Abstract
Microviridins are ribosomally synthesized tricyclic depsipeptides produced by different genera of cyanobacteria. The prevalence of the microviridin gene clusters and the natural diversity of microviridin precursor sequences are currently unknown. Screening of laboratory strains and field samples of the bloom-forming freshwater cyanobacterium Microcystis via PCR revealed global occurrence of the microviridin pathway and an unexpected natural variety. We could detect 15 new variants of the precursor gene mdnA encoding microviridin backbones that differ in up to 4 amino acid positions from known isoforms of the peptide. The survey not only provides insights into the versatility of the biosynthetic enzymes in a closely related group of cyanobacteria, but also facilitates the discovery and characterization of cryptic microviridin variants. This is demonstrated for microviridin L in Microcystis aeruginosa strain NIES843 and heterologously produced variants.
Bloom-forming freshwater cyanobacteria are a rich source of natural products (30). They flourish in lakes and ponds of different temperate zones, where they can eventually form thick scums at the surface. Microcystis is one of the predominant genera composing these blooms, particularly in lakes suffering from eutrophication (17). Like other bloom-forming species, it is infamous for the production of the hepatotoxin microcystin, a nonribosomal peptide toxin that inhibits protein phosphatases in a broad range of eukaryotes from zooplankton to humans (1, 9). Moreover, Microcystis is known to produce a multitude of other peptides that are considered to be potential drug leads (14, 30).
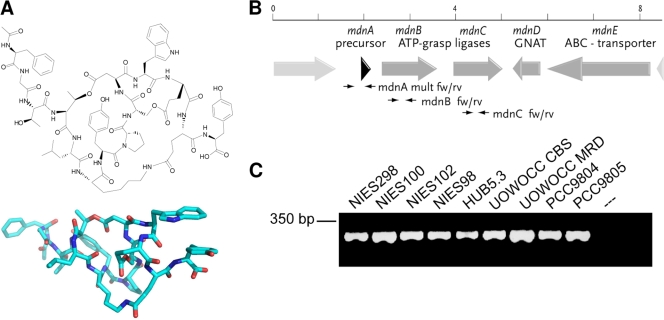
Microviridins form one of the most intriguing classes of peptides, since they feature a cage-like structure (e.g., microviridin B) (Fig. 1A). The highly unusual tricyclic architecture results from ω-ester and ω-amide bonds between amino acid side chains. The past few years have afforded 13 variants of the peptide from both field samples and laboratory strains (5, 8, 13, 15, 21, 22). Most of the variants show inhibitory activities against serine-type proteases, most notably against elastase, which is a target enzyme in the treatment of lung emphysema (28). One of the peptide isoforms, microviridin J, has been shown to inhibit the molting process of Daphnia, leading to death of the animals (23).
FIG. 1.
(A) Representative structure of microviridin B and three-dimensional model. (B) Schematic representation of the microviridin gene cluster of M. aeruginosa NIES298. The positions of primers for the amplification of mdnA, -B, and -C are indicated. GNAT, GCN5-related N-acetyltransferase. (C) PCR gel picture showing the amplification of mdnB from a selection of laboratory strains.
Recent studies have revealed a unique biosynthetic mechanism for microviridins in Microcystis and the filamentous cyanobacterial genus Planktothrix (18, 31). The 14-amino-acid (aa) peptide sequence is encoded at the C terminus of the ribosomal precursor peptide MdnA (31). Macrocyclization of the peptide depends on the activities of two ATP grasp-type ligases, MdnB and -C, that are encoded downstream of the precursor gene. Further enzymes encoded by the cluster include the N-acetyltransferase MdnD and the putative transporter-peptidase MdnE (31) (Fig. 1B). The activities of the enzymes MdnB, -C, and -D were confirmed by heterologous production of microviridins in Escherichia coli (31). The MdnB and -C orthologs of the filamentous cyanobacterial strain Planktothrix agardhii CYA126, MvdC and MvdD, were characterized using synthetic precursor peptides in vitro (18). Furthermore, Philmus et al. have shown limited flexibility of MvdD, catalyzing two rounds of microviridin lactonization to accept substrates with altered amino acid compositions and a stringent ring size requirement (19).
Recent genome-sequencing projects with cyanobacteria have revealed widespread occurrence of microviridin-related gene clusters in various cyanobacterial genera, although the production of microviridins by these strains remains to be shown (18, 24, 31). Here, we present data showing an unprecedented natural diversity of microviridin precursor genes in a set of closely related Microcystis laboratory strains and field samples. The insights gained from our survey provide lessons about the flexibility of the microviridin ligases in a small group of cyanobacteria and an estimate of the size of the natural microviridin library that still awaits discovery. The detection of novel natural microviridin prepeptides from Microcystis can ultimately guide the heterologous production of novel microviridins in E. coli. As proof of principle, we have identified and characterized the new variant microviridin L from the strain Microcystis aeruginosa NIES843.
