Abstract
Vinyl chloride (VC) is a toxic groundwater pollutant associated with plastic manufacture and chlorinated solvent use. Aerobic bacteria that grow on VC as a carbon and energy source can evolve in the laboratory from bacteria that grow on ethene, but the genetic changes involved are unknown. We investigated VC adaptation in two variants (JS623-E and JS623-T) of the ethene-oxidizing Mycobacterium strain JS623. Missense mutations in the EtnE gene developed at two positions (W243 and R257) in cultures exposed to VC but not in cultures maintained on ethene. Epoxyalkane-coenzyme M transferase (EaCoMT) activities in cell extracts of JS623-E and JS623-T (150 and 645 nmol/min/mg protein, respectively) were higher than that of wild-type JS623 (74 nmol/min/mg protein), and in both variant cultures epoxyethane no longer accumulated during growth on ethene. The heterologous expression of two variant etnE alleles (W243G [etnE1] and R257L [etnE2]) from strain JS623 in Mycobacterium smegmatis showed that they had 42 to 59% higher activities than the wild type. Recombinant JS623 cultures containing mutant EtnE genes cloned in the vector pMV261 adapted to growth on VC more rapidly than the wild-type JS623 strain, with incubation times of 60 days (wild type), 1 day (pMVetnE1), and 35 days (pMVetnE2). The JS623(pMVetnE) culture did not adapt to VC after more than 60 days of incubation. Adaptation to VC in strain JS623 is consistently associated with two particular missense mutations in the etnE gene that lead to higher EaCoMT activity. This is the first report to pinpoint a genetic change associated with the transition from cometabolic to growth-linked VC oxidation in bacteria.
Bacteria that biodegrade pollutants are useful for the cleanup of contaminated sites (i.e., bioremediation) and are interesting as models of evolutionary processes (21, 38, 40). Understanding the molecular genetic and evolutionary basis of biodegradation processes allows improved monitoring and predictions of bacterial activities in situ (39) and promises the development of improved strains and enzymes with increased specific activity (3), increased substrate affinity (16), extended substrate range (3, 16, 21, 37), extended inducer range (30, 31), or constitutive expression (39). Missense mutations in catabolic enzymes or regulatory proteins commonly lead to these changes (43), although other important mechanisms include duplication, deletion, and inversion (38-40).
Vinyl chloride (VC) is a common groundwater pollutant (35) and known human carcinogen (24), and it poses a health risk to exposed populations. Although trace amounts (e.g., parts per trillion) of VC have been detected in uncontaminated soil (23), higher concentrations are found only associated with human industry, particularly the manufacture of polyvinylchloride (PVC) plastic and the chlorinated solvents trichloroethene (TCE) and perchloroethene (PCE) (4). Aerobic bacteria that grow on VC as a sole carbon and energy source are diverse, including strains of Mycobacterium (8, 17, 18), Nocardioides (8), Pseudomonas (11, 41, 42), Ochrobactrum (11), and Ralstonia (13, 33). The relative ease of the isolation of VC assimilators from chlorinated ethene-contaminated sites suggests that such bacteria are influential in the natural attenuation of VC, but this interpretation is complicated by the fact that VC-assimilating bacteria are closely related to ethene-assimilating bacteria (8-10, 29) and cannot yet be distinguished from them by molecular tests.
The VC and ethene pathway and genes are homologous to some extent with the propene assimilation pathway and genes in Xanthobacter Py2 and Gordonia B-276. The comparison of the genomes of the VC-assimilating Nocardioides JS614 and the propene-assimilating Xanthobacter Py2 indicates that growth on alkenes requires about 20 kb of alkene/epoxide catabolic genes and approximately 7 kb of coenzyme M (CoM) biosynthesis genes. The oxidation of VC and ethene is initiated by an alkene monooxygenase (AkMO; EtnABCD) (8-10, 29), which yields epoxyethane from ethene and chlorooxirane from VC (8, 17). An epoxyalkane-coenzyme M transferase (EaCoMT) enzyme, EtnE, acts upon these reactive, toxic, and mutagenic epoxides (2, 19), converting them to hydroxyalkyl-CoM derivatives. The remainder of the VC/ethene pathway is unclear. The JS614 genome indicates further homology with propene oxidizers, in that a reductase/carboxylase and SDR family dehydrogenase are present, but that other aspects of the VC/ethene pathway gene cluster are unique (e.g., the presence of a semialdehyde dehydrogenase [5] and a disulfide reductase-like gene [GenBank accession no. NC_008697]).
The EtnE enzyme and the homologous XecA enzyme that acts on epoxypropane in Xanthobacter Py2 and Gordonia B-276 (9, 10, 12, 29) are unusual in their requirement for CoM as a cofactor. The C2- and C3-alkene oxidizers are the only Eubacteria known to biosynthesize and require CoM, which is otherwise found only in methanogenic Archaea. The XecA protein of Py2 has been purified and shown to be a Zn-dependent enzyme (1, 14, 26, 44). Based on sequence homology and the presence of the Cys-X-His-Xn-Cys motif (see Fig. S1 in the supplemental material), the EtnE enzymes also are likely to be Zn-dependent enzymes. Heterologous expression systems for XecA and EtnE have been developed (9, 25), but no crystal structures are available yet for EaCoMT from any source.
