudoalteromonas sp. 3J6 tagging.
Chloramphenicol-resistant plasmid pCJS10 (30) carrying the gfpmut3 gene, which encodes a GFP (3), was introduced into Pseudoalteromonas sp. 3J6 (streptomycin resistant) by conjugation using E. coli DH5α(pCJS10) and E. coli DH5α(pRK2013) as donor and helper strains, respectively. Triparental conjugation was carried out as described previously by Rao et al. (30), and Pseudoalteromonas sp. 3J6 transconjugants were isolated on VNSS agar plates containing chloramphenicol and streptomycin.
Biofilm formation.
Biofilms were grown in continuous-culture three-channel flow cells (channel dimensions, 1 by 4 by 40 mm). The system was assembled and prepared as previously described (36). The substratum was an st1 microscope coverslip (VWR International, Fontenay sous Bois, France). Channels were inoculated with bacterial cultures grown overnight, which were diluted to a final optical density at 600 nm (OD600) of 0.25 after three washes in filtered marine water (FMW) for marine bacteria or in 0.9% NaCl for the other strains. To obtain FMW, seawater from Kernevel Harbor (Larmor Plage, France) was sterilized first by filtration through a membrane with pores of a 0.22-μm diameter and then by autoclaving at 121°C for 20 min. Bacteria were allowed to attach to the glass surface during 2 h at 20°C under static conditions. Biofilm growth was then performed under a constant flow of VNSS or LB medium (0.2 mm s−1) for 48 h at 20°C.
In the case of two-species biofilms, channels were inoculated with a mixture of Pseudoalteromonas sp. 3J6(pCJS10) and one of the other strains in a ratio leading to a similar attachment of each strain to the glass surface (i.e., similar percentages of the glass surface covered by each strain) after the 2-h adhesion step. This ratio was 1:1 for all strain couples except for Pseudoalteromonas sp. 3J6(pCJS10) and Bacillus sp. strain 4J6, which needed a 1:4 ratio.
To investigate the effects of Pseudoalteromonas sp. 3J6 exoproducts on adhesion and biofilm formation by other bacteria, these bacteria were diluted to an OD600 of 0.25 in SN3J6. The latter is a supernatant of a liquid culture of Pseudoalteromonas sp. 3J6 grown overnight that was sterilized by filtration through a membrane with pores with a 0.22-μm diameter. Bacteria diluted in SN3J6 were then inoculated in flow cell channels, and the 2-h adhesion step under static conditions followed by biofilm growth under a flow of fresh VNSS medium was performed as described above. For a better understanding of the SN3J6 action, we modified our procedure as follows: (i) bacteria at an OD600 of 0.25 were incubated with SN3J6 outside flow cell channels for 2 h at 20°C with shaking at 120 rpm, were washed twice in FMW, and were then inoculated into channels, or (ii) SN3J6 was injected without any bacterium into flow cell channels, was left for 2 h at 20°C to coat the glass surface, and the channels were then rinsed with FMW before inoculating bacteria.
Confocal laser scanning microscopy (CLSM) and image analyses.
Microscopic observations were performed with a TCS-SP2 system (Leica Microsystems, Germany) by using a 63× oil immersion objective. To study the attachment of each strain to the glass surface after the 2-h adhesion step, a VNSS flow (0.2 mm s−1) was applied for 15 min to remove planktonic cells, and attached bacteria were observed. The glass surfaces covered by bacteria were evaluated by using Image Tool software (University of Texas Health Science Center at San Antonio). Biofilms were observed by monitoring the GFP fluorescence of Pseudoalteromonas sp. 3J6(pCJS10) (excitation wavelength [λ], 488 nm; emission λ, 510 nm) or after a 10-min incubation with 5 μM Syto 61 red nucleic acid stain (excitation and emission λ, 633 and 645 nm, respectively). For cell viability assays, bacteria were stained with 5 μM Syto 61 red and 0.5 μM Sytox green (excitation and emission λ, 488 and 510 nm, respectively). The fluorescences of P. aeruginosa PAO1YFP, E. coli CC118DsRed, and S. enterica MB1409HcRed were detected at 535, 590, and 647 nm, respectively. Images were obtained by using Leica confocal software, and biofilm stacks were analyzed with COMSTAT software (12). The calculated parameters were the maximal and average thicknesses of the biofilms and the biovolume, which is the volume of bacteria (in μm3) per μm2 of glass surface. The results are representative of at least two independent experiments from which a total of 12 to 18 image stacks were obtained.
Determination of strain proportions in two-species biofilms by qPCR.
Two-species biofilms were grown as described above except that we used BST FC 81 flow cells (BioSurface Technologies, MT) with channel dimensions of 50.8 by 12.7 by 2.54 mm. These flow cells were chosen because they were easily disassembled, allowing total bacterial DNA extraction from biofilms using the DNeasy tissue kit (Qiagen, Germany). The same kit was used to extract bacterial DNA from planktonic cells for control experiments. Quantitative PCR (qPCR) was performed by using primers targeting the 16S rRNA gene sequences (Table 2). The PCR products were detected with SYBR green stain or with TaqMan probes (Table 2). Primers and TaqMan probes were custom synthesized by Eurogentec (Belgium) and Applied Biosystems (Foster City, CA), respectively. The reactions (25-μl reaction mixtures) were performed in triplicate with the 7300 real-time PCR system apparatus (Applied Biosystems). The reaction mixture contained between 0.001 and 10 ng of DNA, 300 nM each primer, and either 12.5 μl of SYBR green PCR master mix (including AmpliTaq gold DNA polymerase; Applied Biosystems) or 200 nM TaqMan probes and 12.5 μl of the TaqMan Universal PCR master mix No AmpErase UNG (Applied Biosystems). The PCR conditions were 50°C for 2 min and 95°C for 10 min for polymerase activation, followed by 40 cycles at 95°C and 60°C for 15 and 60 s, respectively. ROX dye (Applied Biosystems) was used as a passive reference to normalize the non-PCR-related fluorescence variations. Each qPCR was performed in triplicate, and the standard deviations were lower than a CT (threshold cycle) value of 0.15. Each primer pair was validated by verifying that the PCR efficiency was above 0.95 and that a single PCR product with the expected melting temperature (Tm) was obtained. The DNA of a given strain in the sample was quantified by reporting the CT on a standard curve [CT = f(log DNA amount)] obtained with serial dilutions of total DNA extracted from a pure culture of the corresponding strain (22). For each analysis, total DNAs were extracted from two different biofilms, and the two samples were analyzed independently.
