Abstract
In the framework of the Micro-Ecological Life Support System Alternative (MELiSSA) project, a pilot study was performed to identify the effects of triclosan on the MELiSSA carbon-mineralizing microorganism Rhodospirillum rubrum S1H. Triclosan is a biocide that is commonly found in human excrement and is considered an emerging pollutant in wastewater and the environment. Chronic exposure to MELiSSA-relevant concentrations (≥25 μg liter−1) of triclosan resulted in a significant extension of the lag phase of this organism but hardly affected the growth rate. Analytical determinations gave no indication of triclosan biodegradation during the growth experiment, and flow cytometric viability analyses revealed that triclosan is bacteriostatic and only slightly toxic to R. rubrum S1H. Using microarray analyses, the genetic mechanisms supporting the reversibility of triclosan-induced inhibition were scrutinized. An extremely triclosan-responsive cluster of four small adjacent genes was identified, for which there was up to 34-fold induction with 25 μg liter−1 triclosan. These four genes, for which the designation muf (micropollutant-upregulated factor) is proposed, appear to be unique to R. rubrum and are shown here for the first time to be involved in the response to stress. Moreover, numerous other systems that are associated with the proton motive force were shown to be responsive to triclosan, but they were never as highly upregulated as the muf genes. In response to triclosan, R. rubrum S1H induced transcription of the phage shock protein operon (pspABC), numerous efflux systems, cell envelope consolidation mechanisms, the oxidative stress response, beta-oxidation, and carbonic anhydrase, while there was downregulation of bacterial conjugation and carboxysome synthesis genes. The muf genes and three efflux-related genes showed the most potential to be low-dose biomarkers.
Controlled ecological life support systems (CELSS) provide a (bio)technological answer to some of the practical issues associated with the colonization of hostile and isolated environments, such as the moon or Mars, by including cradle-to-cradle handling of all resources (42). The Micro-Ecological Life Support System Alternative (MELiSSA) is a gnotobiotic microorganism-based recycling system that could become an integrated part of a CELSS, since it was specifically designed to allow a small crew to survive at a lunar outpost (35). The current design of the MELiSSA allows recycling of plant biomass, black water, and carbon dioxide to obtain oxygen, potable water, comestible plants, and cyanobacteria. This MELiSSA is a closed-loop system consisting of five serially interconnected compartments, four of which are microbial bioreactors and one of which is a higher plant chamber (28). The raw waste is liquefied and mineralized in the first three compartments of the system, which yields a nutrient solution that is fed to the plant cultivars and edible cyanobacteria in the fourth compartment. A MELiSSA pilot plant is currently being assembled and tested at the Autonomous University of Barcelona (20).
Despite the many advantages of this system (28), there could be an inherent problem since the organic waste that is treated by the MELiSSA can contain a number of micropollutants, such as endocrine-disrupting compounds, pharmaceuticals, and biocides. These compounds are bioactive at low concentrations, and their accumulation in the closed system might have adverse effects on the efficiency of the system and/or crew health. Moreover, the MELiSSA is prone to accumulating (bioactive) pollutants since it is a closed ecosystem with minimized aquatic buffers and limited species richness (29). Whereas the first compartment is inhabited by a complex mixed culture of anaerobic thermophilic microorganisms that liquefy and acidify the incoming organic waste material to form volatile fatty acids (VFA), transformation of the VFA occurs in an axenic anaerobic photoheterotrophic suspension culture of Rhodospirillum rubrum S1H in compartment II and in other axenic cultures in compartments III and IV. Accordingly, compartment II of the MELiSSA appears to be the most fragile component of the loop because of the transition from high biodiversity to restricted biodiversity and because of its proximity to the MELiSSA influent (29). Nevertheless, if R. rubrum S1H had a high degree of resilience with micropollutants, some of these pollutants could pass through this compartment and be biologically removed in the aerobic nitrifying third compartment without impacting the overall loop efficiency. Aerobic nitrifiers were previously shown to biodegrade aromatic hydrocarbons, endocrine-disrupting compounds, and the biocide triclosan (TCS) (12, 15, 58, 65). Therefore, the sensitivity of the MELiSSA bacteria to deleterious micropollutants must be assessed and the response to these compounds must be studied.
The antibacterial and antifungal compound 2,4,4′-trichloro-2′-hydroxydiphenylether, also known as triclosan (TCS) or Irgasan, was chosen as the first contaminant whose impact on R. rubrum S1H in compartment II was studied. TCS is frequently included in personal care products (such as toothpaste, shampoo, and soaps), where it is used at concentrations around 1 g liter−1 (32). When personal care products are used, TCS is absorbed by the body, after which it is rapidly metabolized and excreted in the urine and feces (10, 56, 59). It has even been detected in three of five human milk samples in Sweden (1). Due to its widespread incorporation into products and its chemical rigidity, TCS and its derivatives have been detected in the effluent of wastewater treatment plants across the globe and also in the receiving waters and the surrounding environment (1, 21, 34, 60, 77). TCS is known to adsorb to microbial biomass due to its hydrophobic nature, as shown by its log octanol-water partition coefficient (Kow), which is 4.8 (26, 27). As a biocide, TCS can act on different targets in the microbial cell. At concentrations in the pico- to nanomolar range, TCS has been reported to inhibit type II fatty acid biosynthesis through inhibition of the enoyl-acyl carrier protein reductase (FabI) enzyme in numerous Gram-negative and Gram-positive bacteria (24, 30, 44, 62). This inhibition occurs when TCS forms a noncovalent complex with NAD+ in the FabI active site (55). FabI is required for cellular division, cellular maintenance, and quorum sensing in bacteria (30). Nevertheless, there are TCS-insensitive homologs, such as FabI2 in the closely related bacterium Rhodobacter sphaeroides, that can remain functional in an environment with TCS (25, 36). At concentrations above the nanomolar range, TCS increases the permeability of the cellular envelope by incorporation into the phospholipid membrane, which results in cell lysis (23, 66, 73).
The present study was performed to measure the risks and effects associated with entry of TCS into compartment II of the MELiSSA and to assess the functional robustness of this compartment when it is exposed to a known deleterious compound. Therefore, the response of R. rubrum S1H to TCS was studied at the phenotypic, physiologic, metabolic, and transcriptome levels. Using transcriptome analysis, this study aimed to identify the TCS-responsive mechanisms of R. rubrum S1H and determine the supporting cellular mechanisms, which have not been described yet. Examining chronic exposure of R. rubrum S1H to TCS at environmentally relevant concentrations might provide novel insights in the toxicogenomic effects of TCS. Identification of low-dose response mechanisms by whole-genome profiling might allow selection of TCS-responsive biomarker genes, which could be very important in the development of a biosensor in the MELiSSA. As a pilot investigation, this study focused on the effects of TCS under chemoorganotrophic conditions, while future research will focus on the photoheterotrophic metabolism in a continuously stirred tank reactor (CSTR) (i.e., genuine MELiSSA conditions).
MATERIALS AND METHODS
Strain and growth conditions.
R. rubrum S1H (ATCC 25903) (48) was obtained from the American Type Culture Collection and grown in Sistrom medium A (SIS) with 0.2% (wt/vol) succinate as the sole carbon source under aerobic conditions in the dark at 30°C and 50 rpm (61). Early-stationary-phase cells were inoculated (2%, vol/vol) into 50 ml SIS. Prior to inoculation, SIS was spiked with TCS using a TCS stock solution (97.0%; HPLC grade; Fluka) in methanol, and different samples were supplemented with equal final volumes of methanol to eliminate the potential effects of different solvent concentrations. In addition, a solvent control (with methanol) and a growth control (without methanol) were prepared. Subsequently, SIS with and without TCS was inoculated with early-stationary-phase cells.
Growth kinetics.
Growth experiments were performed using five parallel cultures for each test condition. At fixed intervals, the optical density at 680 nm (OD680) was measured photospectrometrically using an Ultrospec 3000 (Pharmacia-Biotech). The generation time (μm) and duration of the lag phase (A) were determined by determining the mathematical averages for the five biological replicates. Differences in growth kinetics were assessed using the Student t test and were considered significant only if the P value was <0.01. During the experiment, aliquots were removed for analytical determination of the TCS concentration, bacterial viability analysis, and transcriptome analysis. To ascertain that all of the replicate cultures that were used for the transcriptome study were homogeneous suspensions of triclosan-adapted cells, all replicate cultures (with and without TCS) were screened for decreased susceptibility to TCS. In the screening procedure, triclosan-exposed early-stationary-phase cells were washed twice in a 0.85% NaCl solution, and then they were subcultured twice for 48 h at 30°C in triclosan-free SIS. After the second subculture, the early-stationary-phase cells were spotted on solid SIS with and without triclosan using a replica plater and incubated for 7 days at 30°C. Subsequently, their phenotypic sensitivities to triclosan were compared to those of the other replicates and unexposed R. rubrum S1H.
Analytical determination.
