Abstract
The impact of nonstarch polysaccharides (NSP) differing in their functional properties on intestinal bacterial community composition, prevalence of butyrate production pathway genes, and occurrence of Escherichia coli virulence factors was studied for eight ileum-cannulated growing pigs by use of terminal restriction fragment length polymorphism (TRFLP) and quantitative PCR. A cornstarch- and casein-based diet was supplemented with low-viscosity, low-fermentability cellulose (CEL), with high-viscosity, low-fermentability carboxymethylcellulose (CMC), with low-viscosity, high-fermentability oat β-glucan (LG), and with high-viscosity, high-fermentability oat β-glucan (HG). Only minor effects of NSP fractions on the ileal bacterial community were observed, but NSP clearly changed the digestion in the small intestine. Compared to what was observed for CMC, more fermentable substrate was transferred into the large intestine with CEL, LG, and HG, resulting in higher levels of postileal dry-matter disappearance. Linear discriminant analysis of NSP and TRFLP profiles and 16S rRNA gene copy numbers for major bacterial groups revealed that CMC resulted in a distinctive bacterial community in comparison to the other NSP, which was characterized by higher gene copy numbers for total bacteria, Bacteroides-Prevotella-Porphyromonas, Clostridium cluster XIVa, and Enterobacteriaceae and increased prevalences of E. coli virulence factors in feces. The numbers of butyryl-coenzyme A (CoA) CoA transferase gene copies were higher than those of butyrate kinase gene copies in feces, and these quantities were affected by NSP. The present results suggest that the NSP fractions clearly and distinctly affected the taxonomic composition and metabolic features of the fecal microbiota. However, the effects were more linked to the individual NSP and to their effect on nutrient flow into the large intestine than to their shared functional properties.
The porcine intestinal microbiota change in response to dietary carbohydrate composition due to specific substrate preferences of bacteria (6). Therefore, inclusion of specific nonstarch polysaccharides (NSP) in the diet of pigs allows manipulation of the composition of the intestinal microbiota. The NSP can also reduce digestibility of nutrients in the small intestine (8). The resulting changes in nutrient flow alter the availability of fermentable substrate in the different sections of the gut and thus may modify the bacterial community structure. Differences in the fermentability levels of individual NSP may not only affect the kinetics of their degradation by intestinal bacteria but may also change the composition of the fermentation end products (49). Particularly, butyrate is an important metabolite because of its potential to affect gene expression and to improve cellular development in enterocytes (38). The ability of gut microbiota to produce butyrate can vary considerably in response to environmental factors, such as diet composition (3). However, the number of butyrate-producing bacteria in complex fecal samples has been difficult to estimate by targeting the 16S rRNA gene, because these bacteria do not form a homogeneous phylogenetic group, and both butyrate producers and non-butyrate producers are found within the same phylogenetic clusters belonging to Clostridium clusters I, III, IV, XI, XIVa, XV, and XVI (27). Two alternative pathways for butyrate formation in bacteria harboring the rumen and human colon have been described (7, 26). The majority of human colonic butyrate producers use butyryl-coenzyme A (CoA) CoA transferase, whereas soil bacteria mostly utilize the butyrate kinase for the last step of butyrate formation (26, 27). However, information about the butyrate pathways used by intestinal bacteria in pigs is not available.
In addition to the effects of the functional properties of NSP on intestinal physiology and fermentation processes, selection of specific NSP fractions may also prevent or stimulate overgrowth of pathogenic bacteria. For instance, dietary inclusion of highly viscous carboxymethylcellulose (CMC) has been shown to increase fecal shedding of enterotoxigenic Escherichia coli in weaned pigs (15). There is a need to identify those dietary NSP fractions that may either increase or reduce the numbers of potential pathogenic bacteria to formulate diets exerting beneficial effects on gut health, which is particularly important in antibiotic-free feeding regimens.
Most studies pertaining on the effect of diet composition on the bacterial community in pigs have employed natural NSP sources and cereal-based diets, thereby resulting in a mixture of different soluble and insoluble NSP showing considerable interactions and modification of intestinal bacterial ecophysiology (6, 36, 37). Purified NSP fractions are increasingly available from the bioprocessing industry for use in food preparation and potentially in animal feeds, where economics and possible health benefits warrant this use. However, less is known about the fermentative properties of purified NSP fractions than about those of NSP in the grain matrix (37), which may also differ according to their origins.
The aim of the present study was to examine the effects of four purified NSP fractions differing in their functional properties, i.e., viscosity and fermentability, on the ileal and fecal bacterial community, butyrate production pathway genes, and the occurrence of virulence factor genes of swine-pathogenic E. coli, including enterotoxigenic and enteroaggregative E. coli (11, 13).
(This study was presented in part at the 11th Digestive Physiology in Pigs Symposium, Reus, Spain, 19 to 22 May 2009.)
MATERIALS AND METHODS
Animals and diets.
A total of 8 crossbred Duroc-Landrace pigs (average weight, 22 ± 1.4 kg) from the herd of the Swine Research and Technology Centre, Edmonton, AB, Canada, were surgically fitted with a simple T-cannula at the distal ileum (23). The animal protocol was approved by the University of Alberta Animal Care and Use Committee for Livestock and followed the principles established by the Canadian Council on Animal Care (5). The pigs were assigned to one of four diets in a double 4-by-4 Latin square, resulting in 8 observations per diet. A semipurified diet based on cornstarch and casein was formulated to meet or to exceed the nutrient requirements for growing pigs (see Table S1 in the supplemental material) (34). The basal diet was supplemented with 5% active NSP ingredient of four purified NSP fractions: (i) low-fermentability, low-viscosity cellulose (CEL) (TIC pretested Ticacel MCC FG-100; TIC GUMS, White Marsh, MD), (ii) low-fermentability, high-viscosity carboxymethylcellulose (CMC) (TIC pretested Ticalose CMC 6000 F; TIC GUMS), (iii) high-fermentability, low-viscosity oat β-glucan (LG) (OatVantage; GTC Nutrition, Missoula, MT), and (iv) high-fermentability, high-viscosity oat β-glucan (HG) (Viscofiber; Cevena Bioproducts, Edmonton, AB, Canada). To reach 5% of the active NSP in the diet, the inclusion levels of the NSP fractions were 5.20, 6.25, 8.95, and 9.25% for CEL, CMC, LG, and HG, respectively. The NSP fractions were selected based on their in vitro viscosity levels, which were 0.3, 285, 20, and 210 mPa·s for CEL, CMC, LG, and HG, respectively, as determined with 0.5% NSP solution using a rheometer (UDS 200; Paar Physica, Glenn, VA) at a shear rate of 12.9/s and 20°C. The viscosity of β-glucans is linked to their molecular weight and has been reported to increase about 10-fold with a doubling of molecular weight (9). Titanium dioxide was added to the diet as a digestibility marker. The pigs were allowed to consume the experimental diets at a rate of approximately 3% of their maintenance requirement for energy (3 × 110 kcal digestible energy [DE]/kg metabolic body weight [BW0.75]) (34). They were fed two equal meals in mash form twice daily (at 8 a.m. and 4 p.m.).
Collection of intestinal samples.