TABLE 2.
Quantitative PCR primers and TaqMan probes used in this study
| Primer or probe | Sequence (5′→3′)a | Target |
|---|---|---|
| Primers | ||
| 3J6 F | CGAACTGGCAAACTAGAGTGTGAG | Pseudoalteromonas sp. 3J6 |
| 3J6 R | CCGAGGCTCCGAGCTTCTA | |
| D01 F | TGAAACTGGTGAACTAGAGTGCTGT | Vibrio sp. D01 |
| D01 R | CTCAAGGCCACAACCTCCA | |
| 4J6 F | CAACCGTGGAGGGTCATTG | Bacillus sp. 4J6 |
| 4J6 R | GCGGAAACCCTCTAACACCTT | |
| 4M6 F | TGGAACTGCCTTTGAAACTATCAG | Paracoccus sp. 4M6 |
| 4M6 R | CATGCTTGCCGACGTCTG | |
| TaqMan probes | ||
| AD3J6 | VIC-CACTGACGCTCATGTAC-MGB-NFQ | Pseudoalteromonas sp. 3J6 |
| ADD01 | 6-FAM-CTGACACTCAGATGCGA-MGB-NFQ | Vibrio sp. D01 |
| AD4J6 | 6-FAM-TTCGCGCCTCAGTGTCA-MGB-NFQ | Bacillus sp. 4J6 |
VIC and 6-FAM (6-carboxyfluorescein) are fluorescent dyes. MGB, minor-groove binder; NFQ, nonfluorescent quencher.
Liquid cocultures of bacteria and population analyses.
Pseudoalteromonas sp. 3J6(pCJS10) and Vibrio sp. D01 were each inoculated into VNSS medium at an OD600 of 0.25, and bacteria were grown for 24 h at 20°C with shaking. Dilutions were then plated onto VNSS agar without antibiotic (total cell count) and onto VNSS agar containing both streptomycin and chloramphenicol to select Pseudoalteromonas sp. 3J6(pCJS10). The numbers of total CFU and of Pseudoalteromonas sp. 3J6(pCJS10) CFU per ml allowed the calculation of the percentages of each of the two strains in a coculture. Experiments were performed three times.
Concentration of antibiofilm activity.
Two hundred milliliters of SN3J6 was passed through a Sep-Pak Plus C18 cartridge (Waters Corporation, Milford, MA). The cartridge was rinsed three times with water, and the antibiofilm molecules were eluted with 50 ml of 50% acetonitrile. The solvent was evaporated by using a rotary evaporator under a vacuum at 35°C. The residues were dissolved in 2 ml of phosphate-buffered saline (8.0 g liter−1 NaCl, 0.2 g liter−1 KCl, 1.44 g liter−1 Na2HPO4, 0.24 g liter−1 KH2PO4 [pH 7.4]) and sterilized by filtration through a membrane with 0.22-μm pores. A 10−3 dilution of this preparation prevented Paracoccus sp. 4M6 biofilm formation as efficiently as undiluted SN3J6, whereas a 10−1 dilution of SN3J6 displayed a significantly reduced antibiofilm activity. This showed that the antibiofilm activity was concentrated at least 100-fold after elution from the C18 cartridge. No antibiofilm activity was observed with a preparation resulting from the passage of VNSS culture medium on a C18 cartridge.
Bacteriocin-like activity tests.
We used the culture supernatant diffusion method (33), in which SN3J6 was loaded into wells of VNSS agar plates seeded with Vibrio sp. D01 or Paracoccus sp. 4M6. The plates were incubated at 20°C for 24 to 48 h to allow the diffusion of SN3J6 molecules from the wells into the agar medium and the growth of Vibrio sp. D01 or Paracoccus sp. 4M6. The presence of a bacteriocin-like molecule in SN3J6 would lead to clear halos of bacterial growth inhibition around the wells. Alternatively, disks soaked with SN3J6 were placed onto the seeded agar medium instead of using wells. This disk assay was also used to test the antimicrobial activity of antibiofilm molecules, which had been concentrated as described above.
RESULTS
Single-species biofilms of Pseudoalteromonas sp. 3J6 and three other marine bacteria.
We first introduced GFP-encoding plasmid pCJS10 (Table 1) into Pseudoalteromonas sp. 3J6 as described in Materials and Methods. This fluorescent tagging allowed the observation of Pseudoalteromonas sp. 3J6 biofilms by confocal laser scanning microscopy (CLSM) without further staining of the cells. Since bacteria in a biofilm are generally more antibiotic resistant than planktonic cells, Pseudoalteromonas sp. 3J6(pCJS10) biofilms were grown without antibiotic (liquid precultures were performed in the presence of chloramphenicol). This required that pCJS10 was stably maintained in the strain without selective pressure. Biofilms grown in a flow cell under dynamic conditions for 48 h were observed using CLSM by detecting the green fluorescence of GFP (Fig. 1A) and after staining bacteria with Syto 61 red and detecting red fluorescence. When the resulting images were overlaid, GFP-producing cells were yellow (overlay of green and red) or green, whereas GFP-free bacteria were red. Figure 1B shows very few red-stained patches of cells, indicating that the vast majority of bacteria were GFP producers and had thus retained pCJS10. This plasmid is therefore suitable for our study. Pseudoalteromonas sp. 3J6(pCJS10) biofilms were heterogeneous, with the presence of protruding mushroom-like microcolonies, which were up to 21 μm high, whereas the average thickness of the biofilms was 4.8 μm (Fig. 1A and B and Table 3).
FIG. 1.
CLSM images of single- and two-species biofilms. Biofilms were grown on glass surfaces at 20°C for 48 h under a flow of VNSS medium. Single-species biofilms of Pseudoalteromonas sp. 3J6(pCJS10) (A and B), Paracoccus sp. 4M6 (C), Vibrio sp. D01 (E), and Bacillus sp. 4J6 (G) are shown. The Pseudoalteromonas sp. 3J6(pCJS10) biofilm was visualized by detecting only the GFP fluorescence (A) and by staining the cells with Syto 61 red and overlaying the GFP and the Syto 61 red fluorescences (B). The other single-species biofilms were stained with Syto 61 red (C, E, and G). Two-species biofilms resulted from coinoculations of Pseudoalteromonas sp. 3J6(pCJS10) and Paracoccus sp. 4M6 (D), Vibrio sp. D01 (F), or Bacillus sp. 4J6 (H). Overlays of GFP fluorescence from Pseudoalteromonas sp. 3J6(pCJS10) and of Syto 61 red fluorescence from all cells of the biofilms are displayed (D, F, and H). Maximal projections in the xy plane (top of each panel) and three-dimensional (3D) shadow projections (bottom of each panel) are presented. Bars, 30 μm.
