Abstract
Populations of a moderately thermophilic magnetotactic bacterium were discovered in Great Boiling Springs, Nevada, ranging from 32 to 63°C. Cells were small, Gram-negative, vibrioid to helicoid in morphology, and biomineralized a chain of bullet-shaped magnetite magnetosomes. Phylogenetically, based on 16S rRNA gene sequencing, the organism belongs to the phylum Nitrospirae.
Magnetotactic bacteria are a metabolically, morphologically, and phylogenetically heterogeneous group of prokaryotes that passively align and actively swim along magnetic field lines (3). This behavior, called magnetotaxis, is due to the presence of intracellular, membrane-bounded, single-magnetic-domain crystals of magnetite (Fe3O4) and/or greigite (Fe3S4) (3).
Most known cultured magnetotactic bacteria are mesophilic and do not grow much above 30°C (e.g., Magnetospirillum species and Desulfovibrio magneticus strains MV-1 and MC-1 [D. A. Bazylinski, unpublished data]). Uncultured magnetotactic bacteria have been observed in numerous habitats that were mostly at 30°C and below. There is only one report describing thermophilic magnetotactic bacteria despite a number of efforts to look for them (e.g., in hydrothermal vents [D. A. Bazylinski, unpublished data]). Nash (12) reported the presence of thermophilic magnetotactic bacteria in microbial mats at about 45 to 55°C adjacent to the main flow in Little Hot Creek (but not in other springs in the same area at 40 to 80°C) and in microbial mats of other springs in central California at up to 58°C, all on the east side of the Sierra Nevada mountains. Cells biomineralized bullet-shaped crystals of magnetite and were phylogenetically affiliated with the phylum Nitrospirae (12). Few additional details were provided regarding the organisms and their habitat.
In this study, water and surface sediment samples were taken from the Great Boiling Springs (GBS) geothermal field in Gerlach, NV. GBS is a series of hot springs that range from ambient temperature to ∼96°C (2, 5). The geology, chemistry, and microbial ecology of the springs have been described in some detail (2, 5). The pHs of the samples ranged from 6.4 to 7.5, while the salinities were about 4 to 5 ppt, as determined with a handheld Palm Abbe PA203 digital refractometer (MISCO Refractometer, Cleveland, OH). Samples were examined for the presence of magnetotactic bacteria using the hanging drop technique on-site and in the laboratory at room temperature with and without magnetic enrichment of the sample (15). Some samples taken back to the laboratory were kept at an elevated temperature (∼62°C), while others were kept at ambient temperature. There did not appear to be a significant difference in the number of magnetotactic cells in samples taken back to the laboratory and kept at these two temperatures. Only one morphotype of magnetotactic bacteria was found in samples from nine springs whose temperatures ranged from 32 to 63°C, and we estimate their numbers to be between 103 to 105 cells ml−1 in surface sediments in sample bottles. We did not observe magnetotactic cells of this type in a large number of springs or pools that were at <32°C. Only one spring positive for the presence of these magnetotactic bacteria had sediment that was partially covered with a microbial mat, while sediment at most of the springs was dark gray in color. Cells were small (1.8 ± 0.4 by 0.4 ± 0.1 μm; n = 59), Gram negative, vibrioid to helicoid in morphology, and possessed a single polar flagellum (Fig. 1A). Magnetotactic bacteria were not observed in springs that were at 67°C and above, suggesting the maximum survival and perhaps growth temperature for the organism is about 63°C. In the lab, cells remained viable and motile in samples kept at 25 to 62°C for several months. We refer to this organism as strain HSMV-1.
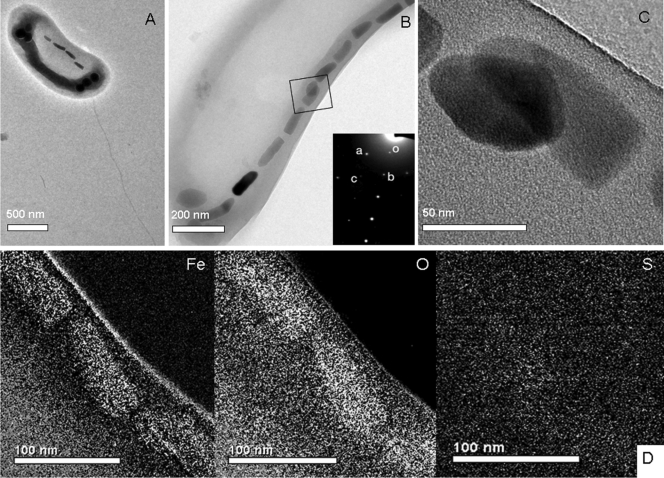
FIG. 1.