Each experimental period comprised 17 days; an adaptation period of 10 days was followed by collection of feces over 3 days and collection of ileal effluent for 8 hours a day, from 8 a.m. to 4 p.m., over 4 days. Feces were collected using plastic bags attached to the skin around the anus (45). The bags were changed each time feces were voided, and after subsamples of fresh feces were taken for analysis of short-chain fatty acids (SCFA), the feces were stored at −20°C until being freeze-dried. For bacterial DNA extraction, subsamples of freshly voided feces were taken and immediately stored at −20°C. The ileal collection procedure was adapted from the method of Li et al. (23), using plastic tubings attached to the barrel of the cannula by elastic bands. Twice during collection of ileal effluent (11 a.m. and 2 p.m.), subsamples of ileal effluent (approximately 50 ml) were collected for bacterial DNA analysis and immediately stored at −20°C. The ileal effluent was pooled within each animal and stored at −20°C. A subsample of fresh ileal effluent was stored separately for SCFA analysis. The remaining ileal and fecal samples were freeze-dried prior to analyses of dry matter and protein.
TRFLP analysis.
Total genomic DNA was extracted from ileal effluent and feces using a FastDNA kit (Qiagen, Mississauga, Ontario, Canada) according to the manufacturer's instructions. Partial fragments of bacterial 16S rRNA genes were amplified by PCR using universal forward primer S-D-Bact-0008-a-S-20 (AGAGTTTGATCMTGGCTCAG), labeled with 6-carboxyflourescein (FAM), and reverse primer S-D-Bact-0926-a-S-20 (CCGTCAATTCATTTGAGTTT) (37). The purified PCR product (200 ng) was digested at 37°C overnight using 15 U of MspI (Fermentas, Burlington, CA) in 2 μl reaction buffer and UV-sterilized Millipore water, made up to 20 μl. Two microliters of the digestion solution was subsequently mixed with 9 μl of formamide and 0.5 μl of an internal size standard (ABI GeneScan 600 LIZ size standard) and denatured at 95°C for 5 min, followed by cooling on ice for 2 min. Fragment sizes were analyzed using an ABI 3130xl genetic analyzer in gene scan mode and GeneMapper version 3.7 software (Applied Biosystems, Ontario, Canada). Fragments that were different at fewer than 3 bp were considered to be identical in accordance with binning criteria.
Genomic DNA extraction for qPCR.
Nucleic acids were extracted from ileal effluents and feces of pigs by use of phenol-chloroform essentially as described by Knarreborg et al. (17). For DNA extraction, 200 mg of sample was weighed into a sterile tube containing 300 to 400 mg of sterile zirconium beads (diameter, 0.1 mm) and suspended in 1 ml of TN150 buffer (10 mM Tris-HCl, 150 mM NaCl [pH 8.0]). The suspension was vortexed and centrifuged at 14,600 × g for 5 min. The pellet was washed twice with 1 ml of TN150 buffer and was resuspended in 1 ml of TN150 buffer. The cells were lysed by physical disruption in a mini-bead beater (Biospec Products, Bartlesville, OK) at 5,000 rpm for 3 min and placed on ice to cool for 5 min. Subsequently, the sample was centrifuged at 14,600 × g for 5 min. A total of 900 μl of the supernatant was extracted twice with 1 ml TE (10 mM Tris, 1 mM EDTA [pH 8.5])-saturated phenol, followed by extraction with an equal volume of chloroform-isoamyl alcohol (24:1). The nucleic acids were precipitated with 2 volumes of cold ethanol (−20°C) and 0.1 volume of 5 M potassium acetate and stored overnight at −20°C. The DNA was collected by centrifugation at 14,600 × g for 20 min at 4°C, dried at room temperature for 1 h, and dissolved in 50 μl TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 7.5]). Prior to quantitative PCR (qPCR), DNA was diluted 5 times with sterilized Millipore water.
Quantitative PCR.
Quantitative PCR was performed on a model 7500 Fast real-time PCR system (Applied Biosystems, Foster City, CA) using the detection software (version 2.01; Applied Biosystems). Each reaction was run in duplicate in a volume of 25 μl in optical reaction plates sealed with optical adhesive film (Applied Biosystems). The reaction mixture consisted of 12.5 μl Fast SYBR green master mix (Applied Biosystems), 1 μl (10 μM) of primers (Table 1), and 1 μl of template DNA of ileal or fecal samples. To account for the degeneracy of the butyryl-CoA CoA transferase and butyrate kinase primers, higher primer concentrations were used in the reaction mixture (20 μM each primer). For amplification of the butyrate kinase gene and virulence factor genes, 12.5 μl QuantiFast SYBR green master mix (Qiagen, Mississauga, ON, Canada) was used. Amplification involved 1 cycle at 95°C for 5 min for initial denaturation, followed by 40 cycles of denaturation at 95°C for 15 s, primer annealing at the optimal temperatures (Table 1) for 30 s, extension at 72°C for 30 s, 1 cycle of 95°C for 1 min, 1 cycle at 55°C for 1 min, and a stepwise increase of the temperature from 55 to 95°C (at 10 s per 0.5°C) to obtain melt curve data. Data were collected at the extension step. Melting curves were checked after amplification in order to ensure correct amplification results. Standard curves were generated using serial dilutions of the purified and quantified PCR products generated by standard PCR using the primers shown in Table 1 and genomic DNA from pig intestinal contents (20). The detection limits were 102, 104, and 103 copies/g wet digesta for the group-specific primers, the butyrate enzyme gene primers, and the E. coli virulence factor primers, respectively.
TABLE 1.
Oligonucleotide primers used to profile intestinal samples
| Targeted bacterial group (primer size [bp]) | Orientationa | Primer sequence (5′-3′) | Annealing temp (°C) | Reference |
|---|---|---|---|---|
| Domain bacteria (200) | F | CGGYCCAGACTCCTACGGG | 60 | 19 |
| R | TTACCGCGGCTGCTGGCAC | |||
| Lactobacillus spp. (341) | F | AGCAGTAGGGAATCTTCCA | 62 | 46 |
| R | CACCGCTACACATGGAG | 14 | ||
| Enterococcus spp. (144) | F | CCCTTATTGTTAGTTGCCATCATT | 60 | 39 |
| R | ACTCGTTGTACTTCCCATTGT | |||
| Bifidobacterium spp. (243) | F | TCGCGTCYGGTGTGAAAG | 60 | 39 |
| R | CCACATCCAGCRTCCAC | |||
| Streptococcus spp. (485) | F | AGAGTTTGATCCTGGCTCAG | 60 | 33 |
| R | GTTAGCCGTCCCTTTCTGG | 10 | ||
| Clostridium cluster XIVa (438-441) | F | AAATGACGGTACCTGACTAA | 60 | 28 |
| R | CTTTGAGTTTCATTCTTGCGAA | |||
| Clostridium cluster IV (130) | F | GCACAAGCAGTGGAGT | 60 | 29 |
| R | CTTCCTCCGTTTTGTCAA | |||
| Clostridium cluster I (120) | F | ATGCAAGTCGAGCGAKG | 60 | 39 |
| R | TATGCGGTATTAATCTYCCTTT | |||
| Bacteroides-Prevotella-Porphyromonas (140) | F | GGTGTCGGCTTAAGTGCCAT | 60 | 39 |
| R | CGGAYGTAAGGGCCGTGC | |||
| Enterobacteriaceae family (195) | F | CATTGACGTTACCCGCAGAAGAAGC | 63 | 4 |
| R | CTCTACGAGACTCAAGCTTGC | |||
| Butyryl-CoA CoA transferase (530) | F | GCIGAICATTTCACITGGAAYWSITGGCAYATG | 53 | 24 |
| R | CCTGCCTTTGCAATRTCIACRAANGC | |||
| Butyrate kinase (301) | F | GTATAGATTACTIRYIATHAAYCCNGG | 53 | 26 |
| R | CAAGCTCRTCIACIACIACNGGRTCNAC | |||
| STa (193) | F | ATGAAAAAGCTAATGTTGGC | 56 | 13 |
| R | TACAACAAAGTTCACAGCAG | |||
| STb (204) | F | AATATCGCATTTCTTCTTGC | 56 | 13 |
| R | GCATCCTTTTGCTGCAAC | |||
| LT (291) | F | CTATTACAGAACTATGTTCGG | 56 | 13 |
| R | TACTGATTGCCGCAATTG | |||
| EAST1 (109) | F | TGCCATCAACACAGTATATCC | 56 | 13 |
| R | GCGAGTGACGGCTTTGT |
F, forward; R, reverse.