For chemical analyses, 5-ml aliquots were removed from two liquid cultures during the growth experiment. The aliquots were frozen at −20°C until solid-phase extraction was performed. Prior to the solid-phase extraction, the samples were defrosted and centrifuged (when required) to precipitate the cells. The supernatants were acidified with H3PO4 and loaded onto Oasis HLB 6cc SPE cartridges (Waters) that were conditioned with 4 ml methanol and 4 ml Milli-Q water at pH 2.0. The cartridges were washed with Milli-Q water at pH 2.0 to remove the salts and then centrifuged until they were dry. TCS was eluted with tetrahydrofuran (THF). The THF extracts were dried with a gentle stream of nitrogen gas in heat blocks at 50°C and redissolved in 2 ml methanol-water (50:50, vol/vol) containing 2% NH3 for injection (10 μl). Liquid chromatographic separation was performed using an Acquity ultraperformance liquid chromatograph (UPLC; Waters). The samples were separated on a Waters Acquity UPLC BEH C18 column (100 mm by 2.1 mm by 1.7 μm) maintained at 40°C. The following gradient was employed (using as solvent A Milli-Q plus 1% NH3 [pH 10.0] and as solvent B acetonitrile): 90% solvent A for 50 s, then 100% solvent B for 3 min, and finally 90% solvent A for 2.5 min. The flow rate was 0.40 ml min−1. The TCS retention time was 2.26 min. Detection was performed with a Quattro Premier XE tandem mass spectrometer (Waters) operated in the negative electrospray ionization mode using selected-ion recording (SIR) (m/z 288.8). The mass spectrometry conditions were as follows: capillary voltage, 3 kV; extractor voltage, 3 V; RF lens, 0.1 V; source temperature, 120°C; desolvation temperature, 350°C; desolvation gas flow rate, 799 liters h−1; cone gas flow rate, 49 liters h−1; dwell time, 0.1 s; span, 0.5 atomic mass unit; and cone voltage, 23 V. Spiking experiments performed with 10 μg liter−1 TCS, 25 μg liter−1 TCS, and 37 μg liter−1 TCS (n = 2) resulted in levels of recovery of 123%, 101 to 108%, and 106 to 109%, respectively, for sterile SIS and 103 to 113%, 99 to 127%, and 90 to 96%, respectively, for noncentrifuged stationary-phase cultures. The limit of detection was 0.5 μg liter−1, and the limit of quantification was 1.2 μg liter−1.
Bacterial viability assay.
Culture aliquots (100 μl) were removed at fixed intervals during growth to assess the toxicity of TCS by flow cytometry. In addition, this technique allowed verification of culture viability homogeneity, which was required for the subsequent transcriptome analysis. Flow cytometric analyses were performed with a Beckham Coulter Epics XL-MCL using a LIVE/DEAD BacLight L-7007 bacterial viability kit (Invitrogen). All analyses were performed by following the manufacturer's instructions, using technical duplicates for each of the five biological replicates. The cutoff was defined as 10,000 recorded events per analysis.
RNA extraction.
Ten milliliters of a cell suspension was harvested at an OD680 of 0.4 from all test cultures with and without TCS, and 20 ml RNAProtect bacterial reagent (Qiagen) was added to preserve the RNA. The fixed cell suspensions were centrifuged at 3,500 rpm for 10 min, the supernatants were removed, and the cell pellets were stored at −80°C. RNA extraction was performed using an RNeasy midi kit (Qiagen) by following the manufacturer's guidelines. RNA quantity and quality were assessed using a Nanodrop ND-1000 and a 2100 bioanalyzer (Agilent) with an RNA 6000 Nano chip kit according to the manufacturer's instructions. Only RNA with an RNA integrity number (RIN) value of 9.5 or higher was used for microarray analysis (57).
Microarray production.
The genome sequence of the parent strain, R. rubrum S1 (53), which comprised a 4.35-Mb chromosome sequence (GenBank accession no. CP000230) and a 53.7-kb plasmid sequence (GenBank accession no. CP000231), was used to design 60-mer aminosilane-modified oligonucleotide probes corresponding to the 3,829 predicted protein-encoding genes. A full description of the array analysis platform is available at the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE14265. The microarray analyses were performed using biological and technical triplicates. The microarray data were analyzed as previously described (39). Only genes with induction and repression ratios of ≥1.50-fold and ≤0.75-fold, respectively, are discussed below. All data presented here within the set thresholds were statistically significant at a P value of <0.001.
RNA labeling and hybridization and data analysis.
Ten micrograms of total RNA was reverse transcribed by following the instructions provided with a Pronto kit (Promega). Two-color labeling, cDNA hybridization, and slide washing were performed as previously described (39).
Genome analysis.
Interpretation of transcriptome data is highly dependent on the quality of genome annotation. Therefore, the automatic annotation of every gene discussed in this paper was scrutinized using the Magnifying Genomes (MaGe) platform (71), where information for R. rubrum S1 is publicly available as part of the “MagnetoScope” project. For promoter prediction, the Berkeley Drosophila Genome Project's Neural Network Promoter Prediction script was used (http://www.fruitfly.org/seq_tools/promoter.html), applying the default prokaryote parameter setting. For regulatory motif searches, Multiple Em for Motif Elicitation (3), Tools for Motif Discovery (49), AlignACE 3.0 (54), and Regulatory Sequence Analysis Tools (72) were used.
Full microarray data set accession number.
The full microarray data set has been deposited at the Gene Expression Omnibus website under accession number GSE16189.
RESULTS
Prediction of TCS entry into the MELiSSA.
By using previously described urinary TCS concentrations (10, 56, 76) and assuming that the average urine production is 1.5 liters day−1 person−1 (67), it was estimated that the range of TCS concentrations in urine is 0.5 to 2.1 mg liter−1. If urine is introduced into the first compartment of the MELiSSA, the parameters are such that it is diluted 14-fold in the final organic waste solution (H. De Wever, unpublished data), resulting in 35 to 150 μg liter−1 TCS in the compartment I input. Since no information on the fate of TCS in anaerobic thermophilic systems is available, it is assumed that removal of TCS in compartment I is restricted to adsorption to the biomass, as demonstrated previously for other anaerobic biotopes (41, 78). TCS removal ratios of 0%, 50%, and 90% due to adsorption were therefore considered in our calculations. Taking into account that there is further 6-fold dilution of the compartment I effluent before it enters compartment II (H. De Wever, unpublished data), the initial TCS concentrations expected for compartment II range from 5 to 25 μg liter−1 for a removal ratio of 0% due to adsorption in compartment I, from 3 to 13 μg liter−1 for a removal ratio of 50%, and from 0.5 to 3 μg liter−1 for a removal ratio of 90%. However, the real TCS concentration ranges could be even higher than the ranges described above, because excretion of TCS in the feces was not taken into account due to the lack of pharmacokinetic data on human subjects (46) and because excretion of TCS could be different in space (22).
Impact of TCS on cell growth and viability.
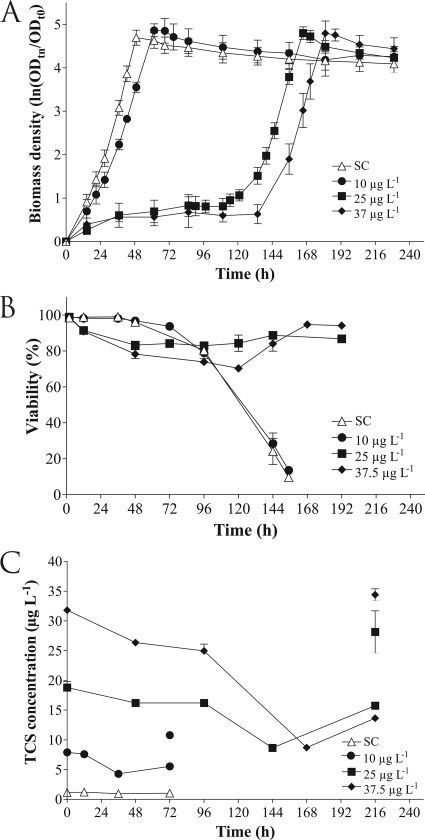
Chronic exposure to 10 μg liter−1 TCS in SIS had a significant impact on the generation time but not on the lag phase. Chronic exposure to 25 μg liter−1 or 37.5 μg liter−1 TCS in liquid medium resulted in an important shift in the lag phase and a small but significant shift in the generation time (Fig. 1A). No significant differences between the solvent control and the growth control were observed (data not shown). By using the flow cytometric viability assay, it was determined that TCS was bacteriostatic rather than bactericidal at concentrations of 25 and 37.5 μg liter−1. Moreover, TCS was only slightly toxic as the culture exposed to 37.5 μg liter−1 TCS still contained 70% viable cells after 120 h of lag phase (Fig. 1B). In addition, it is important to note that the culture viability was near the optimum level when sampling for RNA extraction was performed for all of the TCS concentrations tested. After the maximum optical density was reached (after 48 h for the solvent control and after 60 h for treatment with 10 μg liter−1 TCS), the culture viability decreased to less than 10% within 5 days because of nutrient limitation (Fig. 1B). Identical decreases were observed for the cultures exposed to 25 and 37.5 μg liter−1 TCS (data not shown).
