Abstract
The extracellular medium-chain-length polyhydroxyalkanoate (MCL-PHA) depolymerase of Pseudomonas fluorescens GK13 catalyzes the hydrolysis of poly(3-hydroxyoctanoic acid) [P(3HO)]. Based on the strong tendency of the enzyme to interact with hydrophobic materials, a low-cost method which allows the rapid and easy purification and immobilization of the enzyme has been developed. Thus, the extracellular P(3HO) depolymerase present in the culture broth of cells of P. fluorescens GK13 grown on mineral medium supplemented with P(3HO) as the sole carbon and energy source has been tightly adsorbed onto a commercially available polypropylene support (Accurel MP-1000) with high yield and specificity. The activity of the pure enzyme was enhanced by the presence of detergents and organic solvents, and it was retained after treatment with an SDS-denaturing cocktail under both reducing and nonreducing conditions. The time course of the P(3HO) hydrolysis catalyzed by the soluble and immobilized enzyme has been assessed, and the resulting products have been identified. After 24 h of hydrolysis, the dimeric ester of 3-HO [(R)-3-HO-HO] was obtained as the main product of the soluble enzyme. However, the immobilized enzyme catalyzes almost the complete hydrolysis of P(3HO) polymer to (R)-3-HO monomers under the same conditions.
Polyhydroxyalkanoates (PHAs) are environmentally friendly polyesters that are biosynthesized by numerous microorganisms during unbalanced growth (3, 32). PHAs show material properties similar to those of conventional plastics, having important advantages such as biodegradability, apparent biocompatibility, and the ability to be manufactured from renewable resources (6, 38, 39). According to the number of carbon atoms of the side chain of the monomers, PHAs are classified as short-chain-length (SCL) PHAs (3 to 5 carbon atoms) and medium-chain-length (MCL) PHAs (6 to 14 carbon atoms) (16, 17, 32).
The ability to degrade extracellular PHA in the environment and to use its degradation products as a source of carbon and energy depends on the release of specific extracellular PHA depolymerases (14, 15, 20). Depending on the depolymerase, as a result of enzymatic PHA degradation, the end products are only monomers, both monomers and dimers, or a mixture of oligomers (16). Enantiomer pure (R)-3-hydroxyalkanoic acid [(R)-3-HA] monomers are very attractive building blocks of interest not only in the biomedical and pharmaceutical fields (9, 10) but also for being used as starting materials to obtain other new polyesters (8). Thus, the development of a cost-effective industrial process for the production of both MCL-PHA depolymerase enzyme and (R)-3-HA monomers is of considerable interest.
At present, few extracellular MCL-PHA depolymerases have been purified and characterized (11, 21-24, 33). Traditionally, the purification of microbial depolymerases is achieved by a conventional multistep chromatographic methodology, which includes hydrophobic interaction and size exclusion chromatographies (7, 21, 24, 37). The poly(3-hydroxyoctanoic acid) [P(3HO)] depolymerase from Pseudomonas fluorescens GK13 was the first enzyme purified (37) and characterized at the molecular level (36).
Adsorption of lipases on polypropylene supports has been extensively used for large-scale lipase immobilization (18, 25, 28, 29) since it is a simple and economical method. Moreover, the immobilization of enzyme allows its reusability and increases its operational stability and ease of product recovery (1). Accurel MP-1000 is a commercially available hydrophobic, microporous, low-density polypropylene powder that presents a large surface area for adsorption because of its very small particle size (4). This support has been successfully used for adsorption of lipases and esterases with high yield directly from the fermentation broth (2, 13).
As lipases, MCL-PHA depolymerases are hydrophobic proteins with a tendency to adsorb to hydrophobic supports. In this study we report a novel method for the purification of the P(3HO) depolymerase from P. fluorescens GK13 by adsorption to a polypropylene support as well as some relevant properties of the enzyme. Moreover, this protocol allows the immobilization of the enzyme directly from the culture broth. The immobilized enzyme degrades completely the P(3HO) polymer and releases 3-hydroxyoctanoic acid [(R)-3-HO]. This is the first report describing the immobilization of an extracellular MCL-PHA depolymerase and its potential use in the production of (R)-3-HO chiral monomers.
MATERIALS AND METHODS
Chemicals.
P(3HO) was supplied by Biopolis (Valencia, Spain) and CPI (Newcastle, United Kingdom). The polypropylene support Accurel MP-1000 was purchased from Membrana GmbH (Obenburg, Germany). Chromatography media and equipment were obtained from GE Healthcare (Uppsala, Sweden). Molecular weight standards, p-nitrophenyl (PNP)-alkanoates, and most chemicals were obtained from Sigma-Aldrich (St. Louis, MO). All other chemicals were purchased from Merck (Darmstadt, Germany).
Microorganism and growth conditions.
Pseudomonas fluorescens GK13 (DSM 7139) cells were grown at 30°C in Erlenmeyer flasks containing 100 ml of mineral medium M9 (35) supplemented with a film (0.15 g) of P(3HO) as the sole carbon and energy source.
Preparation of PHA suspensions.
Latex suspensions of P(3HO), composed of 11% 3-hydroxyhexanoate (3-HX) and 89% 3-hydroxyoctanoate (3-HO), were prepared according to the method of Schirmer and Jendrossek (36). Briefly, 4 volumes of cold water was poured into 1 volume of polymer solution in acetone with stirring, and the solvent was then evaporated.
Production of P(3HO) depolymerase.