Analytical methods.
Samples of diets, freeze-dried ileal effluent, and feces were finely ground to pass through a 0.5-mm mesh screen (Lab Retsch Mill, Haan, Germany). Dry matter, crude protein, and titanium dioxide were analyzed according to the method of the AOAC (1). Feces were analyzed for SCFA by gas chromatography as described by Htoo et al. (16).
Data presentation.
Results for terminal restriction fragment length polymorphism (TRFLP) analysis and measurement of gene copy numbers for bacterial groups, butyrate enzymes, E. coli virulence factors, and fecal SCFA are expressed on the basis of wet weight of ileal effluent and feces rather than dry weight to illustrate the actual in situ situation in the gastrointestinal tract. Ileal protein flow was expressed on a dry-matter basis.
Calculations.
Ileal flow of dry matter and crude protein represents the amounts of dry matter and protein present in ileal effluent and was calculated according to the equation DO = AI × (ID/II), where DO is the total output of a nutrient in ileal effluent (g/kg of dry-matter intake), AI is the concentration of a nutrient in ileal effluent (g/kg of dry-matter intake), ID is the marker concentration in the assay diet (g/kg of dry matter), and II is the marker concentration in ileal effluent (g/kg of dry matter). Dry-matter disappearance in the large intestine was calculated as the difference between fecal and ileal dry-matter content levels.
Statistical analysis.
Data were analyzed according to a double 4-by-4 Latin square design using the mixed procedure (PROC MIXED) of the Statistical Analysis System (SAS Institute, Inc., Cary, NC). The fixed effects included animal and treatment effects. Instances of period and animal within a square were considered random effects, assuming a compound symmetry variance-covariance structure. To detect any influential observation on the model, a test was performed using Cook's distance (Cook's D) as a criterion. Any observation with a Cook D greater than 0.5 was considered influential and hence excluded from further analysis. Degrees of freedom were approximated using the Kenward-Rogers method. P values of ≤0.05 were defined as significant, whereas P values that were both >0.05 and ≤0.10 were considered representative of a trend.
The TRFLP results were analyzed using Statistica software (version 6.0; Statsoft, Tulsa, OK). The profiles were normalized, and only fragments with a relative peak area ratio (Pi) of ≥1% were considered for further analyses. The total number of distinct fragments (richness [S]) was counted, and Shannon's index [H′ = −∑i = 1S(Pi)(log Pi)] and Simpson's index [1 − D = ∑i = 1S (Pi2)] were calculated as ecological measures of the relative distribution of bacterial groups in the community. The Shannon and Simpson indices take into account the number of species and the evenness (Shannon's index) or relative distribution (Simpson's index) of the species as represented by terminal restriction fragments (TRFs). The indices are increased either by having additional unique TRFs or by having a greater evenness of TRFs. The mean values for these parameters were compared by analysis of variance (ANOVA), followed by Tukey's honestly significant difference (HSD) test. Individual TRFs were assigned to microbial species by use of the MICA II online analysis tool (http://mica.ibest.uidaho.edu/trflp.php), using the above-mentioned primer set and MspI for an in silico virtual digest against the Ribosomal Database Project (RDP) database.
The discrimination model was developed using linear discriminant analysis in the JMP software program (version 8.0.1; SAS Institute Inc., Cary, NC) to examine potential relationships between NSP fractions and TRFs and NSP fractions and gene copies for bacterial groups. Principal component analysis (PCA) was performed to examine any potential grouping of bacterial group gene copies, butyrate production pathway gene copies, E. coli virulence factors, ileal flow of dry matter, and postileal dry-matter disappearance according to the different diets used in this study by means of JMP software. The loading plot shows the variables responsible for the variation within the data set, and the correlations among individual variables of the first two eigenvalues (i.e., principal component 1 [PC 1] and PC 2). This gives a graphical representation of the extent to which each factor accounts for the variance in the data and shows the relationship between the different variables.
RESULTS
Ileal flow of dry matter and protein, dry-matter disappearances in the large intestine, and concentrations of SCFA in feces.
The dry-matter content levels of ileal effluent and feces were lower (P < 0.01) for CMC than for LG and HG (Table 2). The levels of ileal flow of dry matter and protein were higher (P < 0.01) for CEL, LG, and HG than for CMC. As a result, the levels of postileal dry-matter disappearance were greater (P = 0.01) for CEL, LG, and HG than for CMC. Fecal concentrations of total SCFA, including acetate, propionate, and butyrate, were lower (P < 0.05) in pigs fed the CMC diet than in those fed the CEL, LG, and HG diets. Inclusion of LG and HG additionally increased concentrations of isobutyrate and isovalerate in feces (P < 0.01).
TABLE 2.
Characteristics of ileal effluent, feces, and fecal SCFA of pigs fed diets supplemented with viscous and fermentable nonstarch polysaccharide fractionsa
| Characteristic | Value for indicated supplement |
P | ||||
|---|---|---|---|---|---|---|
| Low fermentability |
High fermentability |
Pooled (SEM) | ||||
| Low-viscosity CEL | High-viscosity CMC | Low-viscosity LG | High-viscosity HG | |||
| Dry-matter content (g/kg [wet wt]) | ||||||
| Ileal effluent | 145† | 55§ | 86‡ | 88‡ | 5.5 | 0.001 |
| Feces | 573† | 268‡ | 571† | 554† | 30.0 | <0.001 |
| Ileal flow rate (g/kg dry-matter intake) | ||||||
| Dry matter | 308† | 165§ | 242‡ | 277†‡ | 17.4 | <0.001 |
| Crude protein | 42†‡ | 26§ | 39‡ | 47† | 2.0 | <0.001 |
| Rate of postileal dry-matter disappearance (g/kg dry matter intake) | 129† | 23‡ | 102† | 126† | 20.3 | 0.010 |
| SCFA concn in feces (μmol/g [wet wt]) | ||||||
| Total | 61† | 28‡ | 69† | 64† | 6.9 | 0.005 |
| Acetate | 43† | 19‡ | 42† | 41† | 4.9 | 0.015 |
| Propionate | 9† | 5‡ | 10† | 9† | 1.2 | 0.038 |
| Butyrate | 5† | 1‡ | 7† | 5† | 0.9 | 0.004 |
| Isobutyrate | 1.4‡ | 0.7§ | 2.8† | 2.3† | 0.22 | <0.001 |
| Valerate | 1.1‡ | 0.5§ | 2.1† | 1.5‡§ | 0.19 | <0.001 |
| Isovalerate | 2.3‡ | 1.1‡ | 4.6† | 3.6† | 0.47 | <0.001 |
| Caproate | 0.2†‡ | 0.2‡ | 0.4† | 0.3† | 0.06 | 0.159 |
Data are presented as least-square means (n = 8). Values within a row not having the same symbol are significantly different (P < 0.05). CEL, cellulose; CMC, carboxymethylcellulose.