TABLE 3.
COMSTAT analyses of single- and two-species biofilms after 48 h of growth
| Biofilm | Max thickness (μm)a | Avg thickness (μm)a | Biovolume (μm3/μm2)a |
|---|---|---|---|
| Single species | |||
| 3J6(pCJS10) | 21 | 4.8 | 2.6 |
| 4M6 | 68 | 8.2 | 2.6 |
| D01 | 34 | 8.7 | 5.4 |
| 4J6 | 76 | 19.0 | 10.4 |
| Two species | |||
| 3J6(pCJS10) + 4M6 | 24 | 2.5 | 2.5 |
| 3J6(pCJS10) + D01 | 44 | 5.8 | 4.6 |
| 3J6(pCJS10) + 4J6 | 92 | 12.0 | 17.6 |
Averages of data from three independent experiments, with standard deviations lower than 10% of each value.
We failed to introduce pCJS10 and other plasmids encoding fluorescent proteins into our model strains Paracoccus sp. 4M6, Vibrio sp. D01, and Bacillus sp. 4J6. We nevertheless grew single-species biofilms of these strains under the same conditions as those described above and stained them with Syto 61 red prior to CLSM observation (Fig. 1C, E, and G). Each of these three strains led to thicker biofilms than those of Pseudoalteromonas sp. 3J6(pCJS10) (Table 3). Paracoccus sp. 4M6 biofilms were heterogeneous and contained cell aggregates but no mushroom-like structure (Fig. 1C). They were 1.7-fold thicker on average than Pseudoalteromonas sp. 3J6(pCJS10) biofilms, while their biovolumes were identical (Table 3), indicating that cell density is lower in Paracoccus sp. 4M6 biofilms. Although not flat, Vibrio sp. D01 biofilms were less heterogeneous: they were devoid of mushroom-like structure, and cell aggregates were less obvious (Fig. 1E). Their average thickness and biovolume were 1.8- and 2-fold higher, respectively, than those of Pseudoalteromonas sp. 3J6(pCJS10) biofilms (Table 3). The Bacillus sp. 4J6 biofilms were the thickest, which led to the highest biovolume, and they displayed a hairy surface, although they contained homogeneously distributed cells (Fig. 1G and Table 3). Under our conditions, each of the four strains was therefore able to develop a single-species biofilm with a specific structure.
Pseudoalteromonas sp. 3J6 inhibits the development of Paracoccus sp. 4M6 and Vibrio sp. D01 in two-species biofilms.
The GFP tagging of Pseudoalteromonas sp. 3J6 allowed the growing of two-species biofilms with any unlabeled strain and distinguishing between the two strains by staining biofilms with Syto 61 red and overlaying green and red fluorescences. The coinoculation of Pseudoalteromonas sp. 3J6(pCJS10) and Bacillus sp. 4J6 led to biofilms in which each strain was abundant: the biofilms consisted of Pseudoalteromonas sp. 3J6(pCJS10) aggregates (green-yellow) surrounded by Bacillus sp. 4J6 cells (red) (Fig. 1H). Furthermore, the biofilm thickness and biovolume were closer to those of Bacillus sp. 4J6 single-species biofilms than to those of Pseudoalteromonas sp. 3J6(pCJS10) biofilms (Table 3). In contrast, when Pseudoalteromonas sp. 3J6(pCJS10) was coinoculated with either Paracoccus sp. 4M6 or Vibrio sp. D01, the resulting biofilms contained only few red cells and were similar to single-species Pseudoalteromonas sp. 3J6(pCJS10) biofilms, in particular with the presence of mushroom-like structures (compare Fig. 1D and F to B). This suggested that Pseudoalteromonas sp. 3J6(pCJS10) was the predominant strain in these biofilms. This visual impression was confirmed by determining the biovolume of each strain within the two-species biofilms using COMSTAT software: Pseudoalteromonas sp. 3J6(pCJS10) was estimated to account for 76 and 84% of the total biovolumes in mixed biofilms including Vibrio sp. D01 and Paracoccus sp. 4M6, respectively (Table 4). On the other hand, Pseudoalteromonas sp. 3J6(pCJS10) accounted for only 35% of the total biovolume in Pseudoalteromonas sp. 3J6(pCJS10)-Bacillus sp. 4J6 biofilms (Table 4).
TABLE 4.
Quantification of each strain in two-species biofilms
| Strain | Proportion of each strain (%) |
|
|---|---|---|
| Biofilm biovolumea | qPCR analysesb | |
| 3J6(pCJS10)-4M6 biofilms | ||
| 3J6(pCJS10) | 84 | 89 |
| 4M6 | 16 | 11 |
| 3J6(pCJS10)-D01 biofilms | ||
| 3J6(pCJS10) | 76 | 95 |
| D01 | 24 | 5 |
| 3J6(pCJS10)-4J6 biofilms | ||
| 3J6(pCJS10) | 35 | 62 |
| 4J6 | 65 | 38 |
Biovolumes were obtained by COMSTAT analyses performed for three independent experiments, with standard deviations lower than 10% of each value.
For each biofilm analysis, total DNAs were extracted from two different biofilms, and the two samples were analyzed independently, with standard deviations lower than 10% of each value.
To ascertain the proportion of each strain in two-species biofilms, we set up qPCR assays using primers targeting the 16S rRNA genes sequences of the four strains (Table 2). To validate the assays, total DNAs independently extracted from pure liquid cultures of the four strains were mixed in known ratios (1:1 and 9:1), and values close to these ratios had to be obtained by qPCR analyses. When studying a mix of Pseudoalteromonas sp. 3J6(pCJS10) and Paracoccus sp. 4M6 DNAs, the corresponding primers were specific enough for each strain to be detected by using SYBR green dye. Analyses of mixes including Pseudoalteromonas sp. 3J6(pCJS10) DNA and either Vibrio sp. D01 or Bacillus sp. 4J6 DNAs required the use of TaqMan probes (Table 2) to more specifically detect the amplicons resulting from each strain. Two-species biofilms were then grown, total DNAs were extracted from each biofilm, and the proportion of each DNA was determined by qPCR. These assays strengthened the above-described conclusions, confirming (i) the codevelopment of Pseudoalteromonas sp. 3J6(pCJS10) and Bacillus sp. 4J6 in mixed biofilms, since Bacillus DNA represented almost 40% of total DNA, and (ii) that Pseudoalteromonas sp. 3J6(pCJS10) cells were largely prevailing (89 and 95% of total DNA) over Paracoccus sp. 4M6 and Vibrio sp. D01 cells in two-species biofilms (Table 4).