FIG. 1.
(A) Growth curves for R. rubrum S1H with and without TCS. The error bars indicate the standard deviations of five biological replicates. ODt0, optical density at 680 nm when the culture was inoculated; ODtn, optical density at 680 nm at sampling time tn, where n is the number of hours. (B) Bacteriostatic effect and toxicity of TCS in R. rubrum S1H determined by a flow cytometric viability assay during the course of the growth experiment. Viability is expressed as the percentage of total cells. (C) Determination of the TCS concentration in solution during the course of the growth experiment. The first value was obtained 2 h after inoculation of the cultures to allow adsorption to equilibrate. The separate values at 72 h and 216 h indicate the total amounts of TCS recovered at the end of the growth experiment by extraction of the TCS from both the liquid and the particulate phase. ▵, solvent control samples with methanol (SC); •, samples chronically exposed to 10 μg liter−1 TCS; ▪, samples chronically exposed to 25 μg liter−1 TCS; ⧫, samples chronically exposed to 37.5 μg liter−1 TCS.
In the triclosan susceptibility screening assay, all replicate cultures that were originally exposed to either 10 μg/liter or 25 μg/liter triclosan retained the wild-type phenotype following cultivation in medium without selective pressure. No differences in the MIC between the replicate cultures were observed (data not shown). This indicated that both the cells exposed to 10 μg/liter triclosan and the cells exposed to 25 μg/liter triclosan could be used in the transcriptome study as homogeneous suspensions of triclosan-exposed R. rubrum S1H cells or that the number of (potentially) triclosan-resistant mutant cells was below the detection limit of the replica plating assay.
Analytical determination.
Chemical analysis showed that the dissolved TCS concentration gradually decreased over time for all TCS concentrations and that there was a dip at the mid-exponential phase (Fig. 1C). At the beginning of the stationary phase, the dissolved TCS concentration increased again, suggesting that there were differences in the adsorption properties of the cell wall during the different growth phases. At the end of the experiment, TCS was extracted from both the particulates (cells) and the liquid phase; hence, all TCS was recovered from the cultures. This indicated that the steady decline in the level of dissolved TCS was not due to biodegradation (Fig. 1C).
Whole-genome transcriptome survey.
There are 3,829 protein-encoding sequences in the R. rubrum genome, and 10 μg liter−1 TCS induced the expression of 20 genes more than 2.00-fold and 166 genes more than 1.50-fold; 25 μg liter−1 TCS induced the expression of 33 genes more than 2.00-fold and 259 genes more than 1.50-fold. Table 1 shows an overview of the 10 genes that were most strongly induced with 25 μg liter−1 TCS, as well as the induction values for 10 μg liter−1 TCS. Most of the genes upregulated in the presence of 10 μg liter−1 TCS were also found to be induced by 25 μg liter−1 TCS (data not shown). Hence, these findings indicated that there was a regulatory hierarchy for the genes that were induced with 10 μg liter−1 (minor inhibition) and 25 μg liter−1 TCS (severe inhibition). With 25 μg liter−1 TCS, nearly two-thirds of the upregulated genes were associated with either efflux mechanisms or stress response mechanisms. The other genes were associated with biosynthesis pathways, plasmid maintenance, or cell envelope consolidation.
TABLE 1.
Ten most strongly induced genes of R. rubrum S1H after chronic exposure to 25 μg liter−1 TCSa
| Locus tag | Gene | Annotated protein function | Fold change with: |
|
|---|---|---|---|---|
| 10 μg liter−1 TCS | 25 μg liter−1 TCS | |||
| Rru_A0425b | mufA2 | Micropollutant-upregulated factor A2c | NDRd | 34.54 |
| Rru_A0426b | mufA1 | Micropollutant-upregulated factor A1c | 0.65 | 15.26 |
| Rru_A0423b | mufB | Micropollutant-upregulated factor Bc | NDR | 7.42 |
| Rru_A0424b | mufM | Micropollutant-upregulated factor Mc | NDR | 6.74 |
| Rru_A3164b | Major facilitator transport, putative permeasec | NDR | 4.41 | |
| Rru_A1218 | pspA | Phage shock protein A | NDR | 3.90 |
| Rru_A0156b | EmrA family secretion protein | NDR | 3.77 | |
| Rru_A3700b | RND efflux system, membrane fusion protein | 2.54 | 3.38 | |
| Rru_A1216b | pspC | Phage shock protein Cc | NDR | 3.23 |
| Rru_A0020 | gpo | Glutathione peroxidase | NDR | 2.82 |
The genes are listed in order based on the fold induction values with 25 μg liter−1 TCS. The fold induction values obtained with 10 μg liter−1 TCS are also shown.
Gene that was reannotated in MaGe.
Putative function of a hypothetical gene.
NDR, gene was not differentially regulated.
(i) Micropollutant-upregulated factors.
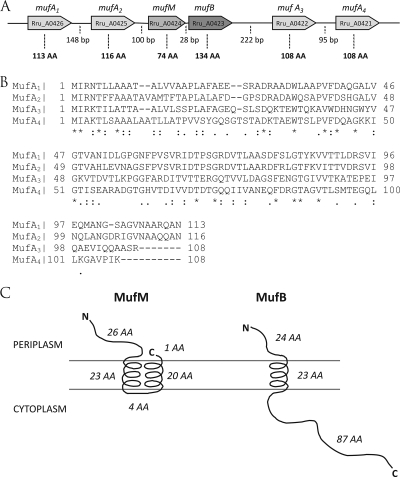
Upon exposure to 25 μg liter−1 TCS, four adjacent hypothetical genes (Rru_A0423, Rru_A0424, Rru_A0425, and Rru_A0426) were the most strongly upregulated genes, with up to 34.54-fold induction (Table 1). These clustered genes appeared to have no known homologs in any other bacterium and are unique to R. rubrum. Sequence homology analysis provided evidence that the cluster most likely extends downstream and thus includes the adjacent genes Rru_A0422 and Rru_A0421 (40% and 31% identity with Rru_A0426, respectively). The two latter genes were found to be homologous to Rru_A0425 and Rru_A0426, but they did not respond to TCS. Given the extreme responsiveness of the first four genes to TCS, this novel gene cluster was referred to as the micropollutant-upregulated factor (muf) cluster (Fig. 2A). The loci in this cluster are Rru_A0426 (mufA1), Rru_A0425 (mufA2), Rru_A0424 (mufM), Rru_A0423 (mufB), Rru_A422 (mufA3), and Rru_A0421 (mufA4). Sequence homology data provided evidence that there is a fifth mufA paralog elsewhere in the R. rubrum genome, namely, Rru_A0565. The muf cluster thus comprises six adjacent genes, four of which (mufA1, mufA2, mufA3, and mufA4) exhibit high levels of sequence similarity (31 to 67% identity) (Fig. 2B). These four paralogs are separated in pairs which are located at the extremities of the cluster. The mufA1 and mufA2 genes appeared to be highly inducible upon exposure to TCS, while the mufA3 gene appeared to be responsive to the nonsteroid anti-inflammatory drug diclofenac (50). Hence, it is possible that the different mufA paralogs exhibit some signal specificity, which could be supported by the fact that all MufA proteins exhibit small sequence variations, primarily at their C termini (Fig. 2B). These sequence variations could represent small unconserved regions, which are not subject to natural selection; on the other hand, the C-terminal variations in the mufA genes might result in condition-specific induction properties of the proteins. Moreover, all MufA homologs possess signal peptides that allow them to be transported to the periplasm. From the multiple-sequence alignment, it was also apparent that the mufA3 sequence had an additional 20 amino acids at the N terminus compared to the four other paralogs (data not shown). The Artemis script in MaGe revealed the presence of a Shine-Dalgarno sequence upstream of the 21st amino acid, thus confirming an error in the automatic start codon attribution. Accordingly, the mufA3 start codon was altered in MaGe to correspond to the start codons of the other mufA paralogs (Fig. 2B).
FIG. 2.
(A) Schematic diagram of the micropollutant-upregulated factor (muf) cluster in the R. rubrum S1 genome. The intergenic region lengths are indicated by the numbers of base pairs, and the protein lengths are indicated by the numbers of amino acids (AA). (B) Multiple alignment of the four MufA paralogs encoded by genes in the muf cluster in the R. rubrum S1 genome, constructed using the clustalW2 algorithm. Only mufA1 and mufA2 appeared to be responsive to TCS. Asterisks indicate conserved amino acids; colons indicate highly conserved amino acids; and periods indicate slightly conserved amino acids. (C) Diagram of the cellular localization of the MufM and MufB proteins as predicted by MaGe. The solenoid shapes represent membrane-encompassing helices. N, N terminus of the protein; C, C terminus of the protein; AA, amino acids.