TRFLP analysis.
The TRFLP analysis of ileal samples showed patterns dominated by relatively few major TRFs, whereas a total of 75 different TRFs were observed in feces (see Tables S2 and S3 in the supplemental material). Measures of species richness (4.1 ± 1.8 versus 14.5 ± 4.3), and Shannon's (0.34 ± 0.21 versus 0.86 ± 0.22) and Simpson's (0.40 ± 0.24 versus 0.76 ± 0.13) diversity were higher in feces than in ileum (Table 3). Among diets, diversity indices were not significantly different. However, a trend (P < 0.10) toward higher levels of diversity for CMC than for the other treatments was observed, whereas CEL showed relatively low diversity indices (Table 3), and similar trends for ecological measures were observed in ileal effluents and fecal samples.
TABLE 3.
Species richness levels and Shannon and Simpson indices of diversity as calculated from normalized TRFLP profiles for ileal effluents and feces of pigs fed diets supplemented with viscous and fermentable nonstarch polysaccharide fractionsa
| Characteristic | Value for indicated supplement |
P | ||||
|---|---|---|---|---|---|---|
| Low fermentability |
High fermentability |
Pooled (SEM) | ||||
| Low-viscosity CEL | High-viscosity CMC | Low-viscosity LG | High-viscosity HG | |||
| Ileal effluent | ||||||
| Species richness | 3.50 | 4.25 | 3.50 | 5.00 | 1.78 | 0.288 |
| Shannon index | 0.23 | 0.42 | 0.29 | 0.43 | 0.20 | 0.160 |
| Simpson index | 0.26 | 0.50 | 0.35 | 0.48 | 0.23 | 0.156 |
| Feces | ||||||
| Species richness | 11.60 | 16.25 | 13.67 | 15.14 | 4.19 | 0.277 |
| Shannon index | 0.68 | 0.98 | 0.80 | 0.90 | 0.21 | 0.086 |
| Simpson index | 0.65 | 0.83 | 0.72 | 0.79 | 0.12 | 0.076 |
Data are presented as least-square means (n = 8). CEL, cellulose; CMC, carboxymethylcellulose.
Terminal restriction fragments identified in ileal effluent could be assigned to Streptococcus/Lactococcus spp., to Lactobacillus spp., to Clostridium clusters I, XIVa, and XVIII, and to Fibrobacter spp. Ileal communities were dominated by TRFs representing Streptococcus agalactiae-like species in almost all animals. In feces, the following bacterial groups and clusters could be identified: Streptococcus spp., Lactobacillus/Enterococcus/Oenococcus spp., Clostridium clusters I, IV, XI, XIVa, and XVIII. Furthermore, TRFs that contributed only about 1 to 2% to all TRFs were Corynebacterium, Collinsella, Fibrobacter, Selenomonas, and Desulfovibrio species-like phylotypes. Two TRFs in ileal effluent and 21 TRFs in feces could not be assigned to known species.
Bacterial populations.
A set of 10 group-specific primers was employed to quantify bacterial populations in ileal effluent and feces. Bacterial populations in ileal effluent were only slightly affected by the NSP fractions (Table 4). The number of total bacterial gene copies in the distal ileum was lower (P < 0.05) for LG than for CEL, CMC, and HG. Clostridium cluster I was detectable only in the ileal effluents of pigs fed the CEL diet. Supplementation with CMC resulted in the highest number of Enterobacteriaceae family gene copies in ileal effluent (P < 0.05). In feces, the number of total bacterial gene copies was highest for CMC and lowest for LG (P < 0.05). The number of Clostridium cluster XIVa gene copies in feces was increased by CMC and reduced by LG in comparison to the number obtained with CEL, whereas the numbers of Clostridium cluster IV gene copies in feces were lower for both CMC and LG than for CEL (P < 0.05). Additionally, CMC and LG caused lower (P < 0.05) numbers of Clostridium cluster I gene copies in feces than CEL and HG. CMC increased the number of Bacteroides-Prevotella-Porphyromonas group gene copies in feces in comparison to the other NSP fractions, whereas HG reduced the number of gene copies in feces in comparison to CEL and CMC (P < 0.05). Finally, the number of Enterobacteriaceae family gene copies in feces was distinctly higher (P < 0.05) for CMC than for LG and HG.
TABLE 4.
Bacterial groups in ileal effluent and feces of pigs fed diets supplemented with viscous and fermentable nonstarch polysaccharide fractionsa
| Bacterial group | Value for indicated diet |
P | ||||
|---|---|---|---|---|---|---|
| Low fermentability |
High fermentability |
Pooled (SEM) | ||||
| Low-viscosity CEL | High-viscosity CMC | Low-viscosity LG | High-viscosity HG | |||
| Ileal effluent | ||||||
| Total bacteria | 9.5† | 9.8† | 8.5‡ | 9.6† | 0.18 | 0.001 |
| Lactobacillus spp. | 7.9 | 8.3 | 7.7 | 8.3 | 0.24 | 0.184 |
| Enterococcus spp. | 8.1†‡ | 8.0†‡ | 7.6‡ | 8.5† | 0.28 | 0.227 |
| Streptococcus spp. | 8.1† | 8.2† | 7.8‡ | 8.2† | 0.08 | 0.006 |
| Bifidobacterium spp. | 6.9 | 7.1 | 6.9 | 7.5 | 0.37 | 0.655 |
| Clostridium cluster XIVa | 6.8 | 7.3 | 6.4 | 6.9 | 0.36 | 0.387 |
| Clostridium cluster IV | 5.2 | 3.0 | 4.5 | 5.1 | 0.94 | 0.363 |
| Clostridium cluster Ib | 6.3 ± 0.34 | <2 | <2 | <2 | ||
| Bacteroides-Prevotella-Porphyromonas | 5.6 | 6.3 | 5.5 | 6.4 | 0.35 | 0.197 |
| Enterobacteriaceae family | 8.2‡ | 9.1† | 7.6§ | 8.8†‡ | 0.27 | 0.006 |
| Feces | ||||||
| Total bacteria | 10.1‡ | 11.3† | 9.2§ | 9.7‡§ | 0.25 | <0.001 |
| Lactobacillus spp. | 6.9† | 5.6‡ | 6.2†‡ | 6.1†‡ | 0.41 | 0.202 |
| Enterococcus spp. | 8.1† | 6.9‡§ | 6.6§ | 7.4†‡ | 0.29 | 0.010 |
| Streptococcus spp. | 7.8‡ | 8.5† | 7.4‡ | 7.7‡ | 0.18 | 0.004 |
| Bifidobacterium spp. | 7.5 | 7.0 | 7.2 | 7.8 | 0.39 | 0.525 |
| Clostridium cluster XIVa | 7.7‡ | 8.7† | 6.4§ | 7.1‡§ | 0.40 | 0.007 |
| Clostridium cluster IV | 7.8† | 6.5‡ | 6.6‡ | 7.1†‡ | 0.33 | 0.059 |
| Clostridium cluster I | 7.2† | 2.5‡ | 3.3‡ | 6.5† | 0.93 | 0.005 |
| Bacteroides-Prevotella-Porphyromonas | 8.9‡ | 10.5† | 8.0§ | 8.4‡§ | 0.20 | <0.001 |
| Enterobacteriaceae family | 9.0†‡ | 10.3† | 7.0§ | 7.6‡§ | 0.58 | 0.004 |
Data are presented as least-square means (n = 8). Values are log10 numbers of 16S rRNA gene copies/g (wet weight). Values within a row not having the same symbol are significantly different (P < 0.05). CEL, cellulose; CMC, carboxymethylcellulose.