A 2-liter Biostat A fermentor (B. Braun Biotech, Melsungen, Germany) containing 1.5 liters of mineral medium M9 (35) supplemented with a film of P(3HO) (0.15%, wt/vol) was inoculated with seed cultures of P. fluorescens GK13 until the optical density at 600 nm (OD600) was 0.2. The film was obtained by dissolving the polymer in chloroform in a beaker. Then, the solvent was evaporated under vacuum in a fume cupboard and the resulting film was stripped off from the bottom, cut in small pieces, and added to the culture medium in the fermentor before autoclaving. The temperature and pH controls were automatically adjusted at 30°C and 8.0, respectively, and O2 saturation was maintained over 40% (vol/vol). The agitation speed was controlled in the range 100 to 200 rpm. To evaluate the production of the enzyme, 4.5 ml of culture broth was concentrated 90-fold by precipitation with 10% (wt/vol) trichloroacetic acid (TCA) at different times and analyzed by SDS-PAGE. Simultaneously, the activity of the culture broth was semiquantitatively evaluated using a drop test on P(3HO) agarose plates (36).
Purification of P(3HO) depolymerase.
The enzyme was purified from the broth of cultures of P. fluorescens GK13 grown for 72 h under the conditions described above. Cells were harvested by centrifugation (10 min, 8,000 × g, 4°C) in a Kubota 7820 centrifuge with an RA-6 rotor (Kubota, Tokyo, Japan). The supernatant was saved and adjusted to 200 mM phosphate with 1 M phosphate buffer, pH 8.0. Then, 1.5 g of Accurel MP-1000, prewetted with ethanol, was added. The preparation was stirred gently for 15 h at 4°C, and the liquid phase was discarded. The polypropylene support was washed thoroughly with 200 mM phosphate buffer (pH 8.0) and finally packed in a chromatography column (20 cm × 2.6 cm). The adsorbed protein was eluted using a linear gradient of 2-propanol (from 0 to 80% [vol/vol]) in 5 mM phosphate buffer (pH 8.0). Further purification was achieved when the active fractions were concentrated (about 110-fold) by ultrafiltration (YM30 membranes; Millipore, Billerica, MA) and resolved in a Sephacryl S-200 (30 cm × 1.5 cm) column equilibrated and eluted (at a 1-ml/min flow rate) with 50 mM glycine-NaOH buffer (pH 9.0) supplemented with 0.15 M KCl. Fractions of 0.5 ml were collected using an Äkta fast protein liquid chromatograph (FPLC; GE Healthcare, Uppsala, Sweden). The purification progress was assessed by SDS-PAGE analysis of fractions and by assaying the P(3HO) depolymerase activity in P(3HO) agarose plates.
Immobilization of P(3HO) depolymerase.
The enzyme was immobilized directly from the culture broth by adsorption onto a polypropylene matrix using the same conditions as those described above for the purification. The time course of adsorption of the enzyme to the support was assessed by measuring the remaining soluble activity in the liquid phase by using P(3HO) agarose plates. To calculate the depolymerase theoretical loading, the enzyme adsorbed in 0.5 g of polypropylene was eluted with 80% (vol/vol) 2-propanol in 5 mM phosphate buffer, pH 8.0, and the protein concentration was determined.
SDS-PAGE analysis of the immobilized preparation.
Samples of the adsorbed enzyme (50 mg of wet support) were boiled for 5 min in 110 μl of sample buffer in the presence of 2% (wt/vol) SDS and 5% (vol/vol) 2-mercaptoethanol and centrifuged (13,000 ×g, 15 min, 4°C), and the supernatants were analyzed by SDS-PAGE. This treatment released any protein molecule physically bound to the support.
Enzyme assays.
Routinely, the P(3HO) depolymerase activity was assayed in a drop test on P(3HO) agarose plates (36) with minor modifications. Five milliliters of a 1% (wt/vol) emulsion of P(3HO) was mixed with 5 ml of 1% (wt/vol) agarose in 0.2 M Tris-HCl buffer, pH 8.5, and poured on a glass plate. Samples (20 μl) were loaded in (5-mm-diameter) holes made in the gel and incubated at 30°C for 24 h. The P(3HO) depolymerase activity was determined by measuring the diameter of the resulting clear zone. The esterase activity was assayed using a number of p-nitrophenyl (PNP)-alkanoates as substrate. The reaction mixture contained 25 mU of the enzyme preparation in 700 μl of 200 mM bicarbonate-NaOH buffer (pH 9.5). To start the reaction, 21 μl of a 10 mM solution of the respective PNP-alkanoate in 95% (vol/vol) ethanol was added. Controls without the enzyme were carried out in parallel to ascertain the possible nonenzymatic hydrolysis of substrate. One unit of esterase activity was the amount of enzyme that released 1 μmol of p-nitrophenol per min under standard conditions. An extinction coefficient (ɛ) for PNP at pH 9.5 of 16.635 mM−1·cm−1 was assumed.
Alternatively, P(3HO) depolymerase activity was determined by measuring for 5 min at 650 nm and 30°C the turbidity decrease of P(3HO) suspensions. The standard assay mixture consisted of 500 μg of P(3HO) in 200 mM Tris-HCl (pH 8.0) in a total volume of 1 ml. After incubation at 30°C for 10 min, the reaction was started by the addition of the purified enzyme. When the immobilized enzyme was assayed, the polypropylene-adsorbed protein was added to the reaction mixture and incubated for 15 min in a gyratory shaker (at 100 rpm) at room temperature. One unit of enzyme activity was the amount of enzyme which catalyzes the decrease of 1 absorbance unit per min at 650 nm.
Enzymatic hydrolysis of P(3HO) polymer and identification of products.