Pure cultures of ethene-assimilating bacteria are capable of spontaneously adapting to growth on VC as a carbon source (22, 42), but the molecular basis of this phenomenon is not clear. This knowledge gap confounds the development of molecular probes specific for VC-assimilating bacteria. Pseudomonas aeruginosa strain DL1 shifted from cometabolism to growth on VC after more than 40 days of incubation (42), while Mycobacterium strains JS622, JS623, JS624, and JS625 took between 55 and 476 days to adapt to VC (22). The VC-adapted phenotype in Mycobacterium strains was not lost after growth in nonselective medium, suggesting a genetic change rather than a physiological adaptation (22).
Here, we tested the hypothesis that mutations in the alkene/epoxide catabolic genes are responsible for VC adaptation. This was done by sequencing EtnEABCD genes in fosmid clones from cultures before and after VC adaptation, by sequencing etnE PCR products at different time points during VC adaptation, and by examining the EtnE enzyme activity in VC-adapted strains and recombinant strains carrying evolved etnE alleles.
MATERIALS AND METHODS
Chemicals, media, bacterial strains, and culture conditions.
Vinyl chloride (VC) (99.5%) was from Fluka, and ethene (99%) was from Airgas. Epoxyethane (≥99.5%) and 2-mercaptoethanesulfonate (CoM) were from Sigma. All other chemicals either were reagent or molecular biology grade. The general-purpose media used were LB (32) for Escherichia coli and 0.1× Trypticase soy glucose (designated 0.1TSG) medium for Mycobacterium strains (22). Growth on gaseous substrates was tested in minimal salts medium (MSM) (7) broths in gas-tight bottles (i.e., sealed with butyl rubber stoppers [Wheaton] and aluminum crimp caps), using 72, 250, or 500 ml medium in 160-ml serum bottles (Wheaton) or 1- or 2-liter modified Erlenmeyer flasks (28), respectively. Cultures were fed with either filter-sterilized ethene (0.25 mM predicted in aqueous phase) or VC (0.77 mM predicted in aqueous phase). All cultures were grown aerobically, with broths shaken at 200 rpm. The incubation of E. coli was at 37°C, while the incubation of Mycobacterium strains was at 30°C for growth on plates or at ambient temperature (21 to 23°C) for broths.
Cultures used here included the wild-type ethene-assimilating Mycobacterium strain JS623 (9) and two VC-adapted variants, JS623-T and JS623-E (22). Other strains used include Mycobacterium smegmatis mc2155 (34), Nocardioides JS614 (8), and E. coli strains TOP10 (Invitrogen), DH5α F′Iq (New England Biolabs), and EPI300 (Epicentre). Recombinants of TOP10, mc2155, and JS623 containing pMV261 were made as described below and were grown on LB plus 50 μg/ml Km (E. coli), LB plus 30 μg/ml Km (mc2155), or MSM-VC/ethene plus 30 μg/ml Km (JS623). E. coli EPI300 fosmid clones were grown in LB plus 12.5 μg/ml chloramphenicol. E. coli DH5α F′Iq pDRIVE clones were grown in LB plus 50 μg/ml Km.
Preparation and screening of fosmid clone libraries.
Genomic DNA was extracted from cultures of ethene-grown wild-type JS623 and VC-grown JS623-T (at 259 days of VC exposure) as described previously (22) and cloned into the pCC1FOS vector (Epicentre) in E. coli EPI300 as described previously (9, 29). Fosmid clones were pooled in sets of eight and PCR screened for the presence of etnE with CoMF1L and CoMR2E primers (29). Fosmid DNA was extracted from one etnE-positive clone derived from wild-type JS623 and one etnE-positive clone from JS623-T, and the genes etnEABCD in both were sequenced by primer walking at The University of Iowa DNA facility using the primers described in Table S1 in the supplemental material.
Construction of etnE clone libraries.
The etnE gene was amplified using Taq polymerase as described previously (15), except that WEtnEF and WEtnER primers were used (see Table S1 in the supplemental material). The etnE PCR products were cloned into the pDRIVE vector (Qiagen) and transformed into competent E. coli DH5α F′Iq cultures (New England BioLabs) according to the manufacturer's protocols. Recombinant plasmids were purified from overnight E. coli cultures with the QIAprep spin Miniprep kit (Qiagen) and sequenced with JS623PMD and JS623WEtnE primers (see Table S1 in the supplemental material).
Construction of recombinant mc2155 and JS623 cultures containing EtnE genes.