MATERIALS AND METHODS
Cultivation of strains and DNA isolation.
M. aeruginosa strains were obtained from the Pasteur Culture Collection (PCC) and the National Institute for Environmental Studies (NIES). DNA from laboratory strains was isolated as described previously (2). DNA of additional Microcystis strains was provided by K. Sivonen (Helsinki, Finland). For DNA isolation from field samples, 500 ml of mixed lake water was filtered onto 0.2-μm polycarbonate filters. The DNA was isolated according to acetyltrimethylammonium bromide (CTAB) protocol, as described previously (20).
PCR amplification of microviridin genes.
DNA fragments were amplified by PCR using the Taq DNA Polymerase System (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. The primer pairs mdnB fw (5′-TTGGCTGGTTTTTGGGATAG-3′) and mdnB rv (5′-CGATCGCATTGGAAATAGGT-3′), mdn mult fw (5′-TCACTCGAAATTACCAGAGGAA-3′) and mdn mult rv (5′-CGGTGTAATCAAGAAAAGTGCT-3′), and mdnC fw (5′-GAAGGTTTGCAATTTTGTCCA-3′) and mdnC rv (5′-CGCCAACGGGATTAATTTCT-3′) were used to amplify the mdnB, mdnC, and mdnA genes, respectively, from laboratory strains and field samples. Standard reactions involved initial denaturation at 95°C for 3 min, followed by 35 cycles of 20 s at 95°C for denaturation, 20 s at the respective annealing temperature (according to the melting temperature of the primer pair used), and 30 s to 2 min at 72°C (depending on the product size; 1 min per kilobase). An additional elongation step at 72°C for 10 min completed the reaction.
The resulting PCR products were cloned into the pDrive cloning system (Qiagen, Hilden, Germany) following the standard protocol. Positive clones were sequenced at SmolBio (Berlin, Germany) and analyzed using the ClustalX program version 1.83 with the BLOSUM 62 protein weight matrix.
Fosmid library preparation and screening.
DNA fragments of approximately 30 to 40 kb obtained from M. aeruginosa NIES843 were directly ligated to the pCC1FOS vector (Epicentre Technologies) following the manufacturer's instructions.
Screening of the library was conducted by a PCR-based strategy using the Qiagen Taq DNA Polymerase kit (Hilden, Germany) and the primer pair mdnB fw and mdnB rv as described above. A total of 518 clones were first analyzed in pools of 12 overnight cultures of E. coli EPI300-T1R carrying fosmids. PCR pools were made using 1 μl from each of 12 overnight cultures; 2.5 μl of these pools was used in each pool PCR. If primers amplified a product in a pool PCR, each of the corresponding individual overnight cultures was analyzed separately using 2.5 μl culture as a template in a single-clone PCR. Applying this technique, fosmid Fos303N843, carrying the genes for MdnL production, was identified. DNA sequencing was carried out commercially at SmolBio (Berlin, Germany). Sequence data were analyzed using the NCBI BLAST server.
Microviridin production and mass spectrometric analysis.
E. coli clones carrying the selected fosmid from M. aeruginosa NIES843 were induced to yield high fosmid copy numbers according to the suggestions of the manufacturer (Epicentre Technologies). The cells were grown for 5 h at 37°C and subsequently harvested by centrifugation at 4,000 × g for 10 min. For peptide isolation, cells were washed once with Tris-HCl, pH 7.5; resuspended in deionized water; and lysed by sonication. The cellular debris was removed by centrifugation. The resulting supernatant was loaded on a Sep-Pak cartridge (Waters, Eschborn, Germany), which was then washed with 5% methanol. The components of interest were eluted with 100% methanol and concentrated in vacuo. Dried extracts were resuspended in 200 μl of 50% methanol and filtered (Acrodisc 4-mm syringe filter; 0.45-μm nylon membrane; Pall). High-performance liquid chromatography (HPLC) separation of cell extract and supernatant samples was conducted on a Shimadzu HPLC unit comprising the system controller SCL-10AVP, the pump LC-10Ai, the autosampler SIL-10A, the fraction collector FRC-10A, and the photo diode array detector SPD-M-10AVP. Separation was carried out on a SymmetryShield RP18 column (Waters, Eschborn, Germany) with a particle size of 3.5 μm, a 4.6-mm inner diameter, and a 100-mm length and a precolumn (3.9 by 20 mm) with an identical sorbent. The following gradient system was used at a flow rate of 1.0 ml min−1: 1 min of loading (20% solvent B), a linear gradient up to 52% solvent B within 30 min, followed by a linear gradient to 100% solvent B in 1 min, and then holding 100% B for 3 min (solvent A, 0.1% trifluoroacetic acid [TFA] in H2O; solvent B, acetonitrile, 0.1%TFA). Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) and TOF/TOF (postsource decay [PSD]) were performed with a Bruker Ultraflex (Bruker Daltonics).