The in vitro hydrolysis of P(3HO) latex catalyzed by P. fluorescens GK13 depolymerase was assessed at 30°C and with 180-rpm stirring for various time intervals (30 min, 2 h, 4 h, and 24 h). The reaction mixtures contained 8 mg of polymer latex in 20 mM phosphate buffer, pH 8.0. The reaction was started by adding 160 μg of the purified enzyme, either in soluble form or immobilized on Accurel. The enzymatic reaction was stopped by heating samples for 5 min at 100°C, and then the reaction mixture was centrifuged for 60 min at 14,000 × g, 4°C. The hydrolysis products in supernatants were identified after derivatization (12) with bromophenacyl bromide (BPB). Then, samples (10 μl) of the derivatization mixture were loaded onto a reverse-phase C18 high-performance liquid chromatography (HPLC) column (Tracer Lichrosorb RP18; 10 μm, 250 mm × 4 mm) in a Waters Corporation chromatograph (Milford, MA). Analytes were eluted at a 1-ml/min flow rate as reported elsewhere (12). The detection of the BPB derivatives was monitored at 254 nm by a photodiode array (PDA) detector, the excess of unreacted BPB being eluted at 24.8 min.
The peaks detected at 254 nm were identified by HPLC-mass spectrometry (HPLC-MS). Mass spectra were obtained on a Micromass (Milford, MA) Quattro micro-triple-quadrupole mass spectrometer coupled at the exit of the diode array detector and equipped with a Z-spray electrospray ionization (ESI) source. A 200-μl/min flow rate from the detector eluent was directed to the ESI interface using a flow splitter. Nitrogen was used as desolvation gas, at 300°C and a flow rate of 450 liters/h, and no cone gas was used. A potential of 3.2 kV was used on the capillary for positive-ion mode. The source block temperature was held at 120°C. MS spectra, within the m/z range 50 to 1,000 atomic mass units, were produced in the positive mode at different cone voltages (10, 20, 30, and 40 V).
Other analytical methods.
SDS-PAGE was performed as described by Laemmli (27). Two-dimensional electrophoresis was performed by isoelectric focusing using immobilized pH gradient (IPG) strips (pH 3 to 10) (first dimension) and SDS-polyacrylamide gel electrophoresis (second dimension). Protein concentration was determined by the method of Peterson (31) using bovine serum albumin as the standard.
RESULTS
Production of P(3HO) depolymerase.
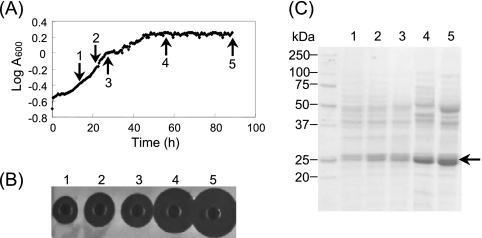
The time course of extracellular production of the P. fluorescens GK13 depolymerase was evaluated in a 2-liter fermentor containing 1.5 liters of mineral medium M9 (35) supplemented with P(3HO) (0.15% [wt/vol]). High enzyme activity was detected only at the end of the exponential phase of growth or after its cessation (Fig. 1). Moreover, the depolymerase activity could be detected only when cells were grown in mineral media containing P(3HO), not when they were grown in media supplemented with either octanoate, lactate, or pyruvate as the sole carbon source (data not shown). These results confirm that the synthesis of PHA depolymerases in bacteria is generally repressed if suitable soluble carbon sources, such as glucose or organic acids, are present (16).
FIG. 1.
Time course appearance of P(3HO) depolymerase in the culture broth of P. fluorescens GK13 cells growing in a fermentor containing mineral medium supplemented with P(3HO) as the sole carbon and energy source. (A) Growth curve of cells in basal medium containing 0.15% (wt/vol) P(3HO). (B) Enzyme activity (after 12 h) measured in P(3HO) agarose plates using 20 μl of culture broth. (C) SDS-PAGE analysis of samples concentrated (90-fold) by precipitation with TCA in a homogeneous 12% (wt/vol) acrylamide gel revealed with Coomassie brilliant blue R-250. Broth samples (5 ml) were withdrawn at the times (arrows) indicated in panel A. Plates 1 to 5 in panel B and lanes 1 to 5 in panel C correspond to numbers 1 to 5 in panel A. Arrow in panel C shows the position of the enzyme.
Purification of P(3HO) depolymerase.
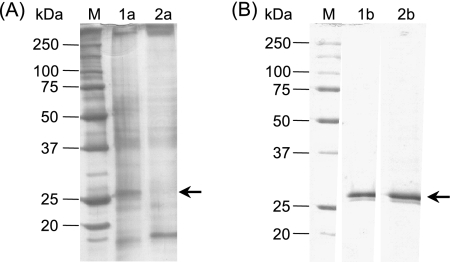
The extracellular P(3HO) depolymerase present in the culture broth was adsorbed directly to the microporous polypropylene Accurel MP-1000 resin with high yield and specificity. Due to the strong adsorption of the enzyme to hydrophobic materials, about 80% of the P(3HO) depolymerase activity was immobilized onto the polypropylene matrix while only 35% of the total protein present in the culture broth was retained (Fig. 2A). The strongly hydrophobically bound proteins were desorbed and analyzed by SDS-PAGE. Thus, samples of the supernatant obtained after boiling 50 mg (wet weight) of Accurel bearing the adsorbed enzyme in the presence of SDS and 2-mercaptoethanol showed negligible amounts of immobilized contaminating proteins which were undetectable in polyacrylamide gels revealed with Coomassie blue R-250 (Fig. 2B). The negligible presence of contaminants in the desorbed physically bound proteins indicated that the adsorption of the enzyme to the polypropylene support is highly specific.
FIG. 2.