The value shown for CEL is the mean ± standard error (SE). The detection limit was 2 log10 16S rRNA gene copies/g (wet weight).
Multivariate analysis.
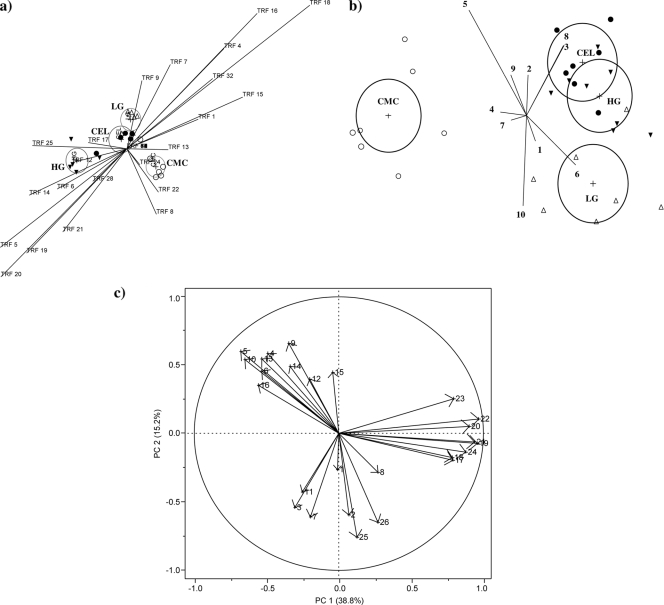
Linear discriminant analysis of NSP fractions and TRFs (Fig. 1 a) divided the effects of NSP fractions into three clusters, comprising the CEL and LG diets (overlapping in their 95% confidence intervals), the CMC diet, and the HG diet. The CEL and LG diets were more related to TRF9 (Bacteroides species-like phylotype), whereas the CMC diet was linked to TRF22 (Clostridium polysaccharolyticum-like phylotype). The HG diet was more related to TRF14 (Clostridium innocuum-like phylotypes). In accordance with the linear discriminant analysis of NSP fractions and gene copy numbers for bacterial groups (Fig. 1b), samples were also divided into three clusters; however, CEL and HG formed a single cluster, as indicated by the intersecting 95% confidence intervals. The results for the LG diet were different from those for the CEL, HG, and CMC diets, whereas the results for CMC were drastically different from those for the other NSP fractions. Here, the gene copy numbers for Bifidobacterium spp. and Clostridium cluster I discriminated best for CEL and HG, whereas those for Clostridium cluster IV and Streptococcus spp. discriminated best for CMC and those for Clostridium cluster XIVa for LG.
FIG. 1.
(a, b) Linear discriminant analysis of the NSP fractions and TRFs (a) and gene copies for bacterial groups (b): cellulose (CEL; •), carboxymethylcellulose (CMC; Ο), low-viscosity oat β-glucan (LG; ▵), and high-viscosity oat β-glucan (HG; ▾). (c) Loading plot showing the correlations among gene copies for bacterial groups in feces, E. coli virulence factors in feces, butyryl-coenzyme A (CoA) CoA transferase and butyrate kinase in feces, fecal SCFA and ileal flow, and disappearance of fermentable substrate in the large intestines of the first two eigenvalues (PC 1 and PC 2). 1, Lactobacillus spp.; 2, Enterococcus spp.; 3, Bifidobacterium spp.; 4, Streptococcus spp.; 5, Bacteroides-Prevotella-Porphyromonas; 6, Clostridium cluster XIVa; 7, Clostridium cluster IV; 8, Clostridium cluster I; 9, Enterobacteriaceae; 10, total bacteria; 11, butyryl-CoA CoA transferase; 12, butyrate kinase; 13, EAST1; 14, STa; 15, STb; 16, LT; 17, acetate; 18, propionate; 19, iso-butyrate; 20, butyrate; 21, isovalerate; 22, valerate; 23, caproate; 24, total SCFA; 25, ileal flow of dry matter; and 26, postileal dry-matter disappearance.
Figure 1c depicts the loading plot showing the individual qPCR data, fecal SCFA, ileal flow, and disappearance of dry matter in the large intestine responsible for the variation of the first two eigenvalues (PC 1 and PC 2) and the correlations among individual variables. The first PC component accounted for 38.8% of the variation, and PC 2 explained 15.2%. The SCFA formed a cluster located on the right part of the graph and were highly influenced by PC 1. Lactobacillus spp., Bifidobacterium spp., Enterococcus spp., Clostridium clusters I and IV, and butyryl-CoA CoA transferase formed a second cluster at the bottom of the loading plot and were negatively correlated with PC 2. A third cluster was formed by total bacteria, Enterobacteriaceae, the Bacteroides-Prevotella-Porphyromonas group, Streptococcus spp., Clostridium cluster XIVa, E. coli virulence factors, and butyrate kinase, which were influenced by both PC 1 and PC 2 and were negatively correlated with the other two clusters (with angles of >90° between the arrows for these clusters). Variables within these three clusters were positively related among each other, as indicated by the small angles between the arrows for these variables (<90°).
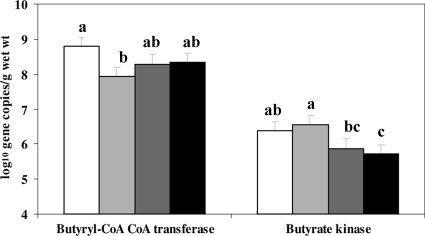
Butyrate production pathway genes.
Butyrate-producing bacteria in feces were determined by targeting genes for the enzymes butyryl-CoA CoA transferase and butyrate kinase (Fig. 2). The gene copy numbers for butyryl-CoA CoA transferase were higher (7.9 to 8.8 log10 DNA gene copies/g [wet weight]) than those for the butyrate kinase (5.7 to 6.6 log10 DNA gene copies/g [wet weight]). Dietary supplementation with CMC resulted in a lower (P < 0.05) gene copy number for the butyryl-CoA CoA transferase than supplementation with the CEL diet. In contrast, CMC increased (P < 0.05) the gene copy number for the butyrate kinase in comparison to LG and HG.