Purification of microviridin L and variants.
Two liters of E. coli EPI300-T1R 843 cells cultured at 37°C for 5 h was harvested by centrifugation. The pelleted cells were resuspended in 80 ml of methanol and then lysed by sonication. After removal of the solvent by evaporation, the residues were dissolved in methanol-H2O (1:2). The membrane-filtered samples were subjected to reverse-phase high-performance liquid chromatography (RP-HPLC) (Supercosil LC18; 21.2 by 250 mm; 5 μm; flow rate, 10 ml min−1) using a gradient system of 20% solvent B for 10 min to 100% solvent B in 40 min (solvent A, 0.1% TFA in H2O; solvent B, acetonitrile-H2O, 83:17) to yield microviridin L1 (1.6 mg), microviridin L (1.1 mg), microviridin L2 (1.5 mg), and microviridin L3 (3.4 mg).
Protease inhibition assays.
Evaluation of serine protease inhibitory activities was carried out by the methods previously described (29). Porcine pancreas trypsin (T7409; Sigma), porcine pancreas elastase (E1250; Sigma), and bovine pancreas chymotrypsin (C4129; Sigma) were used for the assay.
Nucleotide sequence accession numbers.
The mdnB fragment sequences were submitted to the EMBL database under accession numbers FN668540 to FN668556. All mdnA PCR fragments were sequenced, and the corresponding data were deposited in the EMBL database (accession numbers FN667620 to FN667625). The sequences of novel microviridin precursor variants were deposited in the EMBL database under accession numbers FN668693 to FN668700.
RESULTS AND DISCUSSION
To elucidate the distribution of mdn biosynthesic pathways within the genus Microcystis and to discover new microviridin variants, we screened a collection of laboratory strains (listed in Table 1) isolated from various globally distributed bodies of water for orthologous gene clusters. Using two mdn biosynthetic gene clusters from Microcystis species, we designed PCR primers for the detection of genes with high similarity to the microviridin ligase genes mdnB and mdnC. Using these primers, we obtained partial gene fragments (298 bp and 309 bp) of mdnB and mdnC, respectively, for all 16 of the Microcystis strains investigated (Fig. 1C shows selected data). This suggests a global occurrence of the microviridin biosynthetic genes rather than a sporadic distribution, as shown for microcystin and other nonribosomal peptide biosynthetic genes in strains of Microcystis (11, 12, 25). An additional PCR approach using an mdnB forward primer with an mdnC reverse primer revealed that the two genes were generally clustered in all strains analyzed (data not shown). mdnB fragments were sequenced and revealed between 96 and 99% identity to the mdnB sequence of M. aeruginosa NIES298 (see Fig. S1 in the supplemental material for an alignment). The high sequence similarity is characteristic of Microcystis strains that are closely related according to the analysis of their 16S-23S intergenic spacer regions (16). It can be anticipated that microviridin gene clusters of Microcystis are derived from a common ancestor and were not horizontally acquired from other bacterial groups. Prior to this study, microviridins were only incidentally discovered in mass spectrometric surveys of Microcystis cell extracts (12). The PCR approach revealed a far more widespread prevalence of microviridin genes than expected and provided a more comprehensive picture than chemical analysis alone.
TABLE 1.
Presence of mdnB, mdnC, and mdnA amplicons in Microcystis strains with different geographic origins
| Microcystis strain | Geographic origin | Presencea of: |
|
|---|---|---|---|
| mdnB/C | mdnA | ||
| HUB5.3 | Lake Pehlitzsee, Germany | +/+ | − |
| Izancya | Lake Mira, Portugal | +/+ | + |
| 199 | Lake Rusutjärvi, Finland | +/+ | + |
| 269 | River Raisionjoki, Finland | +/+ | − |
| NIES100 | Lake Kasumigaura, Japan | +/+ | + |
| NIES101 | Lake Suwa, Japan | +/+ | − |
| NIES102 | Lake Kasumigaura, Japan | +/+ | + |
| NIES843 | Lake Kasumigaura, Japan | +/+ | + |
| PCC7005 | Lake Mendota, United States | +/+ | + |
| PCC7806 | Braakman Reservoir, Netherlands | +/+ | − |
| PCC9354 | Little Rideau Lake, Canada | +/+ | − |
| PCC9603 | Okesund Dam, Sweden | +/+ | − |
| PCC9804 | Canberra, Australia | +/+ | − |
| PCC9805 | Canberra, Australia | +/+ | + |
| PCC9812 | Lake Mendota, United States | +/+ | − |
| UOWOCC MRD | Malpas Dam, Australia | +/+ | + |
| UOWOCC CBS | Malpas Dam, Australia | +/+ | + |
+, present; −, absent.