Pseudoalteromonas sp. 3J6(pCJS10) therefore inhibited the growth of Vibrio sp. D01 and Paracoccus sp. 4M6, but not that of Bacillus sp. 4J6, in two-species biofilms. This could not be correlated simply to the growth parameters of the strains: Pseudoalteromonas sp. 3J6(pCJS10) displayed a doubling time of 53 min in liquid VNSS culture at 20°C, whereas doubling times of Vibrio sp. D01, Paracoccus sp. 4M6, and Bacillus sp. 4J6 were 55, 160, and 80 min, respectively. Under our conditions, Pseudoalteromonas sp. 3J6(pCJS10) was therefore unlikely to outcompete Vibrio sp. D01 because of faster growth. This was confirmed by liquid cocultures of Pseudoalteromonas sp. 3J6(pCJS10) and Vibrio sp. D01: after 24 h of growth, Pseudoalteromonas sp. 3J6(pCJS10) accounted for about 50% of the total cultivable cell population. This furthermore indicated that the negative effect of Pseudoalteromonas sp. 3J6 on Vibrio sp. D01 growth occurred specifically in biofilms.
Pseudoalteromonas sp. 3J6 exoproducts inhibit the formation of Paracoccus sp. 4M6 and Vibrio sp. D01 biofilms.
We examined whether the supernatants of liquid Pseudoalteromonas sp. 3J6(pCJS10) cultures (SN3J6) affected Vibrio sp. D01 and Paracoccus sp. 4M6 biofilm formation. These two strains were independently resuspended either in filtered marine water (FMW) as positive controls or in SN3J6. The bacterial suspensions were introduced into flow cell chambers and incubated for 2 h at 20°C to allow bacterial attachment onto the glass surface (a control experiment in which FMW was replaced with fresh VNSS medium showed that the VNSS components of SN3J6 did not affect bacterial attachment). Flows of VNSS medium free of Pseudoalteromonas sp. 3J6 exoproducts were then applied for 48 h to allow biofilm formation. The incubation of Vibrio sp. D01 or Paracoccus sp. 4M6 cells with SN3J6 strongly impaired the ability of Paracoccus sp. 4M6 and Vibrio sp. D01 to develop single-species biofilms (compare Fig. 2C and D to A and B): the residual biovolumes were about 14 and 5%, respectively, of the SN3J6-free positive-control biovolumes (Table 5). In subsequent experiments, we attempted to identify at which level(s) SN3J6 is acting.
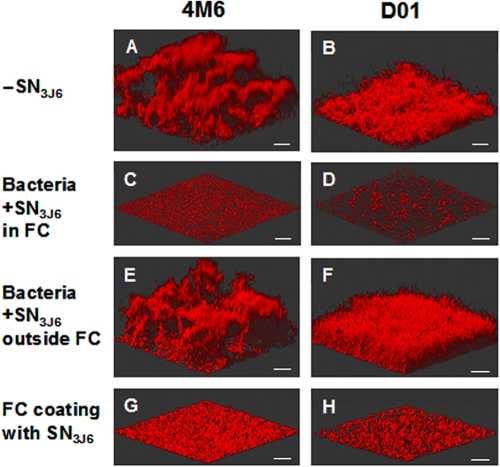
FIG. 2.
Activity of Pseudoalteromonas sp. 3J6 culture supernatant (SN3J6) on biofilm formation by Paracoccus sp. 4M6 and Vibrio sp. D01. (A to D) Biofilms of Paracoccus sp. 4M6 (left) and Vibrio sp. D01 (right) were grown in flow cells (FC) for 48 h after a 2-h adhesion step in flow cells without SN3J6 (A and B) or in the presence of SN3J6 (C and D). (E to H) Alternatively, bacteria were incubated with SN3J6 outside the flow cell and washed prior to inoculation (E and F), or the flow cell glass coverslip was coated with SN3J6 and washed before inoculation of SN3J6-free bacteria (G and H). Biofilms were stained with Syto 61 red. A 3D shadow projection is shown for each biofilm. Bars, 30 μm.
TABLE 5.
Characteristics of single-species biofilms obtained without SN3J6 or in the presence of SN3J6 during the adhesion step
| Group and straina | Max thickness (μm)b |
Biovolume (μm3/μm2)b |
Nonviable cells (%)b |
|||
|---|---|---|---|---|---|---|
| − SN3J6 | + SN3J6 | − SN3J6 | + SN3J6 | − SN3J6 | + SN3J6 | |
| 1 | ||||||
| Algibacter sp. 1M6 | 44.6 | 7.4 | 15.9 | 0.03 | 0.4 | 89 |
| 2 | ||||||
| Paracoccus sp. 4M6 | 95.0 | 13.2 | 6.1 | 0.88 | 1.2 | 100 |
| Vibrio sp. D01 | 45.2 | 16 | 14.2 | 0.68 | 13 | 78 |
| Colwellia sp. 4J3 | 28.7 | 11.3 | 5.9 | 0.27 | 25 | 97 |
| 3 | ||||||
| Sulfitobacter sp. 8J6 | 14.4 | 9.0 | 2.0 | 0.66 | 29 | 8.9 |
| Vibrio sp. D66 | 122 | 46.8 | 15.2 | 1.9 | 3.7 | 2.5 |
| 4 | ||||||
| Alteromonas sp. 1J3 | 14.5 | 16.9 | 3.2 | 2.9 | 8.5 | 80 |
| 5 | ||||||
| Pseudoalteromonas sp. 3J3 | 10.4 | 14.5 | 3.9 | 3.7 | 46 | 36 |
| Pseudoalteromonas sp. D41 | 11.8 | 10.2 | 2.1 | 1.9 | 0.6 | 12.2 |
Groups were defined according to the sensitivity of each strain to SN3J6 (see text for details).
COMSTAT analyses were performed for three independent experiments, and standard deviations were lower than 10% of each value.