Three variants of the EtnE gene (from wild-type JS623, JS623-E, and JS623-T) were amplified with Pfu polymerase using the primers JS623WEtnEF and JS623WEtnER (see Table S1 in the supplemental material). The resulting PCR products (1,312 bp) were cloned into the Mycobacterium-E. coli shuttle vector pMV261 (36) and transformed into E. coli TOP10. Clones with the expected plasmid structure were identified by PCR and grown for large-scale plasmid extraction (32). The resultant pMV261-etnE plasmids were electroporated into Mycobacterium smegmatis mc2155 and wild-type JS623 cultures using essentially the same procedure as that described previously (10, 20), except that an Eppendorf electroporator 2510 (set at 2,500 V) was used. When electroporating JS623 cells, a mixture of pMV261-etnE plasmid DNA (1,000 ng), 50 μl of ethene-grown JS623 cells, and 40 μl of 10% sterile glycerol was used with a 1-mm-gap electroporation cuvette. Cells were recovered for 1 h at 37°C with shaking (300 rpm) in 600 μl of LB (mc2155) and 0.1TSG (JS623). Recombinant mc2155 cultures were grown on LB plus 30-μg/ml Km plates and then LB plus 30-μg/ml Km broths. Recombinant JS623 (R-JS623) cultures were grown on 0.1TSG plus 30-μg/ml Km plates for 2 to 3 weeks and then transferred to MSM broths containing 30 μg/ml Km and either ethene (0.25 mM) or VC (0.76 mM) as carbon sources.
Analytical methods.
Headspace samples of ethene (100 μl), VC (100 μl), and epoxyethane (250 μl) were analyzed via gas chromatography with flame ionization detection, and the peak area was compared to an external standard as described previously (29). Bacterial growth was routinely measured with a spectrophotometer at an optical density at 600 nm (OD600). DNA and protein concentrations were measured by UV spectrophotometry, as described previously (8, 32), or with Qubit fluorometer (Invitrogen) and Quant-iT broad-range DNA assays.
EaCoMT assay.
Cells grown on VC or ethene to an OD600 of ∼0.3 were washed twice with dipotassium phosphate (KP) buffer (20 mM), suspended in 3 ml MGD buffer (50 mM morpholineethanesulfonic acid [MOPS], 10% glycerol, 1 mM dithiothreitol [DTT] [pH 7.2]) to an OD600 of ∼50, and lysed by a mini French pressure cell (124,000 kPa, three cycles) at 4°C. After centrifugation (21,000 × g, 10 min) the supernatant was retained, and samples of this cell extract (0.1 mg protein in 950 μl Tris-HCl buffer [50 mM, pH 8.0]) were sealed in 27-ml serum bottles, epoxyethane (5 μmol) was added, and the bottles were equilibrated (20 min, 30°C, with shaking at 300 rpm). Reactions were started by the addition of 50 μl CoM (to a 10 mM final concentration), and then headspace samples were analyzed at intervals (0, 10, 20, and 30 min). The mean values of abiotic epoxyethane loss were 2.1 nmol/min in Tris-HCl buffer and 17.2 nmol/min in Tris-HCl buffer plus CoM (10 mM).
RESULTS
Independent cultures develop similar etnE mutations during VC adaptation.
The adaptation of strain JS623 to growth on VC occurred reproducibly after an incubation time of approximately 60 days in the case of both ethene-grown and TSAG-grown inocula, which yielded variants JS623-E and JS623-T, respectively (22). Our hypothesis was that VC adaptation was due to a genetic change in one or more of the ethene biodegradation genes. This was tested by sequencing etnEABCD (encoding the first two enzymes in the ethene/VC biodegradation pathway) in fosmid clones derived from the wild-type JS623 culture and from the JS623-T culture grown on VC for 259 days.
The JS623 and JS623-T EtnEABCD genes (GenBank Accession nos. FJ602754 and FJ602755, respectively) had the same arrangement as those of Mycobacterium strain JS60 and Nocardioides sp. strain JS614 (10, 29), and the predicted JS623 polypeptides have 82 to 96% amino acid (aa) identity to those of JS60 and 63 to 84% aa identity to those of JS614. The etnABCD sequences from JS623-T were 100% identical to those of the wild type, indicating that no changes to the ethene monooxygenase sequence are required for it to accept VC as a substrate. The etnE sequence from JS623-T contained a point mutation (G770T) relative to the wild type, which would give the R257L missense mutation in the EtnE protein. The EtnE gene was amplified from JS623-E cultures grown on VC for 178 days, and the PCR product was directly sequenced. The results indicated mutations at two nucleotide positions (727 and 770) (W243 and R257 in the EtnE protein), but the sequence data were ambiguous, possibly due to different etnE alleles existing in the JS623-E culture.
To resolve the sequence ambiguities seen with JS623-E and to determine the frequency of different etnE mutations, we cloned and sequenced approximately 20 PCR-amplified EtnE genes from the strains at multiple time points after VC exposure, i.e., wild-type JS623 (day 0), JS623-E (days 97 and 178), and JS623-T (days 86, 235, and 289). This yielded 138 etnE sequences, 110 of which contained mutations (Table 1). We detected mutations at 57 different positions in etnE, and each mutant clone carried between one and four point mutations. Changes giving rise to missense mutations at positions W243 and R257 dominated the etnE clone libraries from both VC-adapted variants and included W243G (21 instances), W243R (7 instances), W243L (1 instance), and R257L (62 instances) (Table 1). Other potentially significant mutations were seen at V244 (six instances), N98 (three instances), L127 (two instances), S138 (two instances), A166 (two instances), L245 (two instances), A258 (two instances), and N343 (two instances). The remaining 47 mutation types were unique.