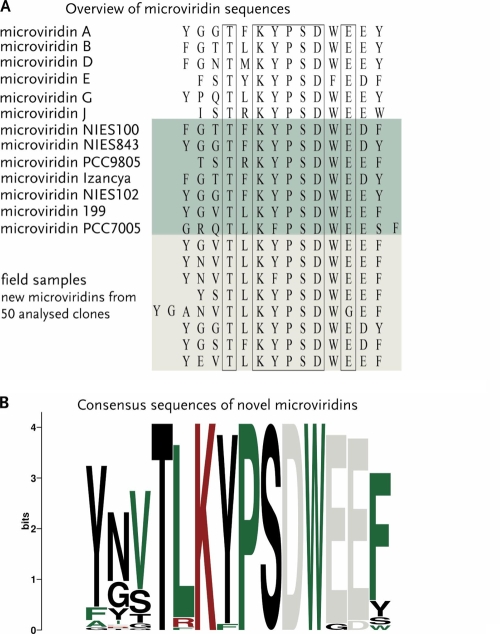
Additionally, we analyzed the same strains in a second PCR approach amplifying the entire precursor gene mdnA. Nine candidate prepeptide genes were identified and sequenced. It was not possible to amplify precursor genes in the other seven Microcystis strains using the applied primers. Due to the small size of the mdnA genes and the prerequisite to include the region encoding the C-terminal precursor sequence, the primers were designed in conserved flanking regions. The fact that no PCR product was obtained thus does not necessarily imply that the strains lack the capacity to produce microviridin-like peptides. Rather, it is possible that the orthologous genes in these strains are in a different order or in an independent position in the genome, as is the case in the strain M. aeruginosa PCC7806. In this genome, mdnB, -C, -D, and -E are clustered, whereas the mdnA gene is located in a completely different region of the genome adjacent to a nonribosomal peptide synthetase gene cluster (3). All mdnA PCR fragments were sequenced. The sequences of the region encoding the N-terminal leader sequence of MdnA were completely identical, whereas remarkable variation was found in the region encoding the amino acid sequences of microviridins at the C terminus (see Fig. S2 in the supplemental material for an alignment). No indication of multiple copies of the mdn gene cassette in a single genome was found. The deduced amino acid sequences of the nine strains yielding a PCR product all show the characteristic KYPSD motif as part of the peptide coding regions at their C termini, except in strain PCC7005, where the tyrosine is replaced by a phenylalanine. Seven of the nine microviridin precursors are novel and differ in up to 4 amino acid positions compared to known variants (Fig. 2A). Remarkably, only one of the precursors occurred twice in the small subset of sequences analyzed, indicating that the combinatorial natural library of microviridins is much larger than expected.
FIG. 2.
Diversity of deduced microviridin precursors in Microcystis. (A) Known and newly discovered amino acid sequences of microviridins in a variety of Microcystis strains. A selection of known microviridin amino acid sequences are shown. Novel microviridin sequences from Microcystis laboratory strains detected in this study are shaded in green and those from field samples in gray-green; conserved amino acids are boxed. (B) The weblogo consists of stacks of symbols, one stack for each position in the sequence. The overall height of the stack indicates the sequence conservation at that position, while the heights of symbols within the stack indicate the relative frequency of each amino acid at that position.
To determine whether the variability of microviridins in a set of closely related laboratory strains is echoed by equal variety in their natural habitats, an additional PCR approach was applied to field samples of Microcystis blooms from the Braakman reservoir (Netherlands) and from a bay on the Baltic Sea in Mecklenburg-Western Pomerania (Germany). To circumvent the time-consuming process of Microcystis strain isolation and culturing, a metagenomic technique was applied. DNA was isolated directly from the filtered field samples and subsequently used for PCR amplification; the PCR products were cloned and transformed into E. coli. Sequencing of approximately 50 clones revealed 9 different microviridin precursor variants (Fig. 2A; see Fig. S2 in the supplemental material), suggesting a wide distribution of microviridin genes in these habitats. Only one of the detected sequences matched a known variant, namely, microviridin B, whereas eight sequences were novel. It can be estimated that the investigation of further field samples and optimization of the PCR approach could dramatically expand the library of natural variants of the tricyclic depsipeptide.
Comparison of microviridin variants described in the literature or predicted in this study revealed a size range from 13 to 16 amino acids and the consensus sequence TXK(Y/W/F)PSDW(E/G)(E/D). To get a graphical display of the conservation of the amino acids, a sequence logo was designed (http://weblogo.berkeley.edu/logo.cgi) displaying the conservation of each position and the relative frequency of the amino acid at that particular position. The microviridin logo (Fig. 2B) clearly shows the highest variability in the first three and the last positions, whereas all precursor peptides contain the microviridin core motif. Bioactivity assays of the microviridin ligases of P. agardhii CYA126/8 with manipulated precursor peptides revealed that the cross-linking of microviridins is strictly ordered. First, the large lactone ring is formed between threonine and aspartate, followed by the second lactone and the lactam ring closure (19). In agreement with these data, threonine and aspartate were found to be strictly conserved in the natural library of microviridins. In the in vitro approach, alanine substitutions in any amino acid of the core motif resulted in the lack of at least the last two cyclizations. The strict conservation of the KYPSD core motif of microviridins may thus reflect the limited flexibility of the ligases MdnB and MdnC; however, it may also have some relevance to the bioactivity and ecological role of the microviridins.