The two other muf genes that were also very responsive to TCS were the mufM and mufB genes. Sequence analysis using the TMHMM tool in MaGe provided insights into the localization of the MufM and MufB proteins and suggested that both of them have an inner membrane-associated conformation (Fig. 2C). The C terminus of MufM (from the 30th amino acid to the 70th amino acid) appeared to be highly conserved in numerous small 50-amino-acid alpha- and betaproteobacterial proteins with 71 to 76% identity, while the sequence of the N-terminal extension that reached the periplasm appeared to be unique to R. rubrum. The MufB protein, on the other hand, possesses only a single transmembrane domain and an 87-amino-acid cytoplasmic tail (Fig. 2C), but no significant homolog was found in related microorganisms. Because only four of the six muf genes were overexpressed in response to TCS, several motif-finding scripts were used to locate recurring regulatory regions upstream of the muf genes, but none were identified. Moreover, these genes do not appear to be transcriptionally coupled based on the intergenic distances (Fig. 2A), the fold induction values, and promoter prediction results; the only exceptions may be mufM and mufB, which are separated by only 28 bp.
(ii) Stress-related proteins.
The highest level of induction for a gene with a known function was the level of induction for the phage shock protein gene pspA, 3.90-fold induction (Table 1). Like the muf cluster, the phage shock protein operon (pspABC) (Rru_A1216, Rru_A1217, and Rru_A1218) was significantly overexpressed only after chronic exposure to 25 μg liter−1 TCS and was not overexpressed in response to 10 μg liter−1 TCS (Table 2). The σ54-dependent pspABC transcriptional activator gene pspF (Rru_A1219) was upregulated at neither concentration. On the contrary, it was slightly repressed by 25 μg liter−1 TCS, as was the σ54-encoding gene itself. TCS also mildly induced several other stress response genes, including genes coding for proteases (Rru_A1550, Rru_A2192, Rru_A2201, Rru_A2912, Rru_A2972, Rru_A2973, Rru_A3600, and Rru_A3710), (deoxy)ribonucleases (Rru_A1700, Rru_A1769, Rru_A2767, Rru_A3209, and Rru_A3696), and helicases (Rru_A1915 and Rru_A2718). One gene of the tellurite resistance (terABC) operon, the TerA-encoding gene (Rru_A0892), was upregulated. The function of this gene, however, has not yet been described (68). The dnaJ gene (Rru_A3559), which encodes the regulator of the DnaK-DnaJ-GrpE protein-folding system, was mildly induced upon exposure to TCS, and so were hrcA and clpP, which encode a heat-inducible transcriptional repressor and a heat shock-related protease, respectively (Table 2). The GroEL system was also significantly induced with 25 μg liter−1 TCS, but the level was below the 1.50-fold induction threshold (data not shown). The formerly hypothetical genes Rru_A0892 (terA), Rru_A1216 (pspC), and Rru_A1219 (pspF) were reannotated in MaGe (Table 2).
TABLE 2.
Genes related to stress response, efflux, cellular envelope consolidation, or other mechanisms that were significantly induced or repressed upon exposure to 25 μg liter−1 TCS (P < 0.001)a
| Locus tagb | Gene | Annotated protein function | Fold change with: |
|
|---|---|---|---|---|
| 10 μg liter−1 TCS | 25 μg liter−1 TCS | |||
| General stress | ||||
| Rru_A0010 | rpoN | RNA polymerase factor sigma-54 | NDRd | 0.63 |
| Rru_A0892c | terA | Tellurite resistance protein A | 1.67 | 1.73 |
| Rru_A1216c | pspC | Phage shock protein Ce | NDR | 3.24 |
| Rru_A1217 | pspB | Phage shock protein B | NDR | 1.69 |
| Rru_A1218 | pspA | Phage shock protein A | NDR | 3.90 |
| Rru_A1219c | pspF | Phage shock protein F (sigma-54 dependent) | NDR | 0.69 |
| Rru_A1550 | clpP | ATP-dependent protease, proteolytic subunit | NDR | 1.60 |
| Rru_A3559 | dnaJ | Heat shock protein | NDR | 1.50 |
| Rru_A3642 | hrcA | Heat-inducible transcription repressor | 1.57 | 1.66 |
| Efflux systems | ||||
| Rru_A0156c | EmrA family secretion protein | NDR | 3.77 | |
| Rru_A0157 | EmrB/QacA family drug resistance transporter | 0.70 | 2.57 | |
| Rru_A0272c | sugE | Small multidrug resistance protein | 1.81 | 1.56 |
| Rru_A0886c | mexF | RND efflux system, inner membrane transporterf | NDR | 1.77 |
| Rru_A0887c | mexE | RND efflux system, membrane fusion protein | 0.50 | 1.16 |
| Rru_A0888 | TetR family transcriptional regulator | NDR | 1.52 | |
| Rru_A1114 | Major facilitator transporter | NDR | 1.50 | |
| Rru_A1798c | Type I secretion, TolC family outer membrane protein | 1.51 | 1.59 | |
| Rru_A2049 | TetR family transcriptional regulator | NDR | 1.83 | |
| Rru_A2050c | mexA | RND efflux system, membrane fusion protein | NDR | 2.30 |
| Rru_A2051c | mexB | RND efflux system, inner membrane transporter | NDR | 2.19 |
| Rru_A2052c | oprM | RND efflux system, outer membrane protein | NDR | 2.58 |
| Rru_A2805c | ABC transporter, putative permeasee | NDR | 1.61 | |
| Rru_A2908c | ABC transporter for organic solvents, putative permeasee | 1.90 | 1.86 | |
| Rru_A2910c | ABC transporter for organic solvents, periplasmic componente | NDR | 1.80 | |
| Rru_A2926c | RND efflux system, outer membrane protein | NDR | 1.51 | |
| Rru_A2928c | mexM | RND efflux system, membrane fusion protein | NDR | 1.62 |
| Rru_A3160c | Drug/metabolite transporter, putative permeasee | 2.31 | 1.88 | |
| Rru_A3164c | Major facilitator transport, putative permeasee | NDR | 4.40 | |
| Rru_A3519 | MarR family transcriptional regulator | 2.53 | 2.09 | |
| Rru_A3520c | Putative permease, conservede | 2.34 | 2.03 | |
| Rru_A3561 | ABC multidrug transport system, ATPase and permease component | NDR | 1.53 | |
| Rru_A3699c | RND efflux system, inner membrane transporter | NDR | 2.40 | |
| Rru_A3700b | RND efflux system, membrane fusion protein | 2.54 | 3.38 | |
| Rru_A3701 | TetR family transcriptional regulator | NDR | 2.31 | |
| Cellular envelope | ||||
| Rru_A0437 | OmpA family protein | NDR | 1.59 | |
| Rru_A1005 | Glycosyl transferase, group 2 | NDR | 1.56 | |
| Rru_A1100 | glmM | Phosphoglucosamine mutase | NDR | 1.50 |
| Rru_A1492c | pssT | Polysaccharide transport system protein Te | 1.58 | 1.58 |
| Rru_A2412c | OmpA family protein | 1.97 | 2.13 | |
| Rru_A2731 | exoD | Exopolysaccharide synthesis protein D | NDR | 1.58 |
| Rru_A2916 | 4′-Phophopantetheinyl transferase | 1.63 | 1.73 | |
| Rru_A3102 | Sugar transferase | 1.73 | 1.50 | |
| Rru_A3125 | Glycosyl transferase, group 1 | 1.56 | 1.83 | |
| Rru_A3192 | Acyl-phosphate glycerol-3-phosphate acyltransferase | NDR | 1.52 | |
| Rru_A3584 | CDP-diacylglycerol-serine O-phosphatidyltransferase | NDR | 1.51 | |
| Rru_A3656 | wbpM | Polysaccharide biosynthesis protein M | 1.77 | 1.63 |
| Rru_A3658 | Glycosyl transferase, group 1 | 1.59 | 1.74 | |
| Conjugation | ||||
| Rru_A0804 | LysR family transcriptional regulator | 0.48 | 0.52 | |
| Rru_A0805 | trbE | Conjugal transfer protein E | 0.66 | 0.66 |
| Rru_A0806 | trbD | Conjugal transfer protein D | 0.36 | 0.39 |
| Rru_A0807 | trbC | Conjugal transfer protein C | 0.46 | 0.43 |
| Rru_A0808 | trbB | Conjugal transfer protein B | 0.73 | 0.69 |
| Rru_A0811 | copG | Helix-turn-helix protein | NDR | 0.72 |
| Rru_A0815c | Conserved hypothetical protein | 0.69 | 0.69 | |
| Rru_A0816c | Conserved hypothetical protein, type IV secretory pathway relaxase componentse | 0.71 | 0.63 | |
| Oxidative response | ||||
| Rru_A0020 | gpo | Glutathione peroxidase | NDR | 2.82 |
| Rru_A0398 | Thiol-disulfide isomerase and thioredoxin-like | 1.81 | 1.74 | |
| Rru_A0580c | osmC | Peroxiredoxin osmotically inducible protein C-like | NDR | 1.74 |
| Rru_A0738 | grxC | Glutaredoxin | 1.55 | 1.63 |
| Rru_A2897 | Alkyl hydroperoxide reductase/thiol-specific antioxidant | 1.75 | 1.69 | |
| Rru_A2898 | trxB | Thioredoxin reductase | 1.50 | 1.55 |
| Rru_A2899 | LysR family transcriptional regulator | 1.74 | NDR | |
| Beta-oxidation | ||||
| Rru_A0729 | Short-chain dehydrogenase/reductase | 1.98 | 1.67 | |
| Rru_A1310 | 3-Ketoacyl-coenzyme A thiolase | 1.85 | 1.79 | |
| Rru_A1834 | fadA | Enoyl-coenzyme A hydratase/isomerase | NDR | 1.52 |
| Rru_A3509 | AMP-dependent synthetase and ligase | 1.73 | NDR | |
| Electron transport | ||||
| Rru_A0362 | fbcC | Cytochrome c1 | NDR | 1.62 |
| Rru_A0363 | fbcB | Cytochrome b/b6-like | NDR | 1.52 |
| Rru_A1224 | ATP synthase F1, alpha subunit | 2.73 | 2.55 | |
| Rru_A1420 | NADH dehydrogenase (quinone) | 1.64 | 1.76 | |
| Rru_A1564 | nuoJ | NADH-ubiquinone/plastoquinone oxidoreductase | NDR | 1.56 |
| Rru_A2267 | fixA | Electron transfer flavoprotein beta subunit | NDR | 1.50 |
| Rru_A2310 | Cytochrome c family protein | NDR | 1.66 | |
| Carbon fixation | ||||
| Rru_A0906 | eutL | Polyhedral shell protein | 0.55 | 0.65 |
| Rru_A0907 | Microcompartment protein | 0.54 | 0.55 | |
| Rru_A0908 | eutM | Microcompartment protein | 0.48 | 0.52 |
| Rru_A0912 | eutN | Carboxysome structural protein | 0.45 | 0.42 |
| Rru_A1725 | acaP | Carbonic anhydrase | NDR | 1.70 |
The cutoff values were ≥1.50-fold for induction and ≤0.75-fold for repression. The fold induction values obtained with 10 μg liter−1 TCS are also shown.