FIG. 2.
Gene copy numbers for butyryl-coenzyme A (CoA) CoA-transferase and butyrate kinase in feces of pigs fed diets supplemented with cellulose (white bars), carboxymethylcellulose (light gray bars), low-viscosity oat β-glucan (dark gray bars), or high-viscosity oat β-glucan (black bars). The detection limit was 4 log10 gene copies/g (wet weight).
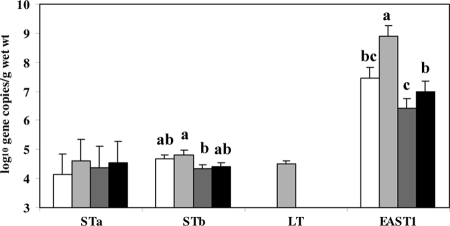
Escherichia coli virulence factors.
The heat-stable enterotoxin of enteroaggregative E. coli was the dominating virulence factor detected in both ileal and fecal samples, ranging from 6.0 to 8.9 log10 DNA gene copies/g (wet weight). In ileal effluent, EAST1 levels were affected by NSP (P = 0.026) and were significantly higher for CMC and HG than for LG (6.8, 7.5, 6.0, and 7.3 ± 0.35 log10 DNA gene copies/g [wet weight] for CEL, CMC, LG, and HG, respectively). The levels of virulence factors of enterotoxigenic E. coli heat-stable toxin A (STa), STb, and heat-labile toxin (LT) were below the detection limits in the ileal effluent. In feces, the EAST1 levels were higher by 1.4 to 2.5 log units for CMC than for the other NSP fractions (P < 0.05) (Fig. 3). The gene copy numbers for STa did not differ in feces, whereas the numbers of STb copies were higher (P < 0.05) for CMC than for LG. The LT was detectable only in the feces of pigs fed the CMC diet.
FIG. 3.
Gene copy numbers for virulence factors (heat-stable enterotoxins [STa and STb], heat-labile enterotoxin [LT] of enterotoxigenic Escherichia coli, and heat-stable enterotoxin [EAST1] of enteroaggregative E. coli) in feces of pigs fed diets supplemented with cellulose (white bars), carboxymethylcellulose (light gray bars), low-viscosity oat β-glucan (dark gray bars), or high-viscosity oat β-glucan (black bars). The detection limit was 3 log10 gene copies/g (wet weight).
DISCUSSION
In the present study, we used a polyphasic approach to study the effects of four purified NSP fractions of low and high viscosity and fermentability on the taxonomic composition of the ileal and fecal microbiota and, at a metabolic level, on butyrate-producing bacteria and E. coli virulence factors, using TRFLP and qPCR. Because purified NSP fractions may affect the bacterial community structure in a different way when added to a cereal-based diet due to the NSP in the grain matrix (36), a semipurified diet was employed in the present experiment.
The NSP fractions differently affected the small intestinal digestion and markedly changed the availability of fermentable substrate in the large intestine. However, there was no evidence that the shared functional properties affected digestive processes and endogenous nitrogen losses (44) consistently among the NSP fractions, suggesting that the specific chemical structures of the NSP are as relevant as shared rheological properties (8, 48). Correspondingly, consistent effects of viscosity and fermentability were not observed for gene copies for bacterial groups in ileal effluent. Cellulose resulted in faster transit than CMC and HG (data not shown); thus, besides the retention time in the small intestine, the accessibility of dietary nutrients appeared to be a critical factor for bacterial growth. Increased digesta viscosity impairs intestinal contractions (21), thereby preventing mixing of digesta and bacteria and thus access of bacteria to new substrate. Low-viscosity CEL, in turn, likely did not impair intestinal contractions and hence digesta mixing. According to the TRFLP profiles, Streptococcus agalactiae-like phylotypes dominated the ileal microbiota, followed by phylotypes belonging to Clostridium cluster XIVa. Surprisingly, TRFs representing Enterobacteriaceae species were not detected in ileal effluent or feces, which can be likely associated with the utilization of only one restriction enzyme (30), whereas the high rRNA gene copy numbers for Enterobacteriaceae produced with qPCR confirmed the prevalence of this bacterial group in the guts of pigs (22, 41).
The effects of the different purified NSP fractions on the formation of fermentation end products and bacterial numbers in feces mostly depended on changes in the ileal flow of dry matter (i.e., NSP fractions and other nondigested dietary ingredients) into the large intestine. In this context, low-fermentability CEL resulted in levels of postileal dry-matter disappearance and SCFA concentrations in feces similar to those obtained with high-fermentability LG and HG and significantly higher than those obtained with low-fermentability CMC.
Diversity indexes indicated that CMC supported a higher level of fecal bacterial diversity than the other NSP. Linear discriminant analysis of qPCR and TRFLP data confirmed that the fecal bacterial community structure in pigs fed the CMC diet differed from that observed in pigs fed the other NSP fractions. For instance, cellulolytic C. polysaccharolyticum-like phylotypes (TRF22) (47) discriminated best for the CMC diet, whereas fibrolytic and amylolytic Bacteroides species-like phylotypes (TRF9) (42) represented the best discrimination variable for CEL and LG. This may indicate that not only the NSP fractions but also the starch content in digesta may have modulated the bacterial community in pigs fed these diets.
Among the NSP fractions, low-fermentability CMC resulted in the highest numbers of total bacterial gene copies in feces. Generally, Clostridium clusters IV and XIVa and the Bacteroides-Prevotella-Porphyromonas group are the dominating strictly anaerobic bacterial groups in the large intestines of pigs (22). Carboxymethylcellulose clearly promoted the growth of Clostridium cluster XIVa and particularly of the Bacteroides-Prevotella-Porphyromonas group and Enterobacteriaceae in comparison to the other NSP. In contrast, the CEL diet favored the growth of Clostridium cluster IV. In addition to cellulose, the availability of other easily fermentable substrates in the ileal effluent, such as starch and protein, may have supported the higher numbers for Clostridium cluster IV, as this cluster contains both fibrolytic and nonfibrolytic species (25, 27), including some butyrate-producing bacterial species, such as Butyrivibrio fibrisolvens (2).
Cellulose, LG, and HG caused higher butyrate concentrations in feces than CMC. However, the measurement of butyrate in colonic digesta and portal blood is insufficient, as butyrate is mainly catabolized by colonozytes (3) and the various Clostridium clusters contain both butyrate producers and non-butyrate producers (27). In humans, butyryl-CoA CoA transferase and butyrate kinase genes are used as marker genes to detect butyrate-producing bacteria in the colon (24, 26). Similar to what was found for human butyrate producers (26), the main route of butyrate formation in the hindguts of pigs is the butyryl-CoA CoA transferase pathway. Moreover, the loading plot of PCA indicated that the ileal flow of dry matter into the large intestine was positively correlated with the number of butyryl-CoA CoA transferase gene copies, suggesting that the availability of not only the NSP fractions but also fermentable substrate was important for butyrate producers using the butyryl-CoA CoA transferase pathway. Similarly, butyryl-CoA CoA transferase correlated with lactic acid-producing groups, such as lactobacilli, bifidobacteria, and enterococci. This may be related to cross-feeding of butyrate-producing bacteria with lactate (27). The butyrate kinase was negatively correlated with the ileal flow of fermentable substrate, and its gene copy numbers were increased by CMC.