SDS-PAGE analysis of samples of P. fluorescens GK13 P(3HO) depolymerase at different stages of purification. (A) Lane M, molecular mass standards; lane 1a, sample of original culture broth (0.75 μg protein); lane 2a, proteins remaining in solution after exposition for 15 h at 4°C of culture broth with Accurel MP-1000. (B) Lane M, molecular mass standards; lane 1b, purified soluble enzyme (2.7 μg of protein) after its desorption from Accurel MP-1000 with 2-propanol; lane 2b, desorbed proteins of a sample of 50 mg of support with immobilized enzyme boiled in the presence of 2% (wt/vol) SDS and 5% (vol/vol) 2-mercaptoethanol. Homogeneous (12% [wt/vol]) acrylamide gels revealed with either silver staining (A) or Coomassie brilliant blue R-250 (B) were used. Arrows show the position of the enzyme.
To recover the enzyme, the adsorbed P(3HO) depolymerase activity was eluted with 80% (vol/vol) 2-propanol (Fig. 2B, lane 1b). Typical results of the enzyme purification protocol are shown in Table 1. This method enables recovery of the majority of the enzyme present in the broth and other proteins, to a lesser extent, which could be detected only in polyacrylamide gels revealed by silver staining. As indicated above, these minor proteins, which do not seem to interfere with the depolymerase activity, could be completely removed by size exclusion chromatography.
TABLE 1.
Purification of the extracellular P(3HO) depolymerase from P. fluorescens GK13 from 1 liter of culture broth of cells grown in mineral medium supplemented with P(3HO) as the sole carbon sourcea
| Purification step | Vol (ml) | Protein (mg) | Activity (U) | Sp act (U/mg protein) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Culture broth | 840 | 41.3 | 756.0 | 18.3 | 1.0 | 100.0 |
| Polypropylene eluate | 35 | 6.3 | 443.7 | 70.1 | 3.8 | 58.7 |
Activity was determined by the standard assay using P(3HO) latex as a substrate.
Characterization of extracellular P(3HO) depolymerase. (i) Molecular mass and pI.
As shown in Fig. 2B, SDS-PAGE analysis of preparations of the purified enzyme revealed a double band with a mass of ∼26 kDa. After analysis of the protein by two-dimensional PAGE, two spots with pIs of 6.45 and 6.95 were detected (data not shown). These pI values are significantly more alkaline than those reported for other MCL-PHA depolymerases so far (19).
(ii) Effects of pH and temperature on enzymatic activity.
The P(3HO) depolymerase showed its maximum activity at pH 9.5 and retained at least 60% of this activity over a pH range from pH 8.0 to 11.0. The enzyme exhibited the highest activity at 40°C in 200 mM bicarbonate-NaOH buffer, pH 9.5, and showed relatively high stability at temperatures below 45°C. The purified enzyme retained at least 75% of its original activity after 5 months of storage at −20 and −80°C.
(iii) Substrate specificity.
P(3HO) depolymerase can hydrolyze various PNP-alkanoates of different chain lengths which are substrates for esterase and lipase activities. The enzyme activity for shorter-chain-length PNP-alkanoates was significantly lower. Among the PNP-alkanoates tested, PNP-tetradecanoate was hydrolyzed most efficiently by the enzyme (Table 2). The apparent Km and Vmax values for p-nitrophenyl octanoate (PNPO) and for P(3HO) were 190 μM and 19.4 U/mg protein and 34 μg/ml and 76 U/mg protein, respectively.
TABLE 2.
Relative activity of pure preparations of P(3HO) depolymerase of P. fluorescens GK13 with several chromogenic substrates assayed at a final concentration of 0.3 mM in all casesa
| Substrate | Relative activity (%) | |
|---|---|---|
| PNP-acetate | 38.9 | |
| PNP-butyrate | 10.7 | |
| PNP-valerate | 44.2 | |
| PNP-octanoate | 82.7 | |
| PNP-decanoate | 88.7 | |
| PNP-dodecanoate | 95.2 | |
| PNP-tetradecanoate | 100.0 | |
| PNP-hexadecanoate | 51.6 | |
| PNP-octadecanoate | 24.7 |
One hundred percent activity corresponded to 11.3 U/mg protein.
(iv) Effects of a number of chemicals on the enzyme activity.
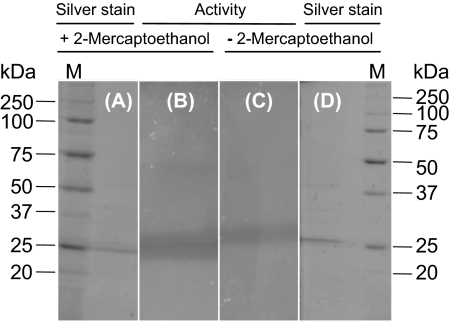
Of the tested compounds (Table 3), only phenylmethylsulfonyl fluoride (PMSF) and EDTA significantly decreased the P(3HO) depolymerase activity assayed with P(3HO) as substrate. Interestingly, the activity was enhanced in the presence of both nonionic and ionic detergents such as 0.1% (wt/vol) Tween 80, 0.005% (wt/vol) Triton X-100, and 0.005% (wt/vol) SDS, as well as in the presence of 5% (vol/vol) 2-propanol. Furthermore, the enzyme retained its activity even after being treated for 15 min at room temperature with the denaturing cocktail and in subsequent analysis by SDS-PAGE under both reducing and nonreducing conditions (Fig. 3). The inactivation of the depolymerase activity by PMSF confirms that the enzyme behaves as a typical serine hydrolase. Moreover, a dependence on reduced thiol groups or essential disulfide bonds is unlikely (37). However, when PNPO was used as a substrate, the enzyme activity was not significantly affected by any of the chemicals shown in Table 3, except by detergents, which appear to markedly diminish the activity.
TABLE 3.