Bold type indicates adjacent loci.
The gene was reannotated in MaGe.
NDR, gene was not differentially regulated.
Putative function of a hypothetical gene.
RND, resistance-nodulation-division.
(iii) Efflux systems.
Besides stress response genes, several efflux-related genes and their transcription regulators were upregulated upon exposure to 10 μg liter−1 and 25 μg liter−1 TCS (Table 2). With 10 μg liter−1 TCS, four efflux-related genes were upregulated at least 2-fold (Table 2), while with 25 μg liter−1 TCS, most of the genes upregulated with 10 μg liter−1 TCS remained important but many additional efflux-related genes, including genes encoding the corresponding regulators, were overexpressed (Table 2). With 25 μg liter−1 TCS, up to 25 efflux-related genes were identified by using a cutoff value 1.50-fold (Table 2). Of these 25 genes, 11 were upregulated at least 2-fold, and most of them encoded resistance-nodulation-cell division (RND) family efflux systems. The hypothetical gene Rru_A3164 showed a high level of induction (4.40-fold) upon exposure to 25 μg liter−1 TCS and was determined to be a putative permease gene using MaGe (Tables 1 and 2). Also, the hypothetical genes Rru_A2805, Rru_A2908, Rru_A2910, Rru_A3160, Rru_A3164, and Rru_A3520 were reannotated and are probably involved in efflux mechanisms (Table 2).
(iv) Cell envelope consolidation.
Several of the genes upregulated with 25 μg liter−1 TCS, such as pssT, exoD, and wbpM, were associated with exopolysaccharide biosynthesis (Table 2). It is interesting that besides wbpM, the upstream CapM-like glycosyl transferase gene (Rru_A3658) was induced in R. rubrum S1H, while the adjacent wbpL homolog (Rru_A3657) was not induced (Table 2). Two genes encoding outer membrane protein A (OmpA) family proteins were found to be upregulated (Table 2).
(v) Other induced pathways.
Exposure of R. rubrum S1H to 25 μg liter−1 TCS also led to induction of genes involved in the oxidative response, and the strongest induction was observed for a glutathione peroxidase gene (Table 2). Other oxidative response genes, such as thiol-disulfide isomerase, glutaredoxin, alkyl hydroperoxide reductase, and thioredoxin reductase genes, were found to be upregulated regardless of the TCS concentration (Table 2). The osmotically inducible peroxiredoxin (OsmC) gene was also induced, but the catalase gene was not induced (Table 2). It was interesting that TCS, a fatty acid biosynthesis-inhibiting compound, also induced the fatty acid beta-oxidation pathway, as well as numerous electron transport chain genes (Table 2). The genes for the horizontal gene transfer machinery were significantly downregulated upon exposure to TCS (Table 2). Many genes involved in carbon metabolism were induced, but complete pathways were never induced, except for the genes involved in the mixed acid fermentation pathway (data not shown). The induction of carbonic anhydrase coincided with downregulation of carboxysomal genes, which aid in concentrating CO2 in intracellular microcompartments (Table 2). The downregulation of the carboxysomal genes might be an indication of an active carbonic anhydrase.
DISCUSSION
In a lunar or Martian habitat, the MELiSSA is meant to treat organic waste that is comprised of feces, urine, unused plant residues, and toilet paper. Thus, micropollutants from personal care products can enter the life support system only as phase II derivatives in urine and feces (2, 46). Prediction of the TCS concentration entering the MELiSSA in compartment I could take only urinary excretion of TCS into account since no data are available for fecal excretion of TCS in humans (10, 56, 76). Hence, the TCS concentrations in the MELiSSA waste could be slightly higher than the concentrations predicted in the present study. Still, it is interesting that the predicted TCS concentration in MELiSSA is similar to the concentrations commonly found in anthropogenic waste streams, such as municipal wastewater (4, 21, 26, 38, 47). Remarkably, acute and chronic human exposure to TCS would result in similar TCS concentrations in urine due to the rapid excretion of TCS (2). This phenomenon is most probably due to the ease with which TCS is absorbed, metabolized, and excreted by the human body. Nevertheless, the pharmacokinetics of TCS could be different in space (22).
According to previous studies, the TCS concentrations found in domestic wastewater do not have a negative impact on wastewater treatment processes (64). This is proven by the fact that at high concentrations (up to 1 mg liter−1), TCS inhibition in continuous systems appears to be reversible (16). In R. rubrum S1H, the reversibility of triclosan inhibition at these elevated concentrations was shown to be at least partially the result of selection of triclosan-resistant mutants (51). It is known that TCS can be biodegraded in aerobic nitrifying systems (65), which might be promising for TCS removal in compartment III of the MELiSSA. Hence, triclosan is not expected to pose a critical issue in the MELiSSA concept, since the present study showed that the wild-type strain R. rubrum S1H was able to circumvent growth inhibition upon exposure to the highest predicted TCS concentrations and triclosan-resistant mutants are readily selected at higher concentrations (51). Regardless, there was a major concentration-dependent extension of the lag phase in R. rubrum S1H when it was exposed to triclosan concentrations greater than 10 μg/liter. This observation was consistent with the results of a previous study of Escherichia coli, Enterococcus hirae, and Staphylococcus aureus (19). In the present study, the sudden growth of R. rubrum S1H in the presence of triclosan coincided with increased transcription of genes involved in stress resistance mechanisms, efflux pumps, and cell wall consolidation. Overall, the transcriptome response to TCS was relatively “mild,” both in terms of the number of genes and in terms of the induction values, and only 0.52% and 0.86% of all genes were induced more than 2-fold upon exposure to 10 μg liter−1 and 25 μg liter−1, respectively. This “mild” response could have been due to (i) concentration-dependent transcriptional regulation, (ii) highly efficient upregulated systems, (iii) the high-level induction of the muf genes, or (iv) a combination of these three possibilities.
The micropollutant-upregulated genes mufA1, mufA2, mufM, and mufB appear to be restricted to R. rubrum and were extremely responsive to TCS. Even though the role of this gene cluster in the resistance to low levels of TCS has not been characterized thus far, these genes were found to be constitutively upregulated in several TCS-resistant mutants (51). The MufA3 protein and mufA3 transcripts have been reported to be overproduced in conditions simulating spaceflight (39) and also in response to the nonsteroidal anti-inflammatory drug diclofenac (50). The mufM gene was also reported to be transcriptionally upregulated in response to both spaceflight and conditions simulating spaceflight (39). Because of the recurring upregulation of mufM in different conditions, along with its conserved domain found in other Proteobacteria and the signal-specific induction of the mufA paralogs, it is hypothesized that the muf genes might be members of a novel stress-regulated cluster in R. rubrum. For development of a micropollutant biosensor, the mufA1 and mufA2 genes have the most potential as biomarker genes, since they are highly overexpressed and they appear to be specific to TCS exposure (50).