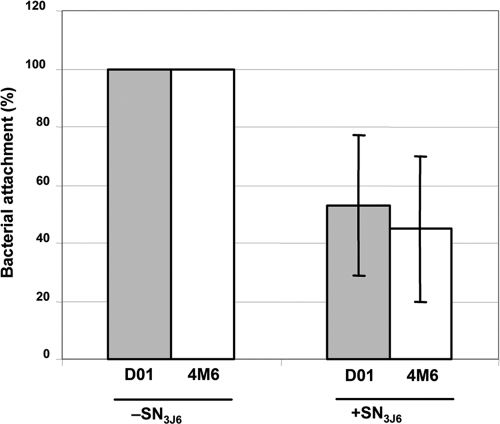
SN3J6 moderately affects bacterial attachment.
The glass surfaces covered by bacteria were determined after a 2-h step of adhesion in the presence or absence of SN3J6. The coverages of these surfaces were about 2-fold lower in the presence of SN3J6 (Fig. 3). However, this effect seems too mild to fully explain the subsequent inability of the attached bacteria to develop biofilms. We furthermore observed that the presence of SN3J6 during the attachment step did not favor the detachment of bacteria after the application of the medium flow (data not shown).
FIG. 3.
Effect of SN3J6 on bacterial adhesion at 20°C. Vibrio sp. D01 and Paracoccus sp. 4M6 were incubated for 2 h under static conditions in flow cell channels in the presence or absence of SN3J6. CLSM images of attached bacteria were recorded, and the glass surfaces covered by attached bacteria were calculated by using Image Tool software and normalized relative to the values obtained in the absence of SN3J6, which were set to 100%. Each experiment was performed twice, and the bacterial attachment was determined for eight different sites of each glass slide. The bars indicate the standard deviations, which were too small to be visible in the absence of SN3J6.
SN3J6 is devoid of bactericidal activity against free-living cells.
We examined whether SN3J6 contained a bacteriocin-like inhibitory substance by using two classical methods (diffusion into agar medium from wells or disks) as described in Materials and Methods. We failed to observe any clear halo of inhibition around the wells or the disks, indicating that SN3J6 was neither bactericidal nor bacteriostatic toward Vibrio sp. D01 and Paracoccus sp. 4M6. Consistently, the addition of 4.5 ml of SN3J6 to 0.5 ml of fresh liquid 10× VNSS medium before the inoculation of Vibrio sp. D01 or Paracoccus sp. 4M6 did not prevent the growth of these two strains. After 24 h of growth, the numbers of Vibrio sp. D01 and Paracoccus sp. 4M6 CFU were 88 and 94%, respectively, of those of control cultures in SN3J6-free VNSS. Furthermore, we used double staining with Syto 61 red (staining of all bacteria) and Sytox green (staining of only bacteria with damaged membranes, which are therefore considered nonviable bacteria) on bacteria attached to the glass surface after a 2-h adhesion step. This showed that the presence of SN3J6 during this step did not decrease the percentage of viable bacteria (about 90% and 100% of viable Vibrio sp. D01 and Paracoccus sp. 4M6, respectively, with or without SN3J6). This set of experiments indicated that the antibiofilm action of SN3J6 did not result from a bactericidal activity exerted during the 2-h contact between SN3J6 and the bacteria (adhesion step). To ascertain this conclusion, Vibrio sp. D01 or Paracoccus sp. 4M6 cells were incubated in SN3J6 for 2 h at 20°C and then washed and resuspended in FMW, and the cell suspensions were injected into flow cell chambers. As in previous experiments, a 2-h adhesion step was then performed, and biofilms were allowed to grow under a VNSS flow for 48 h. This led to full biofilm formation (Fig. 2E and F), showing that the incubation of bacteria with SN3J6 outside the flow cell chamber had no effect on the ability of the bacteria to subsequently form a biofilm. Finally, we used a C18 cartridge to concentrate the antibiofilm activity from SN3J6 at least 100-fold. This preparation also failed to display a bactericidal activity using the disk assay, invalidating the possibility that SN3J6 would contain a subinhibitory concentration of a bacteriocin-like molecule, which would nevertheless be responsible for the antibiofilm activity.
Glass-coating ability of antibiofilm components of SN3J6.
Since SN3J6 did not reduce cell viability during the adhesion step and only mildly affected cell attachment, it was likely to act during biofilm formation, i.e., after applying the flow of VNSS medium, although this flow is removing SN3J6 as well as unattached cells. This suggested that antibiofilm components of SN3J6 could coat the glass surface during the adhesion step. This hypothesis was tested by the incubation of SN3J6 for 2 h in flow cell chambers without bacteria before extensively rinsing the chambers with FMW and inoculating them with Vibrio sp. D01 or Paracoccus sp. 4M6 cells in the absence of SN3J6. This impaired the biofilm formation by the two strains almost as efficiently as the presence of SN3J6 with the bacteria during the adhesion step (compare Fig. 2G and H to C and D). The coating of the glass surface by antibiofilm components of SN3J6 explains how these components can act during biofilm growth, even though SN3J6 was removed from the flow cell chambers after the adhesion step by the medium flow.
SN3J6-dependent reduction of cell viability in biofilms.
As described above, various approaches led to the conclusion that SN3J6 was not bactericidal toward Vibrio sp. D01 and Paracoccus sp. 4M6 and that its antibiofilm action was not resulting from a bactericidal activity exerted during the adhesion step. We therefore expected that SN3J6 would not affect the viability of cells within the subsequently obtained biofilms. We nevertheless assessed the bacterial viability in 48-h biofilms by using Syto 61 red/Sytox green double staining. Whereas control biofilms of Paracoccus sp. 4M6 contained only 1.2% nonviable cells, we were surprised to observe that none of the cells were viable in biofilms obtained after incubation with SN3J6 (Table 5). Although not as pronounced, this effect was also observed with Vibrio sp. D01: control biofilms contained 13% nonviable cells, while this value reached 78% when cells had been incubated with SN3J6 (Table 5). SN3J6 therefore led to a loss of cell viability occurring in a biofilm-specific manner.
SN3J6 antibiofilm activity against other marine bacterial isolates.
We examined whether SN3J6 acts on a larger variety of marine bacteria. Five groups of bacteria could be distinguished (Table 5).
Group 1 contains only the most sensitive strain of the whole study, Algibacter sp. strain 1M6: SN3J6 abolished its ability to develop a biofilm covering the glass surface, and the biovolume of the dispersed cell clusters attached to the glass surface was 530-fold lower than the biovolume of control biofilms. Furthermore, SN3J6 treatment led to a dramatic drop of cell viability after 48 h of biofilm growth.