TABLE 1.
Types and frequencies of etnE mutations in JS623 cultures during VC adaptation
| Culture | Ethene exposure timea,c (days) | VC exposure timeb,c (days) | Predicted EtnE mutant type | No. of clones |
|---|---|---|---|---|
| Wild-type | 21 | 0 | None | 15 |
| JS623 | A60Ad | 1 | ||
| N98I | 1 | |||
| K286R | 1 | |||
| F297S | 1 | |||
| T311T | 1 | |||
| 100 | 0 | None | 13 | |
| V53A, R128L, V304A | 1 | |||
| E210G, W293R | 1 | |||
| H123H, N343N | 1 | |||
| L273S | 1 | |||
| S138N, A258V | 2 | |||
| T268T | 1 | |||
| C214S, A253V | 1 | |||
| JS623-E | 15 | 97 | W243G | 8 |
| R257L | 5 | |||
| V244G | 2 | |||
| W243R | 1 | |||
| W243L, L245V | 1 | |||
| L245P | 1 | |||
| 15 | 178 | W243G | 12 | |
| R257L | 7 | |||
| W243R | 1 | |||
| W243G, R257L | 1 | |||
| JS623-T | 15 | 86 | R257L | 8 |
| R257L, E179G | 1 | |||
| R257L, I165I | 1 | |||
| R257L, E160G, D303G | 1 | |||
| W243R | 1 | |||
| W243R, D68N | 1 | |||
| W243R, S80P | 1 | |||
| W243R, D317Y | 1 | |||
| W243R, F175F, I385V | 1 | |||
| V244G | 1 | |||
| V244G, G223E | 1 | |||
| V244G, N369D | 1 | |||
| V244G, T134T, H352R | 1 | |||
| 15 | 235 | R257L | 10 | |
| R257L, F47S | 1 | |||
| R257L, I82V | 1 | |||
| R257L, T130A | 1 | |||
| R257L, A166T | 1 | |||
| R257L, W293C | 1 | |||
| R257L, N343S | 1 | |||
| R257L, S347S | 1 | |||
| R257L, F37F, L238G | 1 | |||
| R257L, N98S, L127L, F262S | 1 | |||
| 15 | 289 | R257L | 10 | |
| R257L, D25N | 1 | |||
| R257L, D45G | 1 | |||
| R257L, Q58*stop | 1 | |||
| R257L, A79A | 1 | |||
| R257L, I167T | 1 | |||
| R257L, N274S | 1 | |||
| R257L, L127L,H219Y | 1 | |||
| R257L, V218V, S334P | 1 | |||
| R257L, N98D, A166A, T302I | 1 |
Consecutive culturing time on ethene since the strain was received in the Mattes laboratory. VC-adapted variants were cultured for 15 days on ethene before transfer to VC, while the wild-type strain was grown only on ethene and was sampled at 21 and 100 days.
Consecutive culturing time on VC since transfer from ethene (JS623-E) or TSAG (JS623-T) medium. Zero indicates that the wild-type JS623 cultures were not exposed to VC.
Cultures were routinely archived at −80°C after reaching an OD600 of ∼0.3 and were analyzed later for the presence of missense mutations. This experimental design did not allow for the calculation of generation times.
Amino acid location in the EtnE protein sequence (GenBank accession no. FJ602754).
Surprisingly, 5 of 20 etnE clones from the time-zero wild-type JS623 culture deviated from the expected sequence (GenBank accession no. AAO48584). Each of the time-zero mutants was a different point mutation, and none had inferred changes at W243 or R257. To determine if growth on ethene influenced etnE sequences, we grew wild-type JS623 on ethene for a further 79 days and repeated the etnE PCR, cloning, and sequencing. The results (Table 1) were similar to those of time-zero JS623 cultures, with the majority (13/21) of etnE sequences containing no mutations, and the remainder was comprised almost entirely of unique types (including silent and nonsense mutations). No W243 or R257 mutant was seen in cultures maintained on ethene. It must be noted that PCR errors (6) could account for some of the etnE sequence changes seen here. However, these possible artifacts do not change the overall pattern of the data, which indicate that W243 and R257 mutants of EtnE arise and dominate in cultures exposed to VC, while no particular EtnE mutations are selected for by growth on ethene during a comparable time period.
Comparison of EaCoMT activities in VC-adapted variants, recombinants, and wild-type JS623.