Conventional mechanisms for defense against TCS in Gram-negative and Gram-positive bacteria have been extensively studied. The most frequently described Gram-negative mechanisms for defense against TCS are active efflux via pumps belonging to the resistance-nodulation-cell division (RND) family (13, 43) and, to some extent, exclusion by making the cell envelope impermeable (11, 18). Accordingly, the majority of the induced genes in R. rubrum were related to efflux systems. While the MexAB-OprM homolog was the only TCS-induced efflux assembly with a porin in R. rubrum, it is known that the outer membrane component is not required for TCS efflux in Pseudomonas aeruginosa (14). In addition, most of the efflux systems were overexpressed only when R. rubrum was exposed to high concentrations of TCS, demonstrating that there is threshold-dependent regulation. Yet, it is still possible that the cells were able to initiate exponential growth (in the presence of 25 μg liter−1 TCS) merely because of TCS adsorption on the bacterial cell wall. Nevertheless, this study showed that several efflux systems were readily induced upon exposure to TCS. It therefore seems plausible that inhibition was reversed because of the concerted effects of adsorption, efflux, and membrane impermeability.
It is known that cell envelope permeability is an important factor in TCS resistance and that TCS can destabilize the cellular envelope (11, 45). Accordingly, numerous genes that are related to the cell envelope were found to be upregulated in R. rubrum upon exposure to TCS, and these genes included genes encoding OmpA family proteins, PssT, ExoD, and WbpM. The OmpA protein is assumed to maintain the structural link between the membrane and the cell wall. The Pss proteins are associated with exopolysaccharide synthesis, processing, transport, and regulation (40) and were reported previously to be required for tolerance to detergents and antibiotics in Rhizobium leguminosarum (74). More specifically, PssT appears to be linked to alteration of inner and outer membrane permeability (74). The exoD gene is associated with production of acidic exopolysaccharides in Rhizobium meliloti, and a mutation in the exoD gene causes the bacterium to produce less exopolysaccharide than the wild type (52). Finally, the highly conserved wbpML genes are also associated with polysaccharide synthesis (9). The induction of genes associated with exopolysaccharide synthesis (e.g., pssT, exoD, wbpM, and capM) thus might have conferred a certain degree of TCS resistance to R. rubrum by decreasing membrane permeability.
The phage shock proteins were initially discovered in response to phage IV infection, but they appear to be also transiently expressed in E. coli in response to a wide array of physical and chemical stresses, including heat, ethanol, and osmotic shock (8). The present study was the first study to demonstrate that there was induction of the psp operon upon exposure to TCS. Interestingly, PspA is known to negatively regulate σ54-induced transcription of pspF, which is the transcriptional activator of the pspABC operon (6, 17). However, expression of the psp operon has also been found to be independent of alternate sigma factors (8) and thus independent of PspF. These findings were confirmed in the present study since both pspF transcription and rpoN transcription were repressed upon exposure to TCS. The exact role of the Psp proteins in defense against TCS remains to be determined. Nevertheless, a role for PspA in maintaining the proton motive force (PMF) has been postulated previously (33). Because of the central role of the PMF in cell metabolism and the energy balance, it is hypothesized that PspA helps to maintain the PMF upon exposure to TCS, especially given the continuous removal of TCS by proton antiporter-type efflux pumps. The induction of numerous proteases and (deoxy)ribonucleases could have increased the responsiveness of the cell by increasing the rate of turnover of cellular constituents, such as amino acids and nucleotides.
TCS is a chlorinated biphenylether, but in addition to three chlorine residues, it also contains an hydroxyl moiety. Analogous to the findings for exposure of the fungus Saccharomyces cerevisiae to the structurally similar herbicide 2,4-dichlorophenoxyacetic acid (2,4-D), it is predicted that the weakly acidic molecule TCS (pKa 7.8) dissociates upon entry into the cytoplasm (70). As described above, the TCS hydroxyl dissociation in combination with the proton antiporter-mediated efflux could result in significant intracellular acidification. The subsequent increase in the number of intracellular protons could negatively influence the PMF of the bacterium, which is crucial for the energy balance and the efflux of TCS. The induction of carbonic anhydrase, an enzyme that catalyzes the reversible hydration of carbon dioxide and is known to have a pH-regulating function in bacterial cells and blood, could stabilize the intracellular pH (7, 63). Coinciding with this, the genes encoding the CO2-concentrating carboxysome were downregulated. Furthermore, it was shown that exposure to 2,4-D leads to generation of free radicals in a dose- and time course-dependent manner in yeast (70). Analogously, exposure of R. rubrum to TCS induced up to seven genes related to oxidative stress, which confirms the previously proposed prooxidant action of acidic chlorinated aromatics (70). In the present study, genes encoding a glutathione peroxidase and OsmC were found to be induced. These proteins preferentially metabolize organic hydroperoxides instead of inorganic hydrogen peroxide by using their highly reactive cysteine thiol groups to cause hydroperoxide reduction (37). Upregulation of both (peroxisomal) beta-oxidation of fatty acids and (mitochondrial) oxidative phosphorylation, as found in S. cerevisiae upon exposure to 2,4-D, was also observed when R. rubrum was exposed to TCS (69), and this could indicate that reactive oxygen species were formed. A larger amount of electron transport chain proteins might provide a shunt for the excess electrons that become available due to the prooxidant action of TCS. In addition, an upregulated electron transport chain might maintain the proton motive force and yield extra ATP through investment of reducing power in the form of NAD(P)H. The similarity between the observations of Teixeira and colleagues (69) and the observations described in the present study suggests that there is a common interkingdom response (Fungi and Bacteria) to different but chemically related environmental pollutants.
Finally, it was interesting that there was an induction of the chromosomal plasmid maintenance system after severe inhibition by TCS (data not shown), while the horizontal gene transfer machinery was significantly downregulated. TCS did not cause overexpression of its presumed target, FabI1, nor did it induce transcription of the putative TCS-insensitive FabI2 (36) in R. rubrum S1H. This observation was in accordance with the observations in previous studies (5, 31, 75), in which upregulation of the fabI orthologs was not observed in Staphylococcus aureus or Mycobacterium tuberculosis after acute exposure. Numerous hypothetical genes for which homologs with known function were found were reannotated using the experimental evidence obtained in this study.
The present study confirmed the previously established roles of efflux and cell envelope consolidation as mechanisms of defense against TCS (11, 13, 51) and identified the previously hypothetical muf genes as highly triclosan-responsive genes in R. rubrum S1H. Also, the apparent pivotal role of the PMF in protection against TCS was suggested by the results. The latter hypothesis was illustrated by induction of the proton antiporter-type efflux systems, the phage shock protein operon, and the carbonic anhydrase. In the framework of the MELiSSA, TCS appears to be merely a temporary inhibitor, since TCS hardly influences the generation time and it appears to be only slightly toxic. In the future, exposure to TCS (within the concentration limits) might be monitored by using the putative biomarkers that were identified in the present study. Future efforts will focus on characterization of the R. rubrum muf cluster in response to various stressors and the biodegradation of triclosan by the aerobic nitrifying bacteria of MELiSSA, Nitrosomonas europaea and Nitrobacter winogradskyi.
Acknowledgments
We are grateful to Bart Noten and Ann Janssen for technical assistance and to Aurélie Crabbé for critically reading the manuscript.