The CEL and HG diets markedly raised the gene copy numbers for Clostridium cluster I. Although this cluster contains fibrolytic and butyrate-producing bacteria (e.g., Clostridium cellulovorans), other members, such as Clostridium perfringens, may be harmful for the host (32). A TRF representing a C. perfringens-like phylotype was identified in feces, and a TRF was recognized as a Clostridium bifermentans-like phylotype that represents a potential pathogenic bacterium belonging to Clostridium cluster XI (40). However, adverse effects of high-viscosity NSP on gut health are mainly attributed to pathogenic E. coli (18, 43). High-viscosity CMC favored growth of pathogenic E. coli in weaning pigs (15, 31); however, these effects were generally confined to the immediate period after weaning (12, 15). The loose feces in combination with the high numbers of Enterobacteriaceae rRNA gene copies in the feces and ileal effluents of growing pigs used in the present study indicate that older pigs are also susceptible to overgrowth of enteropathogenic bacteria when the diet contains CMC. Quantitative PCR of virulence factors revealed that gene copies for particularly enteroaggregative E. coli bacteria were present in higher numbers in the distal ileum and in feces. Moreover, the heat-labile enterotoxin LT was exclusively detectable in feces of pigs fed the CMC diet and not in those of pigs fed diets supplemented with the other NSP. The similar gene copy numbers for EAST1, STa, and STb in feces of pigs fed CEL, LG, and HG did not cause any signs of diarrhea. The lower numbers observed for Enterobacteriaceae and E. coli virulence factors with the use of high-viscosity HG suggest that factors other than viscosity are involved in the stimulation of pathogenic E. coli. The CMC diet may have influenced the proliferation of pathogenic E. coli through changes in the mucus composition and the amount of mucus produced (35).
In conclusion, this study disclosed that the intestinal bacterial community, genes of alternative pathways of butyrate production, and E. coli virulence factors are specifically modulated by supplementing a semipurified diet with CEL, CMC, LG, or HG. Changes may be attributable to bacterial fermentation of NSP; additionally, NSP altered the ileal flow of nutrients into the large intestine. Effects of the NSP fractions were linked to the individual NSP fractions rather than to their shared functional properties, i.e., viscosity and fermentability. Comparable to what was observed for human colonic microbiota, the gene copy numbers for butyryl-CoA CoA transferase were higher than those for butyrate kinase, indicating that this pathway is the dominant butyrate production pathway in the large intestines of pigs. Although increasing intestinal viscosity was generally associated with impaired gut health (18, 43), only CMC increased the susceptibility of pigs to overgrowth of pathogenic E. coli, suggesting that the use of CMC in diets for growing pigs is detrimental compared to the use of the other NSP fractions investigated.
Supplementary Material
Acknowledgments
We thank the German Research Foundation (ME 3434/2-1), Danisco Animal Nutrition, the Alberta Pulse Growers Commission, and the Agriculture and Food Council of Alberta for funding this study. R. Pieper was funded by a postdoctoral research grant from the German Academic Exchange Service (DAAD, Germany). M. G. Gänzle acknowledges Research Chairs of Canada for funding.
Appreciation is extended to Jason Marshall of the Department of Animal and Poultry Science at the University of Saskatchewan for his assistance in TRFLP analysis.
Footnotes
Published ahead of print on 9 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.AOAC. 2006. Official methods of analysis of the Association of Official Analytical Chemists, 17th ed. AOAC, Arlington, VA.
- 2.Asanuma, N., M. Ishiwata, T. Yoshii, M. Kikuchi, Y. Nishina, and T. Hino. 2005. Characterization and transcription of the genes involved in butyrate production in Butyrivibrio fibrisolvens type I and II strains. Curr. Microbiol. 51:91-94. [DOI] [PubMed] [Google Scholar]
- 3.Bach Knudsen, E. K., A. Serena, N. Canibe, and K. S. Juntunen. 2003. New insight into butyrate metabolism. Proc. Nutr. Soc. 62:81-86. [DOI] [PubMed] [Google Scholar]
- 4.Bartosch, S., A. Fite, G. T. Macfarlane, and M. E. T. McMurdo. 2004. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 70:3575-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canadian Council on Animal Care. 1993. Guide to the care and use of experimental animals, 2nd ed., vol. 1. Ottawa, ON, Canada.
- 6.Castillo, M., G. Skene, M. Roca, M. Anguita, I. Badiola, S. H. Duncan, H. J. Flint, and S. M. Martín-Orúe. 2007. Application of 16S rRNA gene-targetted fluorescence in situ hybridization and restriction fragment length polymorphism to study porcine microbiota along the gastrointestinal tract in response to different sources of dietary fibre. FEMS Microbiol. Ecol. 59:138-146. [DOI] [PubMed] [Google Scholar]
- 7.Diez-Gonzalez, F., D. R. Bond, E. Jennings, and J. B. Russell. 1999. Alternative schemes of butyrate production in Butyrivibrio fibrisolvens and their relationship to acetate utilization, lactate production, and phylogeny. Arch. Microbiol. 171:324-330. [DOI] [PubMed] [Google Scholar]
- 8.Dikeman, C. L., and G. C. Fahey. 2006. Viscosity as related to dietary fiber: a review. Crit. Rev. Food Sci. Nutr. 46:649-663. [DOI] [PubMed] [Google Scholar]
- 9.Eastwood, M. A., and E. R. Morris. 1992. Physical properties of dietary fiber that influence physiological function: a model for polymers along the gastrointestinal tract. Am. J. Clin. Nutr. 55:436-442. [DOI] [PubMed] [Google Scholar]
- 10.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frydendahl, K. 2002. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhoea and edema disease in pigs and a comparison of diagnostic approaches. Vet. Microbiol. 85:169-182. [DOI] [PubMed] [Google Scholar]
- 12.Hampson, D. J. 1994. Postweaning Escherichia coli in pigs, p. 171-191. In C. L. Gyles (ed.), Escherichia coli in domestic animals and humans. CAB International, Wallingford, United Kingdom.