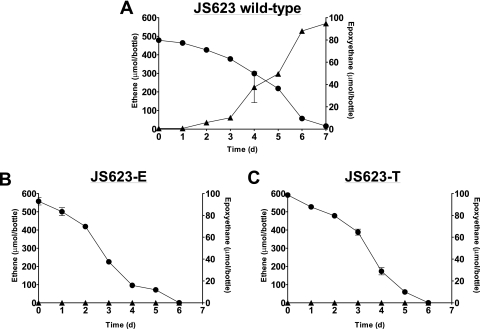
The VC-adapted JS623-E and JS623-T cultures did not accumulate detectable amounts of epoxyethane when grown on ethene (Fig. 1), leading us to hypothesize that EaCoMT activities in the VC-adapted JS623 variants had increased compared to that of the wild type, resulting in more efficient epoxide metabolism. The EaCoMT activities (in nmol/min/mg protein) in JS623 cell extracts (with 10 mM CoM added) were 74 ± 15 (wild type), 150 ± 15 (JS623-E), and 645 ± 248 (JS623-T). The EaCoMT activities of both VC-adapted variants were significantly higher than that of the wild type (P < 0.05). To provide further evidence that the mutant EtnE proteins had higher EaCoMT activity than the wild-type EtnE, we cloned the wild-type etnE and the W243G and R257L mutant genes (hereafter referred to as etnE1 and etnE2, respectively) into the pMV261 expression vector to make the plasmids pMVetnE, pMVetnE1, and pMVetnE2. The plasmids were transformed into M. smegmatis mc2155, and the recombinants were tested for EaCoMT activity. This revealed activities (nmol/min/mg protein) of 39 ± 4 (pMVetnE), 56 ± 5 (pMVetnE1), and 62 ± 4 (pMVetnE2). These EaCoMT activities are 52, 37, and 10% of those observed in wild-type JS623, JS623-E, and JS623-T strains, respectively. The activities of both mutant etnE alleles were significantly higher than that of the wild type (P < 0.005).
FIG. 1.
Ethene (•) biodegradation and epoxyethane (▴) accumulation in (A) ethene-grown, wild-type JS623 cultures, (B) ethene-grown, VC-adapted JS623-E, and (C) ethene-grown, VC-adapted JS623-T. The data points are the averages from analysis of three replicate bottles, and the error bars are the standard deviations. In some cases, the error bars are smaller than the symbols. The behavior of these cultures was confirmed in at least two independent experiments.
To provide another reference point for EaCoMT activity, a pMV261 derivative containing etnE from Nocardioides sp. strain JS614 (GenBank accession no. AY772007) was generated. The JS614 etnE (here designated etnE614) represents a sequence divergent from mycobacterial etnE genes (76% aa identity to that of JS623) and also provides an example from a strain isolated initially on VC. Cells of strain mc2155(pMVetnE614) had very high EaCoMT activities (1,543 ± 90 nmol/min/mg protein), representing ∼38% of the activity in cell extracts from ethene-grown JS614 cells (29). Although the EaCoMT activities in the mc2155 clones typically were 30 to 50% lower than the corresponding activities in the source strains, the level of EaCoMT activity in the clones tended to be representative of the source, validating the use of this system.
Recombinant JS623 cultures containing mutant EtnE genes adapt to VC more rapidly than the wild type.
We asked whether introducing the two most prevalent mutant EtnE alleles (W243G and R257L) into wild-type JS623 cultures would be sufficient for growth on VC. To test this, the plasmids pMVetnE, pMVetnE1, and pMVetnE2 were electroporated into wild-type JS623 to yield three different recombinant strains.
During growth on ethene, wild-type JS623 and recombinant strains JS623(pMVetnE) and JS623(pMVetnE2) accumulated epoxyethane, as has been seen previously for the wild type (9) (see Fig. S2 in the supplemental material). However, the recombinant strain JS623(pMVetnE1) did not accumulate any detectable epoxyethane (see Fig. S2 in the supplemental material). Since the additional gene dosage provided by pMVetnE did not have an obvious effect on ethene or epoxyethane metabolism, the lack of epoxyethane accumulation in JS623(pMVetnE1) cultures can be attributed to the sequence changes in the etnE1 allele.
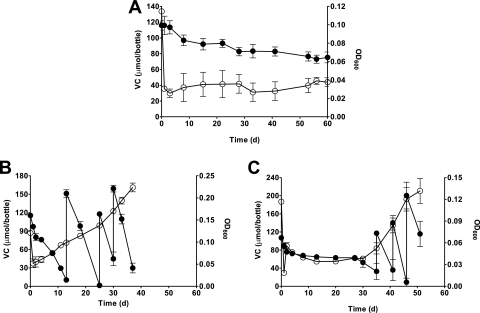
We also examined the behavior of the recombinant JS623 strains during VC adaptation using ethene-grown inoculum cells. Wild-type JS623 typically takes 60 days to adapt to growth on VC (22). Strain JS623(pMVetnE) slowly degraded VC for more than 60 days but did not grow on it (Fig. 2). This is reminiscent of how the TSAG-grown wild-type JS623 strain behaved when incubated with VC (22). A different recombinant JS623 strain that contained only pMV261 did adapt to VC after incubation times of 60 to 65 days (data not shown). In contrast, strain JS623(pMVetnE2) began growing on VC after about 35 days of incubation, while strain JS623(pMVetnE1) displayed almost immediate growth on VC (Fig. 2). We confirmed this behavior in further independent experiments and also confirmed that there was no significant loss of VC in abiotic controls during the same incubation period (data not shown).