This research was funded in part by the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen) through a Ph.D. scholarship to Benny Pycke (grant IWT-SB/53360) and was supported in part by the European Space Agency BELISSIMA project and the Belgian Federal Science Policy.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Adolfsson-Erici, M., M. Pettersson, J. Parkkonen, and J. Sturve. 2002. Triclosan, a commonly used bactericide found in human milk and in the aquatic environment in Sweden. Chemosphere 46:1485-1489. [DOI] [PubMed] [Google Scholar]
- 2.Bagley, D. M., and Y. J. Lin. 2000. Clinical evidence for the lack of triclosan accumulation from daily use in dentifrices. Am. J. Dent. 13:148-152. [PubMed] [Google Scholar]
- 3.Bailey, T. L., and C. Elkan. 1995. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3:21-29. [PubMed] [Google Scholar]
- 4.Bester, K. 2005. Fate of triclosan and triclosan-methyl in sewage treatment plants and surface waters. Arch. Environ. Contam. Toxicol. 49:9-17. [DOI] [PubMed] [Google Scholar]
- 5.Betts, J. C., A. McLaren, M. G. Lennon, F. M. Kelly, P. T. Lukey, S. J. Blakemore, and K. Duncan. 2003. Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:2903-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bose, D., T. Pape, P. C. Burrows, M. Rappas, S. R. Wigneshweraraj, M. Buck, and X. Zhang. 2008. Organization of an activator-bound RNA polymerase holoenzyme. Mol. Cell 32:337-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braus-Stromeyer, S. A., G. Schnappauf, G. H. Braus, A. S. Gossner, and H. L. Drake. 1997. Carbonic anhydrase in Acetobacterium woodii and other acetogenic bacteria. J. Bacteriol. 179:7197-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brissette, J. L., M. Russel, L. Weiner, and P. Model. 1990. Phage shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 87:862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burrows, L. L., R. V. Urbanic, and J. S. Lam. 2000. Functional conservation of the polysaccharide biosynthetic protein WbpM and its homologues in Pseudomonas aeruginosa and other medically significant bacteria. Infect. Immun. 68:931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calafat, A. M., X. Ye, L. Y. Wong, J. A. Reidy, and L. L. Needham. 2008. Urinary concentrations of triclosan in the U.S. population: 2003-2004. Environ. Health Perspect. 116:303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Champlin, F. R., M. L. Ellison, J. W. Bullard, and R. S. Conrad. 2005. Effect of outer membrane permeabilisation on intrinsic resistance to low triclosan levels in Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 26:159-164. [DOI] [PubMed] [Google Scholar]
- 12.Chang, S. W., M. R. Hyman, and K. J. Williamson. 2002. Cooxidation of naphthalene and other polycyclic aromatic hydrocarbons by the nitrifying bacterium, Nitrosomonas europaea. Biodegradation 13:373-381. [DOI] [PubMed] [Google Scholar]
- 13.Chuanchuen, R., R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2003. High-level triclosan resistance in Pseudomonas aeruginosa is solely a result of efflux. Am. J. Infect. Control 31:124-127. [DOI] [PubMed] [Google Scholar]
- 14.Chuanchuen, R., C. T. Narasaki, and H. P. Schweizer. 2002. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J. Bacteriol. 184:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Gusseme, B., B. Pycke, T. Hennebel, A. Marcoen, S. E. Vlaeminck, H. Noppe, N. Boon, and W. Verstraete. 2009. Biological removal of 17α-ethinylestradiol by a nitrifier enrichment culture in a membrane bioreactor. Water Res. 43:2493-2503. [DOI] [PubMed] [Google Scholar]
- 16.Dokianakis, S. N., M. E. Kornaros, and G. Lyberatos. 2004. On the effect of pharmaceuticals on bacterial nitrite oxidation. Water Sci. Technol. 50:341-346. [PubMed] [Google Scholar]
- 17.Dworkin, J., G. Jovanovic, and P. Model. 2000. The PspA protein of Escherichia coli is a negative regulator of σ54-dependent transcription. J. Bacteriol. 182:311-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellison, M. L., A. L. Roberts, and F. R. Champlin. 2007. Susceptibility of compound 48/80-sensitized Pseudomonas aeruginosa to the hydrophobic biocide triclosan. FEMS Microbiol. Lett. 269:295-300. [DOI] [PubMed] [Google Scholar]
- 19.Escalada, M. G., A. D. Russell, J.-Y. Maillard, and D. Ochs. 2005. Triclosan-bacteria interactions: single or multiple target sites? Lett. Appl. Microbiol. 41:476-481. [DOI] [PubMed] [Google Scholar]
- 20.Gòdia, F., J. Albiol, J. Perez, N. Creus, F. Cabello, A. Montras, A. Masot, and C. Lasseur. 2004. The MELISSA pilot plant facility as a integration test-bed for advanced life support systems. Adv. Space Res. 34:1483-1493. [DOI] [PubMed] [Google Scholar]
- 21.Gomez, M. J., M. J. Martinez Bueno, S. Lacorte, A. R. Fernandez-Alba, and A. Aguera. 2007. Pilot survey monitoring pharmaceuticals and related compounds in a sewage treatment plant located on the Mediterranean coast. Chemosphere 66:993-1002. [DOI] [PubMed] [Google Scholar]
- 22.Graebe, A., E. L. Schuck, P. Lensing, L. Putcha, and H. Derendorf. 2004. Physiological, pharmacokinetic, and pharmacodynamic changes in space. J. Clin. Pharmacol. 44:837-853. [DOI] [PubMed] [Google Scholar]
- 23.Guillén, J., A. Bernabeu, S. Shapiro, and J. Villalaín. 2004. Location and orientation of triclosan in phospholipid model membranes. Eur. Biophys. J. 33:448-453. [DOI] [PubMed] [Google Scholar]
- 24.Heath, R. J., J. Li, G. E. Roland, and C. O. Rock. 2000. Inhibition of the Staphylococcus aureus NADPH-dependent enoyl-acyl carrier protein reductase by triclosan and hexachlorophene. J. Biol. Chem. 275:4654-4659. [DOI] [PubMed] [Google Scholar]
- 25.Heath, R. J., and C. O. Rock. 2000. A triclosan-resistant bacterial enzyme. Nature 406:145-146. [DOI] [PubMed] [Google Scholar]
- 26.Heidler, J., and R. U. Halden. 2007. Mass balance assessment of triclosan removal during conventional sewage treatment. Chemosphere 66:362-369. [DOI] [PubMed] [Google Scholar]
- 27.Heidler, J., and R. U. Halden. 2008. Meta-analysis of mass balances examining chemical fate during wastewater treatment. Environ. Sci. Technol. 42:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendrickx, L., H. De Wever, V. Hermans, F. Mastroleo, N. Morin, A. Wilmotte, P. Janssen, and M. Mergeay. 2006. Microbial ecology of the closed artificial ecosystem MELiSSA (Micro-Ecological Life Support System Alternative): reinventing and compartmentalizing the Earth's food and oxygen regeneration system for long-haul space exploration missions. Res. Microbiol. 157:77-86. [DOI] [PubMed] [Google Scholar]
- 29.Hendrickx, L., and M. Mergeay. 2007. From the deep sea to the stars: human life support through minimal communities. Curr. Opin. Microbiol. 10:231-237. [DOI] [PubMed] [Google Scholar]
- 30.Hoang, T. T., and H. P. Schweizer. 1999. Characterization of Pseudomonas aeruginosa enoyl-acyl carrier protein reductase (FabI): a target for the antimicrobial triclosan and its role in acylated homoserine lactone synthesis. J. Bacteriol. 181:5489-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang, H. J., M. W. Chang, F. Toghrol, and W. E. Bentley. 2008. Microarray analysis of toxicogenomic effects of triclosan on Staphylococcus aureus. Appl. Microbiol. Biotechnol. 78:695-707. [DOI] [PubMed] [Google Scholar]
- 32.Jones, R. D., H. B. Jampani, J. L. Newman, and A. S. Lee. 2000. Triclosan: a review of effectiveness and safety in health care settings. Am. J. Infect. Control. 28:184-196. [PubMed] [Google Scholar]
- 33.Kleerebezem, M., W. Crielaard, and J. Tommassen. 1996. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the proton-motive force under stress conditions. EMBO J. 15:162-171. [PMC free article] [PubMed] [Google Scholar]
- 34.Kolpin, D. W., E. T. Furlong, M. T. Meyer, E. M. Thurman, S. D. Zaugg, L. B. Barber, and H. T. Buxton. 2002. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999-2000: a national reconnaissance. Environ. Sci. Technol. 36:1202-1211. [DOI] [PubMed] [Google Scholar]
- 35.Lasseur, C., W. Verstraete, J. B. Gros, G. Dubertret, and F. Rogalla. 1996. MELISSA: a potential experiment for a precursor mission to the Moon. Adv. Space Res. 18:111-117. [DOI] [PubMed] [Google Scholar]
- 36.Lee, I. H., E. J. Kim, Y. H. Cho, and J. K. Lee. 2002. Characterization of a novel enoyl-acyl carrier protein reductase of diazaborine-resistant Rhodobacter sphaeroides mutant. Biochem. Biophys. Res. Commun. 299:621-627. [DOI] [PubMed] [Google Scholar]
- 37.Lesniak, J., W. A. Barton, and D. B. Nikolov. 2003. Structural and functional features of the Escherichia coli hydroperoxide resistance protein OsmC. Protein Sci. 12:2838-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lishman, L., S. A. Smyth, K. Sarafin, S. Kleywegt, J. Toito, T. Peart, B. Lee, M. Servos, M. Beland, and P. Seto. 2006. Occurrence and reductions of pharmaceuticals and personal care products and estrogens by municipal wastewater treatment plants in Ontario, Canada. Sci. Total Environ. 367:544-558. [DOI] [PubMed] [Google Scholar]
- 39.Mastroleo, F., R. Van Houdt, B. Leroy, M. A. Benotmane, A. Janssen, M. Mergeay, F. Vanhavere, L. Hendrickx, R. Wattiez, and N. Leys. 2009. Experimental design and environmental parameters affect Rhodospirillum rubrum S1H response to space flight. ISME J. 3:1402-1419. [DOI] [PubMed] [Google Scholar]
- 40.Mazur, A., J. E. Krol, M. Marczak, and A. Skorupska. 2003. Membrane topology of PssT, the transmembrane protein component of the type I exopolysaccharide transport system in Rhizobium leguminosarum bv. trifolii strain TA1. J. Bacteriol. 185:2503-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McAvoy, D. C., B. Schatowitz, M. Jacob, A. Hauk, and W. S. Eckhoff. 2002. Measurement of triclosan in wastewater treatment systems. Environ. Toxicol. Chem. 21:1323-1329. [PubMed] [Google Scholar]
- 42.McDonough, W., and M. Braungart. 2002. Cradle to cradle. Remaking the way we make things. North Point Press, San Francisco, CA.