Following our survey of the genetic potential for microviridin production in Microcystis, we intended to identify microviridins in selected cyanobacterial strains based on their exact mass as predicted from the corresponding precursor peptides; however, in the three strains tested, we could not detect appropriate mass peaks. Microviridins might be the subject of complex regulatory mechanisms in cyanobacteria or be produced in very small amounts, as indicated from the infrequency with which they are detected in cyanobacterial peptide screenings. Furthermore, cyanobacterial species such as Microcystis are slow growing and poorly amenable to genetic manipulations. We therefore considered the transfer of the microviridin pathways into a suitable heterologous host a more promising approach. We have recently reported the heterologous production of microviridins B and J in E. coli (31) and consider heterologous expression of further cryptic microviridin gene clusters a promising approach for the mining of novel tricyclic depsipeptide variants. As proof of principle, we chose the mdn locus of Microcystis aeruginosa NIES843. The genome of the strain was completely sequenced (10), confirming the presence of the complete set of genes required for the production of correctly processed microviridin, including genes encoding microviridin ligases, the N-acetyltransferase, and the ABC-type transporter in a single gene cluster. According to the amino acid sequence encoded at the C terminus of the NIES843 MdnA precursor peptide, we predicted a tricyclic microviridin variant composed of the amino acid sequence YGGTFKYPSDWEDY, acetylated at its N-terminal tyrosine.
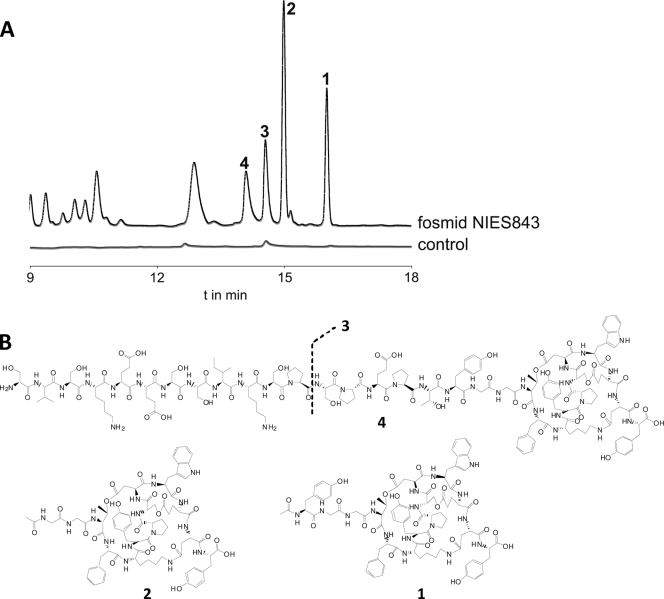
We constructed a fosmid library and screened it for clones carrying the mdn gene cluster. Positive clones were tested for additional peaks in the HPLC profile. The chromatograms showed several peaks in the mdn fosmid cell extracts that were not present in the controls (Fig. 3A and B). MALDI PSD analysis of the fractionated peaks identified them as microviridin-like peptides that all contained the correct tricyclic structure at their C termini (Fig. 3B). The detected monoisotopic ions were in agreement with partial amino acid sequences encoded by mdnA and with predicted cyclizations. The four isolated microviridin isoforms differed in the lengths of their N termini (Fig. 3B; see Fig. S3 and S4 in the supplemental material). More detailed PSD analysis of the tricyclic core of microviridin L variants proposed that all heterologously expressed peptides had the same tricyclized structure as microviridin B and its variants (13) (see Fig. S5 and S6 in the supplemental material). Additionally, microviridin L (Fig. 3, 1) and L1 (2) were not stained by ninhydrin, supporting the notion that the N-terminal amino groups of 3 and 4 were acetylated (data not shown).
FIG. 3.
(A) HPLC profiles of extracts from E. coli cultures harboring the mdn fosmid from NIES843. Shown are microviridins L (1), L1 (2), L2 (3), and L3 (4). (B) Structures of microviridin L (1) and its variants (2 to 4).
The 1,715-Da variant 1 was predicted to be authentic microviridin produced by M. aeruginosa NIES843 based on the analysis of the MdnA coding sequence. We could indeed detect small amounts of this microviridin after thorough analysis using MALDI postsource decay analysis. The corresponding microviridin was named microviridin L, following the common nomenclature for microviridins, and the additional peaks in E. coli were considered falsely processed side products.