Transmission electron microscope (TEM) images of cells and magnetosomes of strain HSMV-1. (A) TEM image of unstained cell of HSMV-1 showing a single polar flagellum and a single chain of bullet-shaped magnetosomes. The electron-dense structures at the poles were found to be phosphorus-rich based on energy-dispersive X-ray analysis (data not shown) and therefore likely represent polyphosphate granules. (B) Higher-magnification TEM image of the magnetosome chain. (C) High-magnification TEM image of magnetosomes from which a selected area electron diffraction (SAED) pattern was obtained (inset of B). The SAED pattern corresponds to the [1 0−1] zone of magnetite, Fe3O4: reflection o, (0 0 0); reflection a, (1 −1 1) (0.48 nm); reflection b, (1 1 1) (0.48 nm); reflection c, (2 0 2) (0.30 nm); angle a-o-b, 70.5°. (D) Iron, sulfur, and oxygen elemental maps, derived from energy-filtering transmission electron microscopy (EFTEM), showing that the positions of the magnetosome crystals correlate with increased concentrations of Fe and O, but not S, consistent with the iron oxide magnetite (Fe3O4).
Cells of HSMV-1 biomineralized a single chain of magnetosomes that traversed the cells along their long axis (Fig. 1A to C). Selected area electron diffraction (SAED) and energy-filtering transmission electron microscopy (EFTEM) elemental maps were determined on magnetosome crystals using a Tecnai model G2 F30 Super-Twin transmission electron microscope (FEI Company, Hillsboro OR). SAED patterns of HSMV-1 magnetosome crystals (Fig. 1B, inset) indicated that they consisted of magnetite, while EFTEM elemental maps (Fe, O and S) (Fig. 1D) clearly showed that the crystals consisted of an iron oxide and not an iron sulfide, again consistent with the mineral magnetite. Cells contained an average of 12 ± 6 magnetosome crystals per cell (n = 15 cells) that averaged 113 ± 34 by 40 ± 5 nm in size (n = 179). A plot of the length of the crystals as a function of the shape factor (width/length ratio) is provided in Figure S1 in the supplemental material and shows that the crystals fit in the theoretical single-magnetic-domain size range (4), along with all known mature magnetosome magnetite crystals from magnetotactic bacteria (3).
Whole-cell PCR amplification of the 16S rRNA gene was performed by first magnetically purifying cells of HSMV-1 using the “capillary racetrack” described by Wolfe et al. (18). Purity of the collected cells was determined by microscopic examination, and contaminating cells were never observed. The 16S rRNA gene was amplified using bacteria-specific primers 27F 5′-AGAGTTTGATCMTGGCTCAG-3′ and 1492R 5′-TACGGHTACCTTGTTACGACTT-3′ (11). PCR products were cloned into pGEM-T Easy vector (Promega Corporation, Madison, WI) and sequenced (Functional Biosciences, Inc., Madison, WI). Six of eight clones sequenced had identical inserts.
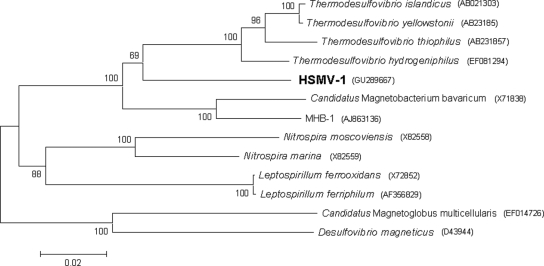
Alignment of 16S rRNA gene sequences was performed using the CLUSTAL W multiple alignment accessory application in the BioEdit sequence alignment editor (7). Phylogenetic trees were constructed using MEGA version 4 (17) by applying the neighbor-joining method (14). Bootstrap values were calculated with 1,000 replicates. The 16S rRNA gene sequence of strain HSMV-1 places the organism in the phylum Nitrospirae (Fig. 2), with its closest relative in culture being Thermodesulfovibrio hydrogeniphilus (87.2% identity) (8). Two other uncultured magnetotactic bacteria are phylogenetically affiliated with the phylum Nitrospirae, including the unnamed rod-shaped bacterium strain MHB-1 (86.5% identity) (6) and the very large Candidatus Magnetobacterium bavaricum (86.4% identity) (16). Interestingly, all the magnetotactic bacteria associated with the phylum Nitrospirae thus far (e.g., Candidatus Magnetobacterium bavaricum) contain bullet-shaped magnetite crystals in their magnetosomes.