Effects of various chemicals on P(3HO) depolymerase activitya
| Chemical and final concn | Relative activity (%) |
|---|---|
| None (control) | 100.0 |
| DTT (10 mM) | 74.5 |
| Sodium azide (10 mM) | 95.7 |
| N-Ethylmaleimide (10 mM) | 85.4 |
| Iodoacetamide (10 mM) | 89.7 |
| Carbodiimide (10 mM) | 87.7 |
| Acetic anhydride (10 mM) | 92.0 |
| PMSF (10 mM) | 5.7 |
| EDTA (10 mM) | 28.9 |
| Triton X-100 (% [wt/vol]) | |
| 0.005 | 132 |
| 0.01 | 108 |
| 0.03 | 0.9 |
| Tween 80 (0.1% [wt/vol]) | 192 |
| SDS (% [wt/vol]) | |
| 0.005 | 128 |
| 0.1 | 0.9 |
| 2-Propanol (% [vol/vol]) | |
| 1 | 117 |
| 5 | 127 |
| 10 | 108 |
The reaction mixture (1 ml), containing 30 μl (122 mU) of the pure enzyme preparation in 200 mM Tris-HCl buffer (pH 8.0), was initially preincubated for 10 min at 30°C with the tested effector, and then the enzymatic reaction was started by adding 50 μl of a 10-mg/ml P(3HO) latex emulsion. One hundred percent activity corresponded to 60 U/mg protein.
FIG. 3.
Assay of P. fluorescens GK13 P(3HO) depolymerase activity in polyacrylamide gels containing 0.1% (wt/vol) SDS. A sample (1.5 μg) of the pure enzyme was incubated with the denaturing cocktail containing 2.5% (vol/vol) 2-mercaptoethanol (A and B) or without 2-mercaptoethanol (C and D) and then analyzed by SDS-PAGE (A and D). Finally, the activity of the depolymerase was assayed by layering the polyacrylamide gel onto an agarose plate with P(3HO) latex (B and C).
Characterization and thermal stability of immobilized P(3HO) depolymerase.
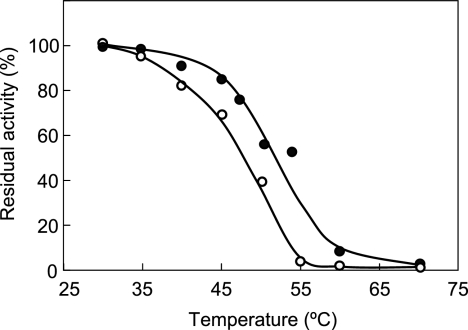
As a result of the adsorption of depolymerase directly from the culture broth, active derivatives (27 U/mg protein) with very low levels of loading (0.21 mg/g of support) were obtained. No change in the optimum pH value for P(3HO) depolymerase activity was observed in the case of the soluble and immobilized forms (data not shown). However, adsorption of the enzyme onto Accurel MP-1000 enhanced the thermal stability of enzyme after 15 min of incubation at different temperatures (Fig. 4).
FIG. 4.
Effect of immobilization on Accurel MP-1000 on thermal stability of pure P. fluorescens GK13 P(3HO) depolymerase. Samples of the soluble (○) or immobilized (•) enzyme were incubated for 15 min at the indicated temperatures and then immediately cooled to 4°C. Finally, the residual activity was determined at 30°C using P(3HO) as substrate.
Hydrolysis of P(3HO) polymer catalyzed by depolymerase in soluble or immobilized forms.
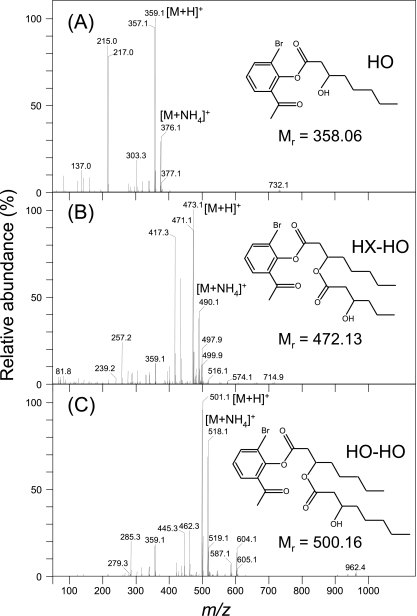
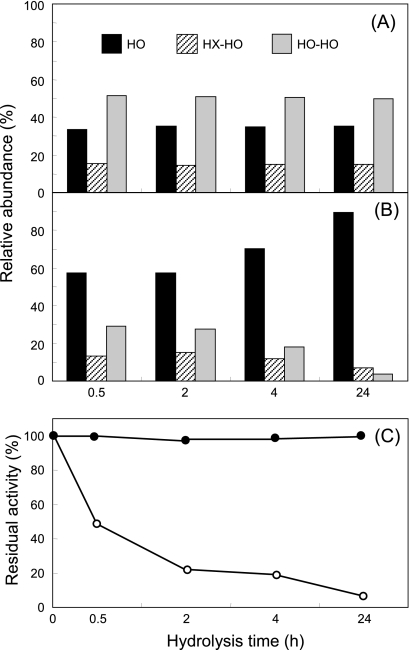
Enzymatic degradation of P(3HO) latex catalyzed by the soluble and immobilized depolymerase was assessed by HPLC-PDA, and the identity of the resulting peaks was determined by HPLC-MS (Fig. 5). When the immobilized enzyme was used to hydrolyze the polymer, both the composition and relative amounts of the resulting hydrolysis products were significantly dependent on the time of hydrolysis (Fig. 6B). During the early enzymatic period (within 30 min), monomers of 3-HO (eluting at 34.6 min), as well as dimers of both 3-hydroxyhexanoate and 3-hydroxyoctanoate (3-HX-HO, eluting at 41.6 min) and 3-hydroxyoctanoate (3-HO-HO, eluting at 46.4 min), were detected. However, after longer periods of incubation, the concentration of the monomer (3-HO) increased, whereas those of dimers were markedly decreased. In fact, after 24 h of enzymatic hydrolysis 90% of 3-HO monomers was obtained (Fig. 6B).