FIG. 2.
VC metabolism in recombinant JS623 strains JS623(pMVetnE) (A), JS623(pMVetnE1) (B), and JS623(pMVetnE2) (C). The data points are the averages from the analysis of three replicate bottles, and the error bars are the standard deviations. In some cases, the error bars are smaller than the symbols. The behavior of these cultures was confirmed in at least two independent experiments.
DISCUSSION
This is the first report of specific molecular changes associated with bacterial adaptation to VC as a sole carbon and energy source. Our data indicate that the EtnE gene is an important target for evolutionary adaptation to VC. Changes to the etnE sequence in Pseudomonas AJ and Ochrobactrum TD cultures grown on different media (VC or LB) were mentioned previously (12), but in that case the details of the changes were not specified. The fact that both our study and that of Danko et al. observed changes in etnE depending on the growth medium suggests that mutations in etnE are significant for the evolution of both Gram-negative and Gram-positive VC-degrading bacteria. The subtle nature of this genetic change will make it challenging to develop molecular probes/primers specific for VC-assimilating bacteria that do not also detect ethene assimilators.
We propose that the W243 and R257 mutations are at least partly responsible for the higher EaCoMT activity of strains JS623-E and JS623-T toward epoxyethane relative to that of wild-type JS623, and further that these specific mutations also raise the activity of EtnE toward chlorooxirane above a critical level that allows growth on this chlorinated epoxide. However, it is important to note that the JS623-E culture contains a mixture of etnE alleles that likely affects the overall EaCoMT activity measured.
We did not directly test the activity of the different EtnE enzymes on chlorooxirane in this study because of the high reactivity and commercial unavailability of this compound. However, the enhanced metabolism of chlorooxirane due to the W243G and R257L mutations can be inferred from the faster adaptation of JS623(pMVetnE1) and JS623(pMVetnE2) cultures to VC relative to that of the wild-type strain and relative to that of JS623(pMVetnE).
In the absence of an EaCoMT crystal structure, it is difficult to predict the structural effects of the W243 and R257 mutations. Although a crystal structure of MetE from Thermotoga is available (NCBI Protein Data Bank no. 1XR2_B), it is not straightforward to extrapolate this to the structure of EtnE because of low overall protein sequence identities (∼10%) and very different protein sizes. Large gaps appear when MetE, XecA, and EtnE are aligned (see Fig. S1 in the supplemental material), and the 243 to 257 region in EtnE and XecA lacks an obviously homologous region in MetE. It is notable, however, that the W243 residue is close to two of the zinc-coordinating residues (H-X-C at position 220) that are likely crucial for the binding of CoM to the enzyme. Since both the W243G and R257L mutations correspond to large residues replacing smaller ones, it is possible that these mutations enhance the access of chlorooxirane (a larger molecule than epoxyethane) to the active site of EtnE. The R257L mutation, which results in the change of a positive residue for a neutral one, might also be of significance for the access of chlorooxirane to the active site, because this molecule will have a different charge distribution than epoxyethane (e.g., the carbon atoms will be more electropositive due to the adjacent chlorine atom).
Some data indicate that other factors apart from changes to EaCoMT are involved in VC adaptation [e.g., the much lower EaCoMT activity of mc2155(pMVetnE2) clones compared to that of the source culture JS623-T]. Mutations in downstream catabolic enzymes, CoM biosynthesis genes, regulatory genes, or the plasmid copy number are possible contributors to VC adaptation (27). A genomic approach using multiple VC- and ethene-assimilating strains is required to further investigate this possibility. The further study of the physiology of the high-activity mc2155(pMVetnE614) clone would be valuable to provide more evidence that changes in etnE alone are necessary and sufficient for adaptation to growth on VC.
The results described here indicate that JS623 is a useful model system for studying microbial enzyme evolution in response to xenobiotic compounds. The case of EaCoMT and epoxides is interesting in an evolutionary sense, because the epoxide substrates are themselves mutagenic and likely will increase mutation rates in cultures exposed to them. Finally, we propose that EaCoMT (and CoM) perform a triple function in the alkene biodegradation pathway: (i) neutralize the toxicity of epoxides, (ii) channel carbon into productive metabolism, and (iii) modulate the mutation rate during growth on alkenes.
Supplementary Material
Acknowledgments
This research was supported by the University of Iowa Center for the Health Effects of Environmental Contamination (CHEEC), the University of Iowa Biological Sciences Funding Program, and University of Iowa start-up funds for T.E.M. Y.O.J. was partially supported by University of Iowa Strategic Initiative Funding. The Coleman laboratory is supported by funds from the Australian Research Council and the University of Sydney.