Although the falsely processed variants of microviridin L account for a large proportion of the microviridin detected in the E. coli cells, 1.1 mg of microviridin L (1) was isolated from only 2 liters of cell culture. Compared to the culturing efforts required for the isolation of other microviridins (Table 2), heterologous expression proved to be extremely effective. Microviridin L (1) and the three major side products (2 to 4) produced in E. coli were tested against elastase, trypsin, and chymotrypsin and compared with known variants (Table 3 shows selected data from the literature). Although the inhibitory activity of the novel microviridin is only in the micromolar range, the high growth rate of the host bacteria and the relatively high production rate offer possibilities for a more systematic screening against a number of additional targets in the future. The detected inhibitory activity of 1 indicated that one of the N-terminal amino acids YGG and/or F at position 11 is crucial for the activities, because microviridin A, which has an amino acid sequence similar to that of 1, does not show any inhibitory activities. Comparison of the data obtained in our study with data from the literature can thus provide valuable insights into the structure-activity relationship (Table 3).
TABLE 2.
Comparison of microviridin yields and culture conditions from the literature and the present study
| Microviridin | Yield/100 g dried cells (mg) | Medium vol (liters) | Cultivation period | Organism | Reference |
|---|---|---|---|---|---|
| A | 87,0 | NDa | 10-14 days | Microcystis | 8 |
| B | 40,0 | 362 | 10-14 days | Microcystis | 15 |
| C | 18,0 | 362 | 10-14 days | Microcystis | 15 |
| D | 6,4 | 400 | 10-14 days | Planktothrix | 29 |
| E | 9,1 | 400 | 10-14 days | Planktothrix | 29 |
| F | 5,5 | 400 | 10-14 days | Planktothrix | 29 |
| G | 3,0 | 590 | 25 days | Nostoc | 13 |
| H | 6,9 | 590 | 25 days | Nostoc | 13 |
| I | 79,1 | ND | ND | Planktothrix | 4 |
| J | 263,3 | ND | ND | Microcystis | 22 |
| SD1684 | 19,9 | ND | ND | Microcystis | 21 |
| SD1634 | 10,0 | ND | ND | Microcystis | 21 |
| SD1652 | 7,8 | ND | ND | Microcystis | 21 |
| L | ∼60-70 | 2 | 5 h | E. coli | This work |
ND, not determined.
TABLE 3.
Compilation of inhibitory activities and amino acid sequences of different microviridins from the literature and the present study
| Microviridin | Mass (Da) | Inhibitory activitya |
Amino acid sequenceb |
Reference | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Elastase | Chymotrypsin | Trypsin | T | X1 | K | Y | P | S | D | X2 | E | E/D | X3 | |||
| A | 1,730 | >58 μM | >58 μM | >58 μM | Est | F | Est | Est | W | Est | E | Y | 8 | |||
| B | 1,722 | 26 nM | 1.5 μM | 34 μM | Est | L | Est | Est | W | Est | E | Y | 15 | |||
| C | 1,754 | 48 nM | 2.8 μM | 18 μM | Est | L | OH | Est | W | Me | E | Y | 15 | |||
| D | 1,802 | 388 nM | 0.7 μM | >55 μM | Est | M | OH | Est | W | Me | D | Y | 29 | |||
| E | 1,666 | 360 nM | 0.7 μM | >60 μM | Est | Y | OH | Est | F | Me | D | F | 29 | |||
| F | 1,682 | 3.4 μM | >59 μM | >59 μM | OH | Y | OH | OH | F | Me | D | F | 29 | |||
| G | 1,805 | 10 nM | 0.78 μM | >55 μM | Est | L | Est | Est | W | Est | E | Y | 13 | |||
| H | 1,837 | 17 nM | 1.6 μM | >54 μM | Est | L | OH | Est | W | Me | E | Y | 13 | |||
| I | 1,764 | 193 nM | ND | ND | Est | L | Est | Est | W | Est | D | Y | 4 | |||
| J | 1,684 | >6.0 μM | 1.7 μM | 20-90 nM | Est | R | Est | Est | W | Est | E | W | 22 | |||
| SD1684 | 1,684 | ND | inactive | inactive | OH | R | OH | OH | W | Me | HO-D | Y | 21 | |||
| SD1634 | 1,634 | ND | 16 μM | 8.2 μM | Est | R | Est | Est | W | Est | HO-D | Y | 21 | |||
| SD1652 | 1,652 | ND | inactive | inactive | OH | R | Est | OH | W | Est | HO-D | Y | 21 | |||
| L | 1,715 | >58 μM | 42 μM | 58 μM | Est | F | Est | Est | W | Est | D | Y | This work | |||
ND, not determined.
The N terminus is on the left. X1, X2, and X3 are variable amino acids. Est, amino acid side chain forms an ester bond; Me, γ-carboxyl group forms methylester; HO-D, β-carboxyl group does not form an amide bond with Lys; OH, Thr and Ser do not form ester bonds with Asp and Glu, respectively.