FIG. 5.
Identification by HPLC-MS of the P(3HO) hydrolysis products catalyzed by the P. fluorescens GK13 P(3HO) depolymerase. (A) HO, hydroxyoctanoate monomer; (B) HX-HO, dimer of hydroxyhexanoate and hydroxyoctanoate; (C) HO-HO, dimer of hydroxyoctanoate. The insets show the structure of the corresponding BPB-derivatized compounds and their corresponding molecular masses.
FIG. 6.
Evolution with the hydrolysis time of the abundance of P(3HO) products catalyzed by the soluble (A) and immobilized (B) P. fluorescens GK13 P(3HO) depolymerase. The stability of the soluble (○) or immobilized (•) enzyme was assessed during the time of hydrolysis (C).
In contrast, when P(3HO) hydrolysis was catalyzed by the soluble enzyme (Fig. 6A), the dimer 3-HO-HO was the main (∼60% of total) hydrolysis product detected, independently of the reaction time used (Fig. 6A). Additionally, in the case of the adsorbed enzyme, the activity remained unaltered until 24 h at 30°C, whereas the soluble counterpart retained only 10% of the initial activity (Fig. 6C).
In order to reuse the enzyme, the long-term stability of the immobilized P(3HO) depolymerase was evaluated too. For this purpose, five consecutive hydrolysis cycles of P(3HO) latex were performed at 30°C for 24 h, with repeated washing with 20 mM phosphate buffer (pH 8.0) after each reaction cycle. In all cases, 90% of 3-HO monomer was obtained (data not shown).
DISCUSSION
Adsorption of P(3HO) depolymerase to hydrophobic polypropylene Accurel MP-1000 support followed by desorption using 2-propanol seems to be an excellent strategy to recover, concentrate, purify, and immobilize P(3HO) depolymerase in a single step from the culture broth of P. fluorescens GK13 cells grown in mineral medium supplemented with P(3HO) polymer. It has been shown that this method is fast, cheap, and simple and provides high recovery. Indeed, following simple adsorption and desorption, an electrophoretically homogeneous enzyme preparation, with a 3.8-fold purification and 58.7% yield, was routinely obtained.
Although the specific activity of the adsorbed enzyme represents only 40% of that determined for the soluble counterpart, the stabilization of the enzyme allows the complete degradation of the substrate polymer to (R)-3HO monomers. In our case, only 0.21 mg of protein was immobilized per g of polypropylene support. Previous studies have shown that, at low levels of loading, the hydrophobic interactions between lipases and polypropylene are strong enough to cause a distortion of the tertiary structure of the protein with a consequent partial inactivation of a significant fraction of the enzyme activity (5, 30, 34).
Moreover, we have successfully reused the immobilized depolymerase for 5 consecutive cycles with activity. Another advantage of this immobilization protocol is that despite the high strength of depolymerase adsorption, it could be fully desorbed from Accurel MP-1000 with 2-propanol, allowing the possibility of recovery and reuse of the support. Besides, as occurred with lipases (18, 25, 28, 29), this procedure could be suitable for easy scale-up.
The P(3HO) depolymerase from P. fluorescens GK13 was first characterized by Schirmer et al. (37), who reported that the enzyme was a dimer consisting of two identical polypeptide chains whose activity was not affected significantly by dithiothreitol (DTT) or PMSF. However, analysis by two-dimensional PAGE of pure preparations of our P(3HO) depolymerase revealed the presence of two spots with distinct molecular masses and pI values (6.45 and 6.95).
In general, extracellular MCL-PHA depolymerases are markedly inhibited by detergents, such as Tween 80 or Triton X-100 (19, 21, 23, 24), except the enzyme of Pseudomonas luteola (33). In our case, a significant enhancement of the activity was observed when the activity was assayed in the presence of low concentrations of nonionic and anionic detergents using P(3HO) as substrate. Similar activation of polyhydroxybutyrate (PHB)-depolymerizing activity was observed too by low concentrations of Triton X-100 in several PHB depolymerases (26). However, these apparent activation effects were not observed when the enzyme was assayed using its PNPO esterase activity. As reported elsewhere (37), no inhibitory effects were observed when our pure enzyme was assayed using this chromogenic substrate in the presence of a number of typical inhibitors. Nevertheless, when the activity was assayed using P(3HO) as the substrate, it was significantly inhibited by PMSF, as occurred in most MCL-PHA depolymerases (11, 19-21).
Interestingly, the P(3HO) depolymerase of P. fluorescens GK13 is still active after denaturing SDS-PAGE analysis under both reducing and nonreducing conditions (Fig. 3). This result indicates that the two subunits of the protein are not maintained by a disulfide bridge and that each subunit is individually active. Apart from this, the remarkable stability of depolymerase in the presence of SDS enables its rapid detection in polyacrylamide gels analyzed by SDS-PAGE.
As shown (Fig. 4 and 6C), the immobilization of P. fluorescens GK13 P(3HO) depolymerase on Accurel MP-1000 resulted in a remarkable enhancement of both the short-term and long-term stability of the enzyme, and this can permit the complete hydrolysis of the P(3HO) polymer into monomeric 3-HO products. Moreover, the possible effect of a favorable substrate partition in the microenvironment of the enzyme, due to the hydrophobic nature of both substrate and immobilization support, as well as the fact that only the end of the P(3HO) would be accessible to the immobilized enzyme, thus favoring the release of monomers, should also be considered.