Group 2 includes strains strongly impaired by SN3J6 in their ability to form biofilms (biovolumes 6.9- to 22-fold lower) and in the viability of biofilm bacteria. This group contains Colwellia sp. strain 4J3 in addition to Paracoccus sp. 4M6 and Vibrio sp. D01.
Group 3 consists of Vibrio sp. D66 and Sulfitobacter sp. strain 8J6, the biofilm formation of which was strongly or mildly inhibited by SN3J6 (biovolumes 3- to 8-fold lower), while SN3J6 did not lead to higher percentages of nonviable cells in biofilms (the percentage of nonviable Sulfitobacter sp. 8J6 cells was even 3.2-fold lower in biofilms obtained after incubation with SN3J6).
Group 4 is defined by Alteromonas sp. strain 1J3, which was unaffected in its ability to form biofilms, although incubation with SN3J6 resulted in a 9.4-fold-higher percentage of nonviable cells within the biofilms.
Group 5 is composed of fully nonsensitive isolates, i.e., the two Pseudoalteromonas sp. strains 3J3 and D41: they were essentially unaffected by SN3J6 in their ability to develop biofilms, and SN3J6 either failed to increase the percentage of nonviable cells in biofilms (3J3) or increased it to only 12% (D41).
Altogether, most of the tested isolates were affected: SN3J6 impaired the biofilm formation of six strains (groups 1, 2, and 3) out of nine and led to an increase in the percentages of nonviable cells from the 0.4-to-29% range to 78 to 100% in biofilms of five strains (groups 1, 2, and 4). Groups 3 and 4 showed that the antibiofilm action of SN3J6 and the loss of cell viability in biofilms can occur independently from each other. Interestingly, the two nonsensitive isolates (group 5) belong to the same genus as strain 3J6, Pseudoalteromonas. Although the number of tested strains remains small, these data suggest that Pseudoalteromonas sp. 3J6 allows the growth of Pseudoalteromonas in marine biofilms while impairing the biofilm development of a variety of other marine bacteria. The sensitive strains indeed displayed various taxonomic relationships with the genus Pseudoalteromonas: they belong to the same class and order (Gammaproteobacteria and Alteromonadales in the cases of Alteromonas sp. 1J3 and Colwellia sp. 4J3), to a different order (Vibrionales for Vibrio sp. D01 and D66) of the same class, to a different class (Alphaproteobacteria for Paracoccus sp. 4M6 and Sulfitobacter sp. 8J6), or even to a different superphylum (Bacteroidetes/Chlorobi group for Algibacter 1M6).
SN3J6 antibiofilm activity against human pathogens.
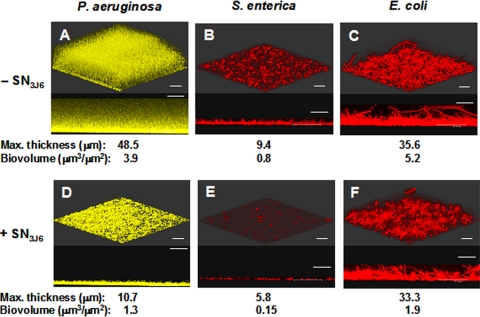
We furthermore examined the effect of SN3J6 on bacteria belonging to the human pathogen species Pseudomonas aeruginosa, Salmonella enterica, and Escherichia coli. These bacteria belong to the same class as Pseudoalteromonas (Gammaproteobacteria) but to different orders (Pseudomonadales for Pseudomonas and Enterobacteriales for Salmonella and Escherichia). We used strains tagged by genes encoding a yellow fluorescent protein (enhanced yellow fluorescent protein [eYFP]) or red fluorescent protein (HcRed or DsRed) in order to visualize biofilms without the addition of fluorescent dye. Consistently with its wide spectrum of action described above, SN3J6 impaired the biofilm formation of all three strains: the biovolumes of P. aeruginosa PAO1YFP, S. enterica MB1409HcRed, and E. coli CC118DsRed biofilms were 3-, 5.3-, and 2.7-fold lower, respectively, when the adhesion step was performed in the presence of SN3J6 (Fig. 4). CLSM images showed that these reductions in biovolume resulted from peculiar effects on P. aeruginosa and E. coli. P. aeruginosa PAO1YFP control biofilms (about 50 μm thick) were not homogeneous with respect to the cell density: it was high in the base of the biofilms, from which it decreased progressively in the upper part (Fig. 4A). This finding suggested that cells were embedded into increasing amounts of extracellular matrix from the base to the top of the biofilms. In contrast, the biofilm resulting from incubation with SN3J6 was thinner (about 5-fold) but homogeneous, with a high cell density, which led to a relatively high biovolume (Fig. 4D). P. aeruginosa PAO1YFP biofilm formation was therefore more impaired by SN3J6 than indicated by the sole biovolume comparison. On the other hand, SN3J6-resulting biofilms of E. coli CC118DsRed differed from control biofilms by their biovolumes, whereas the biofilm structures and maximal thicknesses were similar (Fig. 4C and F). The biovolume reduction in E. coli biofilms obtained after SN3J6 incubation therefore resulted from a lower cell density. In this study, this is the only example of a biovolume reduction that occurred without a lowering of the biofilm maximal thickness.
FIG. 4.
Effect of SN3J6 on biofilm formation by Pseudomonas aeruginosa, Salmonella enterica, and Escherichia coli. Biofilms of P. aeruginosa PAO1YFP (A and D), Salmonella enterica MB1409HcRed (B and E), and Escherichia coli CC118DsRed (C and F) were grown at 20°C for 48 h in LB broth after a 2-h adhesion step without SN3J6 (A, B, and C) or in the presence of SN3J6 (D, E, and F). The yellow (eYFP) or red (HcRed or DsRed) fluorescent protein produced by each strain was detected. A 3D shadow representation and a side-view projection are shown on the top and bottom, respectively, of each panel. For each experiment, maximal thicknesses and biovolumes were calculated from COMSTAT analyses of 12 to 18 image stacks obtained from at least two independent biofilms. Standard deviations were lower than 10% of the values. Bars, 30 μm.