- 13.Han, W., B. Liu, B. Cao, L. Beutin, U. Krüger, H. Liu, Y. Li, Y. Liu, L. Feng, and L. Wang. 2007. DNA microarray-based identification of serogroups and virulence gene patterns of Escherichia coli isolates associated with porcine postweaning diarrhea and edema disease. Appl. Environ. Microbiol. 73:4082-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heilig, H. G. H. J., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. L. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopwood, D. E., D. W. Pethick, and D. J. Hampson. 2002. Increasing the viscosity of the intestinal contents stimulates proliferation of enterotoxigenic Escherichia coli and Brachyspira pilosicoli in weaner pigs. Br. J. Nutr. 88:523-532. [DOI] [PubMed] [Google Scholar]
- 16.Htoo, J. K., B. A. Araiza, W. C. Sauer, M. Rademacher, Y. Zhang, M. Cervantes, and R. T. Zijlstra. 2007. Effect of dietary protein content on ileal amino acid digestibility, growth performance, and formation of microbial metabolites in ileal and cecal digesta of early-weaned pigs. J. Anim. Sci. 85:3303-3312. [DOI] [PubMed] [Google Scholar]
- 17.Knarreborg, A., M. A. Simon, R. M. Engberg, B. B. Jensen, and G. W. Tannock. 2002. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl. Environ. Microbiol. 68:5918-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langhout, D. J., J. B. Schutte, P. Van Leeuwen, J. Wiebenga, and S. Tamminga. 1999. Effect of dietary high- and low-methylated citrus pectin on the activity of the ileal microflora and morphology of the small intestinal wall of broiler chicks. Br. Poult. Sci. 40:340-347. [DOI] [PubMed] [Google Scholar]
- 19.Lee, D.-H., Y.-G. Zo, and S.-J. Kim. 1996. Nonradioactive method to study genetic profiles of natural bacterial communities by PCR-single-strand-conformation polymorphism. Appl. Environ. Microbiol. 62:3112-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, C., J. Kim, S. G. Shin, and S. Hwang. 2006. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J. Biotechnol. 123:273-280. [DOI] [PubMed] [Google Scholar]
- 21.Lentle, R. G., and P. W. M. Jansson. 2008. Physical characteristics of digesta and their influence on flow and mixing in the mammalian intestine: a review. J. Comp. Physiol. B 178:673-690. [DOI] [PubMed] [Google Scholar]
- 22.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Møller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, S., W. C. Sauer, and M. Z. Fan. 1993. The effect of dietary crude protein level on amino acid digestibility in early-weaned pigs. J. Anim. Physiol. Anim. Nutr. 70:26-37. [Google Scholar]
- 24.Louis, P., and H. J. Flint. 2007. Development of a semiquantitative degenerate real-time PCR-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl. Environ. Microbiol. 73:2009-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louis, P., and H. J. Flint. 2009. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294:1-8. [DOI] [PubMed] [Google Scholar]
- 26.Louis, P., S. H. Duncan, S. I. McCrae, J. Millar, M. S. Jackson, and H. J. Flint. 2004. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacteriol. 186:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louis, P., K. P. Scott, S. H. Duncan, and H. J. Flint. 2007. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 102:1197-1208. [DOI] [PubMed] [Google Scholar]
- 28.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Miyamoto, T. Takada, K. Matsumoto, H. Oyaizu, and R. Tanaka. 2002. Development of 16S rRNA-gene targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68:5445-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuki, T., K. Watanabe, J. Fujimoto, T. Takada, and R. Tanaka. 2004. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 70:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto, M., M. Sakamoto, H. Hayashi, and Y. Benno. 2005. Novel phylogenetic assignment database for terminal-restriction fragment length polymorphism analysis of human colonic microbiota. J. Microbiol. Methods 61:305-319. [DOI] [PubMed] [Google Scholar]
- 31.McDonald, D. E., D. W. Pethick, B. P. Mullan, and D. J. Hampson. 2001. Increasing viscosity of the intestinal contents alters small intestinal structure and intestinal growth, and stimulates proliferation of enterotoxigenic Escherichia coli in newly-weaned pigs. Br. J. Nutr. 86:487-498. [DOI] [PubMed] [Google Scholar]
- 32.McLane, B. A., F. A. Uzal, M. E. F. Miyakawa, D. Lyberly, and T. Wilkins. 2006. The enterotoxic clostridia. Prokaryotes 4:698-752. [Google Scholar]
- 33.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nutrient Requirement Council. 1998. Nutrient requirements of swine, 10th ed. National Academic Press, Washington, DC.
- 35.Piel, C., L. Montagne, B. Sève, and J. P. Lallès. 2005. Increasing digesta viscosity using carboxymethylcellulose in weaned piglets stimulates ileal goblet cell numbers and maturation. J. Nutr. 135:86-91. [DOI] [PubMed] [Google Scholar]
- 36.Pieper, R., R. Jha, B. Rossnagel, A. G. Van Kessel, W. B. Souffrant, and P. Leterme. 2008. Effect of barley and oat cultivars with different carbohydrate compositions on the intestinal bacterial communities in weaned piglets. FEMS Microbiol. Ecol. 66:556-566. [DOI] [PubMed] [Google Scholar]
- 37.Pieper, R., J. Bindelle, B. Rossnagel, A. G. Van Kessel, and P. Leterme. 2009. Effect of carbohydrate composition in barley and oat cultivars on microbial ecophysiology and the proliferation of Salmonella enterica in an in vitro model of the porcine gastrointestinal tract. Appl. Environ. Microbiol. 75:7006-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pryde, S. E., S. H. Duncan, G. L. Hold, C. S. Stewart, and H. J. Flint. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217:133-139. [DOI] [PubMed] [Google Scholar]
- 39.Rinttilä, T., A. Kassinen, E. Malinen, L. Krogius, and A. Palva. 2004. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 97:1166-1177. [DOI] [PubMed] [Google Scholar]
- 40.Scanlan, D. R., M. A. Smith, H. D. Isenberg, S. Engrassia, and E. Hilton. 1994. Clostridium bifermentans bacteremia with metastatic osteomyelitis. J. Clin. Microbiol. 32:2867-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scharek, L., J. Guth, K. Reiter, K. D. Weyrauch, D. Taras, P. Schwerk, P. Schierack, M. F. Schmidt, L. H. Wieler, and K. Tedin. 2005. Influence of a probiotic Enterococcus faecium strain on development of the immune system of sows and piglets. Vet. Immunol. Immunopathol. 105:151-161. [DOI] [PubMed] [Google Scholar]
- 42.Smith, C. J., E. R. Rocha, and B. J. Paster. 2006. The medically important Bacteroides spp. in health and disease. Prokaryotes 7:381-427. [Google Scholar]
- 43.Smits, C. H., A. Veldman, H. J. Verkade, and A. C. Beynen. 1998. The inhibitory effect of carboxymethylcellulose with high viscosity on lipid absorption in broiler chickens coincides with reduced bile salt concentration and raised microbial numbers in the small intestine. Poult. Sci. 77:1534-1539. [DOI] [PubMed] [Google Scholar]
- 44.Souffrant, W. B. 2001. Effect of dietary fibre on ileal digestibility and endogenous nitrogen losses in the pig. Anim. Feed. Sci. Technol. 90:93-102. [Google Scholar]
- 45.van Kleef, D. J., K. Deuring, and P. van Leeuwen. 1994. A new method of faeces collection in the pig. Lab. Anim. 28:78-79. [DOI] [PubMed] [Google Scholar]
- 46.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warnick, T. A., B. A. Methé, and S. B. Leschine. 2002. Clostridium phytofermentans sp. nov., a cellulolytic mesophile from forest soil. Int. J. Syst. Evol. Microbiol. 52:1155-1160. [DOI] [PubMed] [Google Scholar]
- 48.Wenk, C. 2001. The role of dietary fibre in the digestive physiology of the pig. Anim. Feed. Sci. Technol. 90:21-33. [Google Scholar]
- 49.Williams, B. A., M. W. Bosch, A. Awati, S. R. Konstantinov, H. Smidt, A. D. L. Akkermans, M. W. A. Verstegen, and S. Tamminga. 2005. In vitro assessment of grastointestinal tract (GIT) fermentation in pigs: fermentable substrates and microbial activity. Anim. Res. 54:191-202. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.