In this study we report a novel method to recover, purify, and immobilize the P(3HO) depolymerase from P. fluorescens GK13 directly from the crude fermentation broth. This immobilization procedure was found to be highly selective, providing a stable enzyme preparation with elevated purity (near to homogeneity) in just one step. Thus, by using a simple and low-cost immobilization procedure it is possible to obtain a new robust immobilized biocatalyst with remarkable stability that can be useful in producing pure chiral (R)-3-HAs.
Acknowledgments
This work was carried out in the framework of the IP project “Sustainable Microbial and Biocatalytic Production of Advanced Functional Materials” (BIOPRODUCTION/NMP-2-CT-2007-026515) funded by the European Commission. M.S. and J.G. were the recipients of scholarships from the Spanish Ministry of Education.
We thank M. A. Prieto (CIB-CSIC, Madrid, Spain) for kindly providing us with a strain of P. fluorescens GK13. P(3HO) substrate was kindly supplied by Biopolis, S.A. (Valencia, Spain), and CPI (Newcastle, United Kingdom). HPLC-MS analysis was performed in the Analysis Central Service of the University of the Basque Country.
Footnotes
Published ahead of print on 16 April 2010.
REFERENCES
- 1.Al-Duri, B., and Y. P. Yong. 2000. Lipase immobilization: an equilibrium study of lipases immobilised on hydrophobic and hydrophilic/hydrophobic supports. Biochem. Eng. J. 4:207-215. [Google Scholar]
- 2.Almeida, R. V., R. V. Branco, B. Peixoto, C. D. S. Lima, S. M. C. Alqueres, O. B. Martins, O. A. C. Antunes, and D. M. G. Freire. 2008. Immobilization of a recombinant thermostable esterase (Pf2001) from Pyrococcus furiosus on microporous polypropylene: isotherms, hyperactivation and purification. Biochem. Eng. J. 39:531-537. [Google Scholar]
- 3.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial use of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balcao, V. M., A. L. Paiva, and F. X. Malcata. 1996. Bioreactors with immobilized lipases: state of the art. Enzyme Microb. Technol. 18:392-416. [DOI] [PubMed] [Google Scholar]
- 5.Bosley, J. A., and A. D. Peillow. 1997. Immobilization of lipases on porous polypropylene: reduction in esterification efficiency at low loading. J. Am. Oil Chem. Soc. 74:107-111. [Google Scholar]
- 6.Braunegg, G., G. Lefebvre, and K. F. Genser. 1998. Polyhydroxyalkanoates, biopolyesters from renewable resources: physiological and engineering aspects. J. Biotechnol. 65:127-161. [DOI] [PubMed] [Google Scholar]
- 7.Calabia, B. P., and Y. Tokiwa. 2006. A novel PHB depolymerase from a thermophilic Streptomyces sp. Biotechnol. Lett. 28:383-388. [DOI] [PubMed] [Google Scholar]
- 8.Chen, G. Q. 2009. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem. Soc. Rev. 38:2434-2446. [DOI] [PubMed] [Google Scholar]
- 9.Chen, G. Q., and Q. Wu. 2005. Microbial production and applications of chiral hydroxyalkanoates. Appl. Microbiol. Biotechnol. 67:592-599. [DOI] [PubMed] [Google Scholar]
- 10.de Roo, G., M. B. Kellerhals, Q. Ren, B. Witholt, and B. Kessler. 2002. Production of chiral acids and R-3-hydroxyalkanoic acid methylesters via hydrolytic degradation of polyhydroxyalkanoates synthesized by pseudomonads. Biotechnol. Bioeng. 77:717-722. [DOI] [PubMed] [Google Scholar]
- 11.Elbanna, K., T. Lütke-Eversloh, D. Jendrossek, H. Luftmann, and A. Steinbüchel. 2004. Studies on the biodegradability of polythioester copolymers and homopolymers by polyhydroxyalkanoate (PHA)-degrading bacteria and PHA depolymerases. Arch. Microbiol. 182:212-225. [DOI] [PubMed] [Google Scholar]
- 12.Gebauer, B., and D. Jendrossek. 2006. Assay of poly(3-hydroxybutyrate) depolymerase activity and product determination. Appl. Environ. Microbiol. 72:6094-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta, N., P. Rathi, R. Singh, V. K. Goswami, and R. Gupta. 2005. Single-step purification of lipase from Burkholderia multivorans using polypropylene matrix. Appl. Microbiol. Biotechnol. 67:648-653. [DOI] [PubMed] [Google Scholar]
- 14.Jaeger, K. E., A. Steinbüchel, and D. Jendrossek. 1995. Substrate specificities of bacterial polyhydroxyalkanoate depolymerases and lipases: bacterial lipases hydrolyze poly(3)-hydroxyalkanoates. Appl. Environ. Microbiol. 61:3113-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jendrossek, D., A. Frisse, A. Behrends, M. Andermann, H. D. Kratzin, T. Stanislawski, and H. G. Schlegel. 1995. Biochemical and molecular characterization of the Pseudomonas lemoignei polyhydroxyalkanoate depolymerase system. J. Bacteriol. 177:596-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jendrossek, D., and R. Handrick. 2002. Microbial degradation of polyhydroxy-alkanoates. Annu. Rev. Microbiol. 56:403-432. [DOI] [PubMed] [Google Scholar]