FIG. 2.
Phylogenetic tree based on 16S rRNA gene sequences showing the phylogenetic position of strain HSMV-1 in the phylum Nitrospirae. Bootstrap values at nodes are percentages of 1,000 replicates. The magnetotactic bacteria Desulfovibrio magneticus and Candidatus Magnetoglobus multicellularis (outgroup; deltaproteobacteria) were used to root the tree. GenBank accession numbers are given in parentheses. Bar represents 2% sequence divergence.
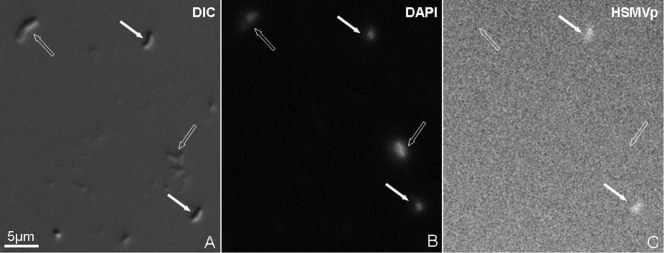
Fluorescent in situ hybridization (FISH) was used to authenticate the 16S rRNA gene sequence. A specific Alexa594-labeled probe for HSMV-1 was designed (HSMVp, 5′-CCTTCGCCACAGGCCTTCTA-3′, complementary to nucleotides 690 to 709 of the 16S rRNA molecule) based on the alignment of 10 of the most similar 16S rRNA gene sequences found in GenBank after BLAST analysis (1) and on cultivated members of the phylum Nitrospirae. FISH with the Alexa594-labeled probe was carried out after fixation of magnetically concentrated cells directly on the wells of gelatin-coated hydrophobic microscope slides with 4% paraformaldehyde. FISH was performed according to the work of Pernthaler et al. (13). The hybridization solution contained 10 ng/ml of the probe, 20% formamide, 0.9 M NaCl, 20 mM Tris-HCl (pH 7.4), 1 mM Na2EDTA, and 0.01% sodium dodecyl sulfate (SDS). Cells of HSMV-1 hybridized to the HSMVp probe, while other cells in the sample did not (Fig. 3), indicating that the 16S rRNA gene sequence we obtained is from the magnetotactic bacterium under study. Strain HSMV-1 clearly represents a new genus (Fig. 2), and based on the phylogeny and what we currently know phenotypically about strain HSMV-1, we propose the name Candidatus Thermomagnetovibrio paiutensis (the GBS site was originally occupied by the Paiute Indian Tribe).
FIG. 3.
Fluorescent in situ hybridization (FISH) of cells of strain HSMV-1 using an HSMV-1-specific oligonucleotide rRNA probe (HSMVp). Cells used for FISH were magnetically concentrated by placing a magnet next to the side of the sample bottle for 30 min and then removed with a Pasteur pipette. This technique was used rather than the magnetic racetrack method in order to have many HSMV-1 cells as well as some other cells that could be used as a negative control. (A) Differential interference contrast (DIC) image of HSMV-1 cells (filled arrows) and other cells (negative control; empty arrows) from hot spring samples; (B) cells stained with 4′,6-diamidino-2-phenylindole (DAPI); (C) cells hybridized with the specific probe HSMVp.
Nash (12) first reported thermophilic magnetotactic bacteria phylogenetically affiliated with the Nitrospirae phylum in hot springs, and it would be interesting and important to compare these organisms and their habitats. However, little can be compared at this time due to lack of information. Nash (12) reported that the one spring at Little Hot Creek was freshwater and that microbial mats were present at all springs where thermophilic magnetotactic bacteria were found. The water at our sampling sites was brackish, not freshwater, and microbial mats were not an important feature of our springs. Thus, it is difficult to determine without knowing the relationship between the organisms found by Nash (12) and strain HSMV-1 what environmental parameters are important to the growth and survival of these bacteria.