The most important advantage of the heterologous-expression approach is the possibility to produce microviridins from metagenomic libraries independently of extensive strain isolation and cultivation. The PCR survey in just two habitats has revealed the potential of such a metagenomic mining approach. It is likely that screening of fosmid libraries constructed from cyanobacterial field samples combined with inhibition assays could yield novel tricyclic depsipeptide structures with a range of interesting bioactivities in the future. In the last few years, enormous progress has been made in establishing new protocols for the discovery of natural products from environmental sources (6, 7, 24, 26). Ribosomal peptide families from cyanobacteria, such as cyanobactins (11, 27, 32) and microviridins (19, 31), are at the forefront of this development and may allow the further expansion and diversification of natural peptide libraries.
Supplementary Material
Acknowledgments
We thank Maria Pötsch for MALDI-TOF MS measurements and Arthur Guljamow for metagenomic DNA isolations.
This work was supported by a grant from the BMBF (02WT0799) to E.D. and by the Pakt für Forschung und Innovation of the Free State of Thuringia and the Federal Ministry of Science and Technology (BMBF, Germany) and the DFG-funded excellence graduate school Jena School for Microbial Communication (JSMC) to C.H.
Footnotes
Published ahead of print on 2 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Dittmann, E., and C. Wiegand. 2006. Cyanobacterial toxins—occurrence, biosynthesis and impact on human affairs. Mol. Nutr. Food Res. 50:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Franche, C., and T. Damerval. 1988. Tests on Nif probes and DNA hybridizations. Methods Enzymol. 167:803-808. [Google Scholar]
- 3.Frangeul, L., P. Quillardet, A. M. Castets, J. F. Humbert, H. C. Matthijs, D. Cortez, A. Tolonen, C. C. Zhang, S. Gribaldo, J. C. Kehr, Y. Zilliges, N. Ziemert, S. Becker, E. Talla, A. Latifi, A. Billault, A. Lepelletier, E. Dittmann, C. Bouchier, and N. T. de Marsac. 2008. Highly plastic genome of Microcystis aeruginosa PCC 7806, a ubiquitous toxic freshwater cyanobacterium. BMC Genomics 9:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujii, K., K. Sivonen, E. Naganawa, and K. Harada. 2000. Non-toxic peptides from toxic cyanobacteria, Oscillatoria agardhii. Tetrahedron 56:725-733. [Google Scholar]
- 5.Gesner-Apter, S., and S. Carmeli. 2009. Protease inhibitors from a water bloom of the cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 72:1429-1436. [DOI] [PubMed] [Google Scholar]
- 6.Gross, H. 2009. Genomic mining—a concept for the discovery of new bioactive natural products. Curr. Opin. Drug Discov. Dev. 12:207-219. [PubMed] [Google Scholar]
- 7.Hertweck, C. 2009. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 48:4688-4716. [DOI] [PubMed] [Google Scholar]
- 8.Ishitsuka, M. O., T. Kusumi, H. Kakisawa, K. Kaya, and M. M. Watanabe. 1990. Microviridin: a novel tricyclic depsipeptide from the toxic cyanobacterium Microcystis viridis. J. Am. Chem. Soc. 112:8180-8182. [Google Scholar]
- 9.Jochimsen, E. M., W. W. Carmichael, J. S. An, D. M. Cardo, S. T. Cookson, C. E. Holmes, M. B. Antunes, D. A. de Melo Filho, T. M. Lyra, V. S. Barreto, S. M. Azevedo, and W. R. Jarvis. 1998. Liver failure and death after exposure to microcystins at a hemodialysis center in Brazil. N. Engl. J. Med. 338:873-878. [DOI] [PubMed] [Google Scholar]
- 10.Kaneko, T., N. Nakajima, S. Okamoto, I. Suzuki, Y. Tanabe, M. Tamaoki, Y. Nakamura, F. Kasai, A. Watanabe, K. Kawashima, Y. Kishida, A. Ono, Y. Shimizu, C. Takahashi, C. Minami, T. Fujishiro, M. Kohara, M. Katoh, N. Nakazaki, S. Nakayama, M. Yamada, S. Tabata, and M. M. Watanabe. 2007. Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Res. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leikoski, N., D. P. Fewer, and K. Sivonen. 2009. Widespread occurrence and lateral transfer of the cyanobactin biosynthesis gene cluster in cyanobacteria. Appl. Environ. Microbiol. 75:853-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martins, J., M. L. Saker, C. Moreira, M. Welker, J. Fastner, and V. M. Vasconcelos. 2009. Peptide diversity in strains of the cyanobacterium Microcystis aeruginosa isolated from Portuguese water supplies. Appl. Microbiol. Biotechnol. 82:951-961. [DOI] [PubMed] [Google Scholar]