- 17.Jendrossek, D., A. Schirmer, and H. G. Schlegel. 1996. Biodegradation of polyhydroxyalcanoic acids. Appl. Microbiol. Biotechnol. 46:451-463. [DOI] [PubMed] [Google Scholar]
- 18.Kaewthong, W., S. Sirisansaneeyakul, P. Prasertsan, and A. H. Kittikun. 2005. Continuous production of monoacylglycerols by glycerolysis of palm olein with immobilized lipase. Process Biochem. 40:1525-1530. [Google Scholar]
- 19.Kim, D. Y., H. C. Kim, S. Y. Kim, and Y. H. Rhee. 2005. Molecular characterization of extracellular medium-chain-length poly(3-hydroxyalkanoate) depolymerase genes from Pseudomonas alcaligenes strains. J. Microbiol. 43:285-294. [PubMed] [Google Scholar]
- 20.Kim, D. Y., H. W. Kim, M. G. Chung, and Y. H. Rhee. 2007. Biosynthesis, modification, and biodegradation of bacterial medium-chain-length polyhydroxyalkanoates. J. Microbiol. 45:87-97. [PubMed] [Google Scholar]
- 21.Kim, D. Y., J. S. Nam, and Y. H. Rhee. 2002. Characterization of an extracellular medium-chain-length poly(3-hydroxyalkanoate) depolymerase from Pseudomonas alcaligenes LB19. Biomacromolecules 3:291-296. [DOI] [PubMed] [Google Scholar]
- 22.Kim, H., H. S. Ju, and S. Kim. 2000. Characterization of an extracellular poly(3-hydroxy-5-phenylvalerate) depolymerase from Xanthomonas sp. JS02. Appl. Microbiol. Biotechnol. 53:323-327. [DOI] [PubMed] [Google Scholar]
- 23.Kim, H. J., D. Y. Kim, J. S. Nam, K. S. Bae, and Y. H. Rhee. 2003. Characterization of an extracellular medium-chain-length poly(3-hydroxyalkanoate) depolymerase from Streptomyces sp. KJ-72. Antonie Van Leeuwenhoek 83:183-189. [DOI] [PubMed] [Google Scholar]
- 24.Kim, H. M., K. E. Ryu, K. S. Bae, and Y. H. Rhee. 2000. Purification and characterization of extracellular medium-chain-length polyhydroxyalkanoate depolymerase from Pseudomonas sp. RY-1. J. Biosci. Bioeng. 89:196-198. [DOI] [PubMed] [Google Scholar]
- 25.Kittikun, A. H., P. Prasertsan, and C. Sungpud. 2000. Continuous production of fatty acids from palm olein by immobilized lipase in a two-phase system. J. Am. Oil Chem. Soc. 77:599-603. [Google Scholar]
- 26.Kobayashi, T., A. Sugiyama, Y. Kawase, T. Saito, J. Mergaert, and J. Swings. 1999. Biochemical and genetic characterization of an extracellular poly(3-hydroxybutyrate) depolymerase from Acidovorax sp. strain TP4. J. Environ. Polym. Degrad. 7:9-18. [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Malcata, F. X., H. R. Reyes, H. S. García, C. G. Hill, Jr., and C. H. Amundson. 1990. Immobilized lipase reactors for modification of fats and oils. A review. J. Am. Oil Chem. Soc. 67:890-910. [Google Scholar]
- 29.Murray, M., D. Rooney, M. van Neikerk, A. Montenegro, and L. R. Weatherley. 1997. Immobilization of lipase onto lipophilic polymer particles and application to oil hydrolysis. Process Biochem. 32:479-486. [Google Scholar]
- 30.Persson, M., E. Wehtje, and P. Adlercreutz. 2002. Factors governing the activity of lyophilised and immobilised lipase preparation in organic solvents. Eur. J. Chem. Biol. 3:566-571. [DOI] [PubMed] [Google Scholar]
- 31.Peterson, G. L. 1983. Determination of total protein. Methods Enzymol. 91:95-119. [DOI] [PubMed] [Google Scholar]
- 32.Prieto, M. A., L. I. de Eugenio, B. Galán, J. M. Luengo, and B. Witholt. 2007. Synthesis and degradation of polyhydroxyalkanoates, p. 397-428. In J. L. Ramos and A. Filloux (ed.), Pseudomonas: a model system in biology, vol. V. Springer Verlag, Dordrecht, Netherlands. [Google Scholar]
- 33.Rhee, Y. H., Y. H. Kim, and K. S. Shin. 2006. Characterization of an extracellular poly(3-hydroxyoctanoate) depolymerase from the marine isolate, Pseudomonas luteola M13-4. Enzyme Microb. Technol. 38:529-535. [Google Scholar]
- 34.Salis, A., M. S. Bhattacharyya, M. Monduzzi, and V. Solinas. 2009. Role of the support surface on the loading and the activity of Pseudomonas fluorescens lipase used for biodiesel synthesis. J. Mol. Catal. B Enzym. 57:262-269. [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Schirmer, A., and D. Jendrossek. 1994. Molecular characterization of the extracellular poly(3-hydroxyoctanoic acid) [P(3HO)] depolymerase gene of Pseudomonas fluorescens GK13 and of its gene product. J. Bacteriol. 176:7065-7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schirmer, A., D. Jendrossek, and H. G. Schlegel. 1993. Degradation of poly(3-hydroxyoctanoic acid) [P(3HO)] by bacteria: purification and properties of a P(3HO) depolymerase from Pseudomonas fluorescens GK13. Appl. Environ. Microbiol. 59:1220-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinbüchel, A., and B. Füchtenbusch. 1998. Bacterial and other biological systems for polyester production. Trends Biotechnol. 16:419-427. [DOI] [PubMed] [Google Scholar]
- 39.Verlinden, R. A., D. J. Hill, M. A. Kenward, C. D. Williams, and I. Radecka. 2007. Bacterial synthesis of biodegradable polyhydroxyalkanoates. J. Appl. Microbiol. 102:1437-1449. [DOI] [PubMed] [Google Scholar]