It is also difficult to determine the temperature ranges for the survival and growth for strain HSMV-1 without having a pure culture. Data presented here suggest that the temperature range for both is quite wide, and this would be important for the continued presence of HSMV-1 at GBS, as temperatures in the hot springs are known to fluctuate greatly (2). Even if the maximum growth temperature of HSMV-1 is slightly lower than the maximum survival temperature (a conservative estimate) that we know of (63°C), it would still be considered a moderately thermophilic bacterium.
The results presented here clearly show that some magnetotactic bacteria can be considered at least moderately thermophilic. They extend the upper temperature limit for environments where magnetotactic bacteria exist and likely grow (∼63°C) and where magnetosome magnetite is deposited, a finding that may prove significant in the study and interpretation of magnetofossils (9, 10).
Nucleotide sequence accession number.
The 16S rRNA gene sequence of strain HSMV-1 was deposited in GenBank under accession number GU289667.
Supplementary Material
Acknowledgments
We thank David and Sandie Jamieson for access to the GBS field site and J. Dodsworth for help with collecting samples.
C.T.L. and D.A.B. were supported by U.S. National Science Foundation (NSF) grant EAR-0715492, B.P.H. was supported by NSF grant MCB-054865, and M.L.S. was the recipient of an award from the NSF Research Experience for Undergraduates (REU) program A Broad View of Environmental Microbiology at UNLV (award NSF-0649267).
Footnotes
Published ahead of print on 9 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J. P. 1978. A geochemical study of the southwest part of the Black Rock Desert and its geothermal areas; Washoe, Pershing, and Humboldt counties, Nevada. Colo. School Mines Q. 73:15-22. [Google Scholar]
- 3.Bazylinski, D. A., and R. B. Frankel. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217-230. [DOI] [PubMed] [Google Scholar]
- 4.Butler, R. F., and S. K. Banerjee. 1975. Theoretical single-domain grain size range in magnetite and titanomagnetite. J. Geophys. Res. 80:4049-4058. [Google Scholar]
- 5.Costa, K. C., J. B. Navarro, E. L. Shock, C. L. Zhang, D. Soukup, and B. P. Hedlund. 2009. Microbiology and geochemistry of great boiling and mud hot springs in the United States Great Basin. Extremophiles 13:447-459. [DOI] [PubMed] [Google Scholar]
- 6.Flies, C. B., J. Peplies, and D. Schüler. 2005. Combined approach for characterization of uncultivated magnetotactic bacteria from various aquatic environments. Appl. Environ. Microbiol. 71:2723-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NY. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 8.Haouari, O., M.-L. Fardeau, J.-L. Cayol, G. Fauque, C. Casiot, F. Elbaz-Poulichet, M. Hamdi, and B. Ollivier. 2008. Thermodesulfovibrio hydrogeniphilus sp. nov., a new thermophilic sulphate-reducing bacterium isolated from a Tunisian hot spring. Syst. Appl. Microbiol. 31:38-42. [DOI] [PubMed] [Google Scholar]
- 9.Jimenez-Lopez, C., C. S. Romanek, and D. A. Bazylinski. 2010. Magnetite as a prokaryotic biomarker: a review. J. Geophys. Res. Biogeosci. 115:G00G03. [Google Scholar]
- 10.Kopp, R. E., and J. L. Kirschvink. 2008. The identification and biogeochemical interpretation of fossil magnetotactic bacteria. Earth Sci. Rev. 86:42-61. [Google Scholar]
- 11.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley & Sons, Chichester, United Kingdom.
- 12.Nash, C. 2008. Ph.D. thesis. California Institute of Technology, Pasadena, CA.
- 13.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization with rRNA-targeted oligonucleotide probes, p. 207-226. In J. Paul (ed.), Methods in microbiology: marine microbiology, vol. 30. Academic Press Ltd., London, United Kingdom.
- 14.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 15.Schüler, D. 2002. The biomineralization of magnetosomes in Magnetospirillum gryphiswaldense. Int. Microbiol. 5:209-214. [DOI] [PubMed] [Google Scholar]
- 16.Spring, S., R. Amann, W. Ludwig, K.-H. Schleifer, H. van Gemerden, and N. Petersen. 1993. Dominating role of an unusual magnetotactic bacterium in the microaerobic zone of a freshwater sediment. Appl. Environ. Microbiol. 59:2397-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 18.Wolfe, R. S., R. K. Thauer, and N. Pfennig. 1987. A “capillary racetrack” method for isolation of magnetotactic bacteria. FEMS Microbiol. Ecol. 45:31-35. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.