- 13.Murakami, M., Q. Sun, K. Ishida, H. Matsuda, T. Okino, and K. Yamaguchi. 1997. Microviridins, elastase inhibitors from the cyanobacterium Nostoc minutum (NIES-26). Phytochemistry 45:1197-1202. [Google Scholar]
- 14.Namikoshi, M., and K. L. Rinehart. 1996. Bioactive compounds produced by cyanobacteria. J. Ind. Microbiol. Biotechnol. 17:373-384. [Google Scholar]
- 15.Okino, T., H. Matsuda, M. Murakami, and K. Yamaguchi. 1995. New microviridins, elastase inhibitors from the blue-green alga Microcystis aeruginosa. Tetrahedron 51:10679-10686. [Google Scholar]
- 16.Otsuka, S., S. Suda, S. Shibata, H. Oyaizu, S. Matsumoto, and M. M. Watanabe. 2001. A proposal for the unification of five species of the cyanobacterial genus Microcystis Kutzing ex Lemmermann 1907 under the rules of the Bacteriological Code. Int. J. Syst. Evol. Microbiol. 51:873-879. [DOI] [PubMed] [Google Scholar]
- 17.Paerl, H. W., R. S. Fulton III, P. H. Moisander, and J. Dyble. 2001. Harmful freshwater algal blooms, with an emphasis on cyanobacteria. ScientificWorldJournal 1:76-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Philmus, B., G. Christiansen, W. Y. Yoshida, and T. K. Hemscheidt. 2008. Post-translational modification in microviridin biosynthesis. Chembiochem. 9:3066-3073. [DOI] [PubMed] [Google Scholar]
- 19.Philmus, B., J. P. Guerrette, and T. K. Hemscheidt. 2009. Substrate specificity and scope of MvdD, a GRASP-like ligase from the microviridin biosynthetic gene cluster. ACS Chem. Biol. 4:429-434. [DOI] [PubMed] [Google Scholar]
- 20.Porebski, S., L. G. Bailey, and B. R. Baum. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15:8-15. [Google Scholar]
- 21.Reshef, V., and S. Carmeli. 2006. New microviridins from a water bloom of the cyanobacterium Microcystis aeruginosa. Tetrahedron 62:7361-7369. [Google Scholar]
- 22.Rohrlack, T., K. Christoffersen, P. E. Hansen, W. Zhang, O. Czarnecki, M. Henning, J. Fastner, M. Erhard, B. A. Neilan, and M. Kaebernick. 2003. Isolation, characterization, and quantitative analysis of microviridin J, a new Microcystis metabolite toxic to Daphnia. J. Chem. Ecol. 29:1757-1770. [DOI] [PubMed] [Google Scholar]
- 23.Rohrlack, T., K. Christoffersen, M. Kaebernick, and B. A. Neilan. 2004. Cyanobacterial protease inhibitor microviridin J causes a lethal molting disruption in Daphnia pulicaria. Appl. Environ. Microbiol. 70:5047-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rounge, T. B., T. Rohrlack, A. J. Nederbragt, T. Kristensen, and K. S. Jakobsen. 2009. A genome-wide analysis of nonribosomal peptide synthetase gene clusters and their peptides in a Planktothrix rubescens strain. BMC Genomics 10:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saker, M. L., M. Welker, and V. M. Vasconcelos. 2007. Multiplex PCR for the detection of toxigenic cyanobacteria in dietary supplements produced for human consumption. Appl. Microbiol. Biotechnol. 73:1136-1142. [DOI] [PubMed] [Google Scholar]
- 26.Scherlach, K., and C. Hertweck. 2009. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 7:1753-1760. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt, E. W., and M. S. Donia. 2009. Cyanobactin ribosomally synthesized peptides-a case of deep metagenome mining. Methods Enzymol. 458:575-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapiro, S. D. 2002. Proteinases in chronic obstructive pulmonary disease. Biochem. Soc. Trans. 30:98-102. [DOI] [PubMed] [Google Scholar]
- 29.Shin, H. J., M. Murakami, H. Matsuda, and K. Yamaguchi. 1996. Microviridins D-F, serine protease inhibitors from the cyanobacterium Oscillatoria agardhii (NIES-204). Tetrahedron 52:8159-8168. [Google Scholar]
- 30.Welker, M., and H. von Döhren. 2006. Cyanobacterial peptides: nature's own combinatorial biosynthesis. FEMS Microbiol. Rev. 30:530-563. [DOI] [PubMed] [Google Scholar]
- 31.Ziemert, N., K. Ishida, A. Liaimer, C. Hertweck, and E. Dittmann. 2008. Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angew. Chem. Int. Ed. Engl. 47:7756-7759. [DOI] [PubMed] [Google Scholar]
- 32.Ziemert, N., K. Ishida, P. Quillardet, C. Bouchier, C. Hertweck, N. T. de Marsac, and E. Dittmann. 2008. Microcyclamide biosynthesis in two strains of Microcystis aeruginosa: from structure to genes and vice versa. Appl. Environ. Microbiol. 74:1791-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.