DISCUSSION
We report here an antibiofilm activity exerted by Pseudoalteromonas sp. 3J6 exoproducts on a wide range of Gram-negative bacteria but not on the other tested strains of the same genus. This activity leads Pseudoalteromonas sp. 3J6 to predominate over other bacterial isolates such as Vibrio sp. D01 and Paracoccus sp. 4M6 in two-species biofilms. Marine Pseudoalteromonas bacteria are known as producers of a variety of biologically active extracellular compounds, including antibacterial agents that can lead to antifouling effects (2, 11, 13, 15). The best-known example is the antibacterial protein AlpP secreted by Pseudoalteromonas tunicata D2 (16). AlpP provides a competitive advantage to the latter for biofilm formation in the marine environment and for the colonization of the surface of the green alga Ulva australis (30, 31). Consistently, AlpP producer strains seem rather widespread in Northern Europe, since P. tunica D2 was isolated from the surface of the tunicate Ciona intestinalis collected from waters off the Swedish west coast (14), and the alpP gene was PCR amplified from marine samples collected on Ciona intestinalis and on the green algae Ulva lactuca and Ulvaria fusca in the waters around Aarhus, Denmark (35). AlpP furthermore mediates cell death in mature biofilms of its own producer strain, leading to the dispersal of surviving cells within the marine environment (23). Pseudoalteromonas sp. 3J6 is unlikely to produce AlpP since we failed to PCR amplify any alpP gene fragment from Pseudoalteromonas sp. 3J6 DNA with four different primer couples (data not shown). Furthermore, the antibiofilm molecule produced by Pseudoalteromonas sp. 3J6 presents the originality of differing from AlpP and other classical antibacterial compounds by its biofilm-specific activity: SN3J6 displayed no antibacterial activity against Paracoccus sp. 4M6 and Vibrio sp. D01 in liquid, on agar plates, or during the adhesion step, whereas SN3J6 impaired biofilm formation by these two strains and, more generally, by a variety of Gram-negative bacteria. Interestingly, this antibiofilm effect was accompanied in most cases by a loss of cell viability in biofilms. We propose the two following hypotheses to explain this observation. SN3J6 would display a direct bactericidal activity against biofilm-included cells but not against free-living cells of the same strains. This could occur if the target of the antibacterial compound is produced (or accessible) only in biofilm cells. The antibacterial agent is unlikely to be a lytic phage released by Pseudoalteromonas sp. 3J6 that targets a biofilm-specific receptor, since we did not observe any phage-like particle in SN3J6 by scanning electron microscopy (data not shown). In an alternative hypothesis, SN3J6 would act indirectly on biofilm cells, perhaps by somehow stimulating the regulated cell death that takes place during biofilm maturation and is thought to be caused by the self-destruction (suicide) of individual cells (1). Cell death contributes to normal biofilm development by leading to the release of genomic DNA, which can be a component of the biofilm extracellular matrix, and to the dispersal of surviving cells (1, 38). Although it is obvious that the loss of cell viability could contribute to the antibiofilm action of SN3J6, these two phenomena occurred independently from each other in three examples (strains of groups 3 and 4) (Table 5), which suggests that none of them is the consequence of the other one.
To our knowledge, very few antibiofilm molecules secreted by bacteria and devoid of antibacterial activity against free-living cells were previously reported. E. coli strains producing group II capsules release a soluble polysaccharide in their environment, preventing biofilm formation by a wide range of Gram-positive and -negative bacteria (37). This polysaccharide acts both on the initial adhesion by weakening cell surface contacts and on the subsequent biofilm formation by reducing cell-cell interactions. In another study, a lipopeptide biosurfactant from a Bacillus circulans strain of marine origin displayed antiadhesive properties against various bacteria, but its effect on biofilm formation was not investigated (4). This finding suggests that the reduced bacterial attachment due to SN3J6 could result from a polysaccharidic molecule or a biosurfactant. However, this adhesion impairment might not fully account for the subsequent biofilm formation defect and does not explain the decrease in cell viability within the biofilms. Quorum sensing (QS) contributes to the control of biofilm formation either negatively in the cases of Vibrio cholerae and Staphylococcus aureus or positively in the case of Pseudomonas aeruginosa (17). QS inhibitors are considered antipathogenic drugs, which can be used to reduce virulence and to increase biofilm susceptibility to antimicrobial compounds and human immunity cells (32), and species-specific antibiofilm molecules (9). QS signal molecules of the homoserine lactone type were previously shown to accumulate in some marine biofilms (6), and the production of a QS inhibitor(s) by Pseudoalteromonas sp. 3J6 is a possibility. It would not, however, fully explain the antibiofilm activity of SN3J6, since all SN3J6-sensitive strains do not share similar QS systems: P. aeruginosa produces several homoserine lactones, whereas E. coli does not use such signaling molecules. Alternatively, the antibiofilm effects of Pseudoalteromonas sp. 3J6 exoproducts could be due to a novel molecule or the complementary actions of several molecules. We are currently undertaking the purification and identification of the active molecule(s) that is required to understand its (their) mode(s) of action and to more precisely define which of the potential applications can be drawn. The latter could extend from the use of Pseudoalteromonas sp. 3J6 to prevent the biofilm formation of undesirable bacteria in aquaculture or fish farming to the use of its exoproducts in antibiofilm strategies. From this point of view, several features of Pseudoalteromonas sp. 3J6 exoproducts are particularly attractive: (i) the fact that, in addition to impairing biofilm growth, they lead to biofilms containing a majority of nonviable cells and (ii) their wide spectrum of action, which does not limit their use to the marine environment. Medical applications could be imagined, since this spectrum includes human pathogens such as P. aeruginosa, the biofilms of which are particularly difficult to reduce during chronic infection of the lungs of cystic fibrosis patients (29).
Acknowledgments
We are grateful to S. Kjelleberg (University of New South Wales, Australia), T. Tolker-Nielsen (Technical University of Denmark), and L. Herman (Institute for Agricultural and Fisheries Research, Melle, Belgium) for kindly providing plasmids and strains. We thank G. Legendre (LBCM, Université de Bretagne-Sud) for HcRed tagging of S. enterica MB1409.