Footnotes
Published ahead of print on 2 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allen, J. R., and S. A. Ensign. 1997. Purification to homogeneity and reconstitution of the individual components of the epoxide carboxylase multiprotein enzyme complex from Xanthobacter strain Py2. J. Biol. Chem. 272:32121-32128. [DOI] [PubMed] [Google Scholar]
- 2.Barbin, A., H. Brésil, A. Croisy, P. Jacquignon, C. Malaveille, R. Montesano, and H. Bartsch. 1975. Liver-microsome-mediated formation of alkylating agents from vinyl bromide and vinyl chloride. Biochem. Biophys. Res. Commun. 67:596-603. [DOI] [PubMed] [Google Scholar]
- 3.Bosma, T., J. Damborsky, G. Stucki, and D. B. Janssen. 2002. Biodegradation of 1,2,3-trichloropropane through directed evolution and heterologous expression of a haloalkane dehalogenase gene. Appl. Environ. Microbiol. 68:3582-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley, P. M. 2003. History and ecology of chloroethene biodegradation: a review. Bioremediat. J. 7:81-109. [Google Scholar]
- 5.Chuang, A. S., and T. E. Mattes. 2007. Identification of polypeptides expressed in response to vinyl chloride, ethene, and epoxyethane in Nocardioides sp. strain JS614 by using peptide mass fingerprinting. Appl. Environ. Microbiol. 73:4368-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cline, J., J. Braman, and H. Hogrefe. 1996. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 24:3546-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman, N. V., T. E. Mattes, J. M. Gossett, and J. C. Spain. 2002. Biodegradation of cis-dichloroethene as the sole carbon source by a beta-proteobacterium. Appl. Environ. Microbiol. 68:2726-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, N. V., T. E. Mattes, J. M. Gossett, and J. C. Spain. 2002. Phylogenetic and kinetic diversity of aerobic vinyl chloride-assimilating bacteria from contaminated sites. Appl. Environ. Microbiol. 68:6162-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman, N. V., and J. C. Spain. 2003. Distribution of the coenzyme M pathway of epoxide metabolism among ethene- and vinyl chloride-degrading Mycobacterium strains. Appl. Environ. Microbiol. 69:6041-6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman, N. V., and J. C. Spain. 2003. Epoxyalkane: coenzyme M transferase in the ethene and vinyl chloride biodegradation pathways of Mycobacterium strain JS60. J. Bacteriol. 185:5536-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danko, A. S., M. Z. Luo, C. E. Bagwell, R. L. Brigmon, and D. L. Freedman. 2004. Involvement of linear plasmids in aerobic biodegradation of vinyl chloride. Appl. Environ. Microbiol. 70:6092-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danko, A. S., C. A. Saski, J. P. Tomkins, and D. L. Freedman. 2006. Involvement of coenzyme M during aerobic biodegradation of vinyl chloride and ethene by Pseudomonas putida strain AJ and Ochrobactrum sp. strain TD. Appl. Environ. Microbiol. 72:3756-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elango, V. K., A. S. Liggenstoffer, and B. Z. Fathepure. 2006. Biodegradation of vinyl chloride and cis-dichloroethene by a Ralstonia sp. strain TRW-1. Appl. Microbiol. Biotechnol. 72:1270-1275. [DOI] [PubMed] [Google Scholar]
- 14.Ensign, S. A. 2001. Microbial metabolism of aliphatic alkenes. Biochemistry 40:5845-5853. [DOI] [PubMed] [Google Scholar]
- 15.Fennell, D. E., A. B. Carroll, J. M. Gossett, and S. H. Zinder. 2001. Assessment of indigenous reductive dechlorinating potential at a TCE-contaminated site using microcosms, polymerase chain reaction analysis, and site data. Environ. Sci. Technol. 35:1830-1839. [DOI] [PubMed] [Google Scholar]
- 16.Furukawa, K., H. Suenaga, and M. Goto. 2004. Biphenyl dioxygenases: functional versatilities and directed evolution. J. Bacteriol. 186:5189-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartmans, S., and J. A. M. de Bont. 1992. Aerobic vinyl chloride metabolism in Mycobacterium aurum L1. Appl. Environ. Microbiol. 58:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartmans, S., J. A. M. de Bont, J. Tramper, and K. C. A. M. Luyben. 1985. Bacterial degradation of vinyl chloride. Biotechnol. Lett. 7:383-388. [Google Scholar]
- 19.Henschler, D. 1994. Toxicity of chlorinated organic-compounds-effects of the introduction of chlorine in organic-molecules. Angew. Chem. Int. Ed. Engl. 33:1920-1935. [Google Scholar]
- 20.Jacobs, W. R., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 21.Janssen, D. B., I. J. T. Dinkla, G. J. Poelarends, and P. Terpstra. 2005. Bacterial degradation of xenobiotic compounds: evolution and distribution of novel enzyme activities. Environ. Microbiol. 7:1868-1882. [DOI] [PubMed] [Google Scholar]