- 43.McMurry, L. M., M. Oethinger, and S. B. Levy. 1998. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol. Lett. 166:305-309. [DOI] [PubMed] [Google Scholar]
- 44.McMurry, L. M., M. Oethinger, and S. B. Levy. 1998. Triclosan targets lipid synthesis. Nature 394:531-532. [DOI] [PubMed] [Google Scholar]
- 45.Meincke, B. E., R. G. Kranz, and D. L. Lynch. 1980. Effect of irgasan on bacterial growth and its adsorption into the cell wall. Microbios 28:133-147. [PubMed] [Google Scholar]
- 46.Moss, T., D. Howes, and F. M. Williams. 2000. Percutaneous penetration and dermal metabolism of triclosan (2,4,4′-trichloro-2′-hydroxydiphenyl ether). Food Chem. Toxicol. 38:361-370. [DOI] [PubMed] [Google Scholar]
- 47.Nakada, N., T. Tanishima, H. Shinohara, K. Kiri, and H. Takada. 2006. Pharmaceutical chemicals and endocrine disrupters in municipal wastewater in Tokyo and their removal during activated sludge treatment. Water Res. 40:3297-3303. [DOI] [PubMed] [Google Scholar]
- 48.Ning, C., and H. Gest. 1966. Regulation of l-isoleucine biosynthesis in the photosynthetic bacterium Rhodospirillum rubrum. Proc. Natl. Acad. Sci. U. S. A. 56:1823-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pavesi, G., P. Mereghetti, F. Zambelli, M. Stefani, G. Mauri, and G. Pesole. 2006. MoD Tools: regulatory motif discovery in nucleotide sequences from co-regulated or homologous genes. Nucleic Acids Res. 34:W566-W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pycke, B. 2009. The fate and effects of micropollutants in a biological life support system. Ph.D. thesis. Ghent University, Ghent, Belgium.
- 51.Pycke, B. F. G., A. Crabbé, W. Verstraete, and N. Leys. 2010. Characterization of triclosan-resistant mutants reveals multiple antimicrobial resistance mechanisms in Rhodospirillum rubrum S1H. Appl. Environ. Microbiol. 76:3116-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reed, J. W., and G. C. Walker. 1991. The exoD gene of Rhizobium meliloti encodes a novel function needed for alfalfa nodule invasion. J. Bacteriol. 173:664-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reslewic, S., S. Zhou, M. Place, Y. Zhang, A. Briska, S. Goldstein, C. Churas, R. Runnheim, D. Forrest, A. Lim, A. Lapidus, C. S. Han, G. P. Roberts, and D. C. Schwartz. 2005. Whole-genome shotgun optical mapping of Rhodospirillum rubrum. Appl. Environ. Microbiol. 71:5511-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roth, F. P., J. D. Hughes, P. W. Estep, and G. M. Church. 1998. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 16:939-945. [DOI] [PubMed] [Google Scholar]
- 55.Roujeinikova, A., C. W. Levy, S. Rowsell, S. Sedelnikova, P. J. Baker, C. A. Minshull, A. Mistry, J. G. Colls, R. Camble, A. R. Stuitje, A. R. Slabas, J. B. Rafferty, R. A. Pauptit, R. Viner, and D. W. Rice. 1999. Crystallographic analysis of triclosan bound to enoyl reductase. J. Mol. Biol. 294:527-535. [DOI] [PubMed] [Google Scholar]
- 56.Sandborgh-Englund, G., M. Adolfsson-Erici, G. Odham, and J. Ekstrand. 2006. Pharmacokinetics of triclosan following oral ingestion in humans. J. Toxicol. Environ. Health A 69:1861-1873. [DOI] [PubMed] [Google Scholar]
- 57.Schroeder, A., O. Mueller, S. Stocker, R. Salowsky, M. Leiber, M. Gassmann, S. Lightfoot, W. Menzel, M. Granzow, and T. Ragg. 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi, J., S. Fujisawa, S. Nakai, and M. Hosomi. 2004. Biodegradation of natural and synthetic estrogens by nitrifying activated sludge and ammonia-oxidizing bacterium Nitrosomonas europaea. Water Res. 38:2322-2329. [DOI] [PubMed] [Google Scholar]
- 59.Siddiqui, W. H., and H. S. Buttar. 1979. Pharmacokinetics of triclosan in rat after intravenous and intravaginal administration. J. Environ. Pathol. Toxicol. 2:861-871. [PubMed] [Google Scholar]
- 60.Singer, H., S. Muller, C. Tixier, and L. Pillonel. 2002. Triclosan: occurrence and fate of a widely used biocide in the aquatic environment: field measurements in wastewater treatment plants, surface waters, and lake sediments. Environ. Sci. Technol. 36:4998-5004. [DOI] [PubMed] [Google Scholar]
- 61.Sistrom, W. R. 1960. A requirement for sodium in the growth of Rhodopseudomonas spheroides. J. Gen. Microbiol. 22:778-785. [DOI] [PubMed] [Google Scholar]
- 62.Slayden, R. A., R. E. Lee, and C. E. Barry III. 2000. Isoniazid affects multiple components of the type II fatty acid synthase system of Mycobacterium tuberculosis. Mol. Microbiol. 38:514-525. [DOI] [PubMed] [Google Scholar]
- 63.Smith, K. S., and J. G. Ferry. 2000. Prokaryotic carbonic anhydrases. FEMS Microbiol. Rev. 24:335-366. [DOI] [PubMed] [Google Scholar]
- 64.Stasinakis, A. S., D. Mamais, N. S. Thomaidis, E. Danika, G. Gatidou, and T. D. Lekkas. 2008. Inhibitory effect of triclosan and nonylphenol on respiration rates and ammonia removal in activated sludge systems. Ecotoxicol. Environ. Saf. 70:199-206. [DOI] [PubMed] [Google Scholar]
- 65.Stasinakis, A. S., A. V. Petalas, D. Mamais, N. S. Thomaidis, G. Gatidou, and T. D. Lekkas. 2007. Investigation of triclosan fate and toxicity in continuous-flow activated sludge systems. Chemosphere 68:375-381. [DOI] [PubMed] [Google Scholar]
- 66.Suller, M. T., and A. D. Russell. 2000. Triclosan and antibiotic resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 46:11-18. [DOI] [PubMed] [Google Scholar]
- 67.Sullivan, L. P., and J. J. Grantham. 1982. Physiology of the kidney, 2nd ed., p. 236. Lea & Febiger, Philadelphia, PA.
- 68.Taylor, D. E. 1999. Bacterial tellurite resistance. Trends Microbiol. 7:111-115. [DOI] [PubMed] [Google Scholar]
- 69.Teixeira, M. C., P. Duque, and I. Sa-Correia. 2007. Environmental genomics: mechanistic insights into toxicity of and resistance to the herbicide 2,4-D. Trends Biotechnol. 25:363-370. [DOI] [PubMed] [Google Scholar]
- 70.Teixeira, M. C., J. P. Telo, N. F. Duarte, and I. Sa-Correia. 2004. The herbicide 2,4-dichlorophenoxyacetic acid induces the generation of free-radicals and associated oxidative stress responses in yeast. Biochem. Biophys. Res. Commun. 324:1101-1107. [DOI] [PubMed] [Google Scholar]
- 71.Vallenet, D., L. Labarre, Z. Rouy, V. Barbe, S. Bocs, S. Cruveiller, A. Lajus, G. Pascal, C. Scarpelli, and C. Medigue. 2006. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 34:53-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Helden, J. 2003. Regulatory sequence analysis tools. Nucleic Acids Res. 31:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Villalaín, J., C. R. Mateo, F. J. Aranda, S. Shapiro, and V. Micol. 2001. Membranotropic effects of the antibacterial agent triclosan. Arch. Biochem. Biophys. 390:128-136. [DOI] [PubMed] [Google Scholar]
- 74.Wielbo, J., A. Mazur, J. E. Krol, M. Marczak, and A. Skorupska. 2004. Environmental modulation of the pssTNOP gene expression in Rhizobium leguminosarum bv. trifolii. Can. J. Microbiol. 50:201-211. [DOI] [PubMed] [Google Scholar]
- 75.Wilson, M., J. DeRisi, H. H. Kristensen, P. Imboden, S. Rane, P. O. Brown, and G. K. Schoolnik. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. U. S. A. 96:12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ye, X., Z. Kuklenyik, L. L. Needham, and A. M. Calafat. 2005. Automated on-line column-switching HPLC-MS/MS method with peak focusing for the determination of nine environmental phenols in urine. Anal. Chem. 77:5407-5413. [DOI] [PubMed] [Google Scholar]
- 77.Ying, G. G., and R. S. Kookana. 2007. Triclosan in wastewaters and biosolids from Australian wastewater treatment plants. Environ. Int. 33:199-205. [DOI] [PubMed] [Google Scholar]
- 78.Ying, G. G., X. Y. Yu, and R. S. Kookana. 2007. Biological degradation of triclocarban and triclosan in a soil under aerobic and anaerobic conditions and comparison with environmental fate modelling. Environ. Pollut. 150:300-305. [DOI] [PubMed] [Google Scholar]