This work was funded by the GIS Europôle Mer, the GDR 2614 Biosalissures Marines, the Ministère de l'Enseignement Supérieur et de la Recherche, France (doctoral fellowship to Alexandra Dheilly), the IFREMER and the Région Bretagne, France (doctoral fellowship to Géraldine L. Klein), and European FEDER.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Bayles, K. W. 2007. The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 5:721-726. [DOI] [PubMed] [Google Scholar]
- 2.Bowman, J. P. 2007. Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar. Drugs 5:220-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 4.Das, P., S. Mukherjee, and R. Sen. 2009. Antiadhesive action of a marine microbial surfactant. Colloids Surf. B Biointerfaces 71:183-186. [DOI] [PubMed] [Google Scholar]
- 5.Dobretsov, S., H.-U. Dahms, and P.-Y. Qian. 2006. Inhibition of biofouling by marine microorganisms and their metabolites. Biofouling 22:43-54. [DOI] [PubMed] [Google Scholar]
- 6.Dobretsov, S., M. Teplitski, and V. Paul. 2009. Quorum sensing in the marine environment and its relationship to biofouling. Biofouling 25:413-427. [DOI] [PubMed] [Google Scholar]
- 7.Donlan, R. M. 2009. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 17:66-72. [DOI] [PubMed] [Google Scholar]
- 8.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estrela, A. B., M. G. Heck, and W. R. Abraham. 2009. Novel approaches to control biofilm infections. Curr. Med. Chem. 16:1512-1530. [DOI] [PubMed] [Google Scholar]
- 10.Grasland, B., R. Briandet, E. Quemener, T. Meylheuc, K. Vallée-Réhel, and D. Haras. 2003. Bacterial biofilm in seawater: cell surface properties of early-attached marine bacteria. Biofouling 19:307-313. [DOI] [PubMed] [Google Scholar]
- 11.Hayashida-Soiza, G., A. Uchida, N. Mori, Y. Kuwahara, and Y. Ishida. 2008. Purification and characterization of antibacterial substances produced by a marine bacterium Pseudoalteromonas haloplanktis strain. J. Appl. Microbiol. 105:1672-1677. [DOI] [PubMed] [Google Scholar]
- 12.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 13.Holmström, C., and S. Kjelleberg. 1999. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol. Ecol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 14.Holmström, C., D. Rittschof, and S. Kjelleberg. 1992. Inhibition of settlement by larvae of Balanus amphitrite and Ciona intestinalis by a surface-colonizing marine bacterium. Appl. Environ. Microbiol. 58:2111-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmström, C., S. Egan, A. Franks, S. McCloy, and S. Kjelleberg. 2002. Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiol. Ecol. 41:47-58. [DOI] [PubMed] [Google Scholar]
- 16.James, S. G., C. Holmström, and S. Kjelleberg. 1996. Purification and characterization of a novel antibacterial protein from the marine bacterium D2. Appl. Environ. Microbiol. 62:2783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karatan, E., and P. Watnick. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73:310-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jørgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 19.Lambertsen, L., C. Sternberg, and S. Molin. 2004. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6:726-732. [DOI] [PubMed] [Google Scholar]
- 20.Leroy, C., C. Delbarre, F. Ghillebaert, C. Compere, and D. Combes. 2008. Influence of subtilisin on the adhesion of a marine bacterium which produces mainly proteins as extracellular polymers. J. Appl. Microbiol. 105:791-799. [DOI] [PubMed] [Google Scholar]
- 21.Leroy, C., C. Delbarre, F. Ghillebaert, C. Compere, and D. Combes. 2008. Effects of commercial enzymes on the adhesion of a marine biofilm-forming bacterium. Biofouling 24:11-22. [DOI] [PubMed] [Google Scholar]
- 22.Mackay, I. M., S. A. Bustin, J. M. Andrade, M. Kubista, and T. P. Sloots. 2007. Quantification of microorganisms: not human, not simple, not quick, p. 133-182. In I. M. Mackay (ed.), Real-time PCR in microbiology: from diagnosis to characterization. Caister Academic Press, Norfolk, United Kingdom.
- 23.Mai-Prochnow, A., F. Evans, D. Dalisay-Saludes, S. Stelzer, S. Egan, S. James, J. S. Webb, and S. Kjelleberg. 2004. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 70:3232-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mårdén, P., A. Tunlid, K. Malmcrona-Friberg, G. Odham, and S. Kjelleberg. 1985. Physiological and morphological changes during short term starvation of marine bacterial isolates. Arch. Microbiol. 142:326-332. [Google Scholar]
- 25.Messens, W., K. Grijspeerdt, and L. Herman. 2006. Eggshell penetration of hen's eggs by Salmonella enterica serovar Enteritidis upon various storage conditions. Br. Poult. Sci. 47:554-560. [DOI] [PubMed] [Google Scholar]
- 26.Monds, R. D., and G. A. O'Toole. 2009. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17:73-87. [DOI] [PubMed] [Google Scholar]
- 27.Pradier, C. M., C. Rubio, C. Poleunis, P. Bertrand, P. Marcus, and C. Compère. 2005. Surface characterization of three marine bacterial strains by Fourier transform IR, X-ray photoelectron spectroscopy, and time-of-flight secondary-ion mass spectrometry, correlation with adhesion on stainless steel surfaces. J. Phys. Chem. B 109:9540-9549. [DOI] [PubMed] [Google Scholar]
- 28.Qin, G., L. Zhu, X. Chen, P. G. Wand, and Y. Zhang. 2007. Structural characterization and ecological roles of a novel exopolysaccharide from the deep-sea psychrotolerant bacterium Pseudoalteromonas sp. SM9913. Microbiology 153:1566-1572. [DOI] [PubMed] [Google Scholar]
- 29.Ramsey, D. M., and D. J. Wozniak. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospect for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56:309-322. [DOI] [PubMed] [Google Scholar]
- 30.Rao, D., J. S. Webb, and S. Kjelleberg. 2005. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudomonas tunicata. Appl. Environ. Microbiol. 71:1729-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao, D., J. S. Webb, and S. Kjelleberg. 2006. Microbial colonization and competition on the marine alga Ulva australis. Appl. Environ. Microbiol. 72:5547-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen, T. B., and M. Givskov. 2006. Quorum-sensing inhibitors as anti-pathogenic drugs. Int. J. Med. Microbiol. 296:149-161. [DOI] [PubMed] [Google Scholar]
- 33.Rincé, A., A. Dufour, P. Uguen, J.-P. Le Pennec, and D. Haras. 1997. Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl. Environ. Microbiol. 63:4252-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Skovhus, T. L., C. Holmström, S. Kjelleberg, and I. Dahllöf. 2007. Molecular investigation of the distribution, abundance and diversity of the genus Pseudoalteromonas in marine samples. FEMS Microbiol. Ecol. 61:348-361. [DOI] [PubMed] [Google Scholar]
- 36.Sternberg, C., B. B. Christensen, T. Johansen, A. Toftgaard Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valle, J., S. Da Re, N. Henry, T. Fontaine, D. Balestrino, P. Latour-Lambert, and J.-M. Ghigo. 2006. Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc. Nat. Acad. Sci. U. S. A. 103:12558-12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]