- 22.Jin, Y. O., and T. E. Mattes. 2008. Adaptation of aerobic, ethene-assimilating Mycobacterium strains to vinyl chloride as a growth substrate. Environ. Sci. Technol. 42:4784-4789. [DOI] [PubMed] [Google Scholar]
- 23.Keppler, F., R. Borchars, J. Pracht, S. Rheinberger, and H. Scholer. 2002. Natural formation of vinyl chloride in the terrestrial environment. Environ. Sci. Technol. 36:2479-2483. [DOI] [PubMed] [Google Scholar]
- 24.Kielhorn, J., C. Melber, U. Wahnschaffe, A. Aitio, and I. Mangelsdorf. 2000. Vinyl chloride: still a cause for concern. Environ. Health Perspect. 108:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krum, J. G., and S. A. Ensign. 2000. Heterologous expression of bacterial epoxyalkane:coenzyme M transferase and inducible coenzyme M biosynthesis in Xanthobacter strain Py2 and Rhodococcus rhodochrous B276. J. Bacteriol. 182:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krum, J. G., H. Ellsworth, R. R. Sargeant, G. Rich, and S. A. Ensign. 2002. Kinetic and microcalorimetric analysis of substrate and cofactor interactions in epoxyalkane:CoM transferase, a zinc-dependent epoxidase. Biochemistry 41:5005-5014. [DOI] [PubMed] [Google Scholar]
- 27.Mattes, T. E., A. K. Alexander, and N. V. Coleman. 8 January 2010, posting date. Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology and evolution. FEMS Microbiol. Rev. doi: 10.1111/j.1574-6976.2010.00210.x. [DOI] [PubMed]
- 28.Mattes, T. E., N. V. Coleman, A. S. Chuang, A. J. Rogers, J. C. Spain, and J. M. Gossett. 2007. Mechanism controlling the extended lag period associated with vinyl chloride starvation in Nocardioides sp. strain JS614. Arch. Microbiol. 187:217-226. [DOI] [PubMed] [Google Scholar]
- 29.Mattes, T. E., N. V. Coleman, J. C. Spain, and J. M. Gossett. 2005. Physiological and molecular genetic analyses of vinyl chloride and ethene biodegradation in Nocardioides sp. strain JS614. Arch. Microbiol. 183:95-106. [DOI] [PubMed] [Google Scholar]
- 30.McGuinness, M., C. Ivory, N. Gilmartin, and D. N. Dowling. 2006. Investigation of substrate specificity of wildtype and mutant BphKLB400 (a glutathione S-transferase) from Burkholderia LB400. Int. Biodeterior. Biodegrad. 58:203-208. [Google Scholar]
- 31.Pries, F., A. J. van den Wijngaard, R. Bos, M. Pentenga, and D. B. Janssen. 1994. The role of spontaneous cap domain mutations in haloalkane dehalogenase specificity and evolution. J. Biol. Chem. 269:17490-17494. [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Singh, H., F. E. Loffler, and B. Z. Fathepure. 2004. Aerobic biodegradation of vinyl chloride by a highly enriched mixed culture. Biodegradation 15:197-204. [DOI] [PubMed] [Google Scholar]
- 34.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 35.Squillace, P. J., M. J. Moran, W. W. Lapham, C. V. Price, R. M. Clawges, and J. S. Zogorski. 1999. Volatile organic compounds in untreated ambient groundwater of the United States, 1985-1995. Environ. Sci. Technol. 33:4176-4187. [Google Scholar]
- 36.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 37.Suenaga, H., M. Mitsuoka, Y. Ura, T. Watanabe, and K. Furukawa. 2001. Directed evolution of biphenyl dioxygenase: emergence of enhanced degradation capacity for benzene, toluene, and alkylbenzenes. J. Bacteriol. 183:5441-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Top, E. M., and D. Springael. 2003. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14:262. [DOI] [PubMed] [Google Scholar]
- 39.van der Meer, J. R. 1997. Evolution of novel metabolic pathways for the degradation of chloroaromatic compounds. Antonie Van Leeuwenhoek 71:159-178. [DOI] [PubMed] [Google Scholar]
- 40.van der Meer, J. R., and V. Sentchilo. 2003. Genomic islands and the evolution of catabolic pathways in bacteria. Curr. Opin. Microbiol. 14:248. [DOI] [PubMed] [Google Scholar]
- 41.Verce, M. F., R. L. Ulrich, and D. L. Freedman. 2000. Characterization of an isolate that uses vinyl chloride as a growth substrate under aerobic conditions. Appl. Environ. Microbiol. 66:3535-3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verce, M. F., R. L. Ulrich, and D. L. Freedman. 2001. Transition from cometabolic to growth-linked biodegradation of vinyl chloride by a Pseudomonas sp. isolated on ethene. Environ. Sci. Technol. 35:4242-4251. [DOI] [PubMed] [Google Scholar]
- 43.Woodford, N., and M. J. Ellington. 2007. The emergence of antibiotic resistance by mutation. Clin. Microbiol. Infect. 13:5-18. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, Z. S., K. Peariso, J. E. Penner-Hahn, and R. G. Matthews. 1999. Identification of the zinc ligands in cobalamin-independent methionine synthase (MetE) from Escherichia coli. Biochemistry 38:15915